Abstract
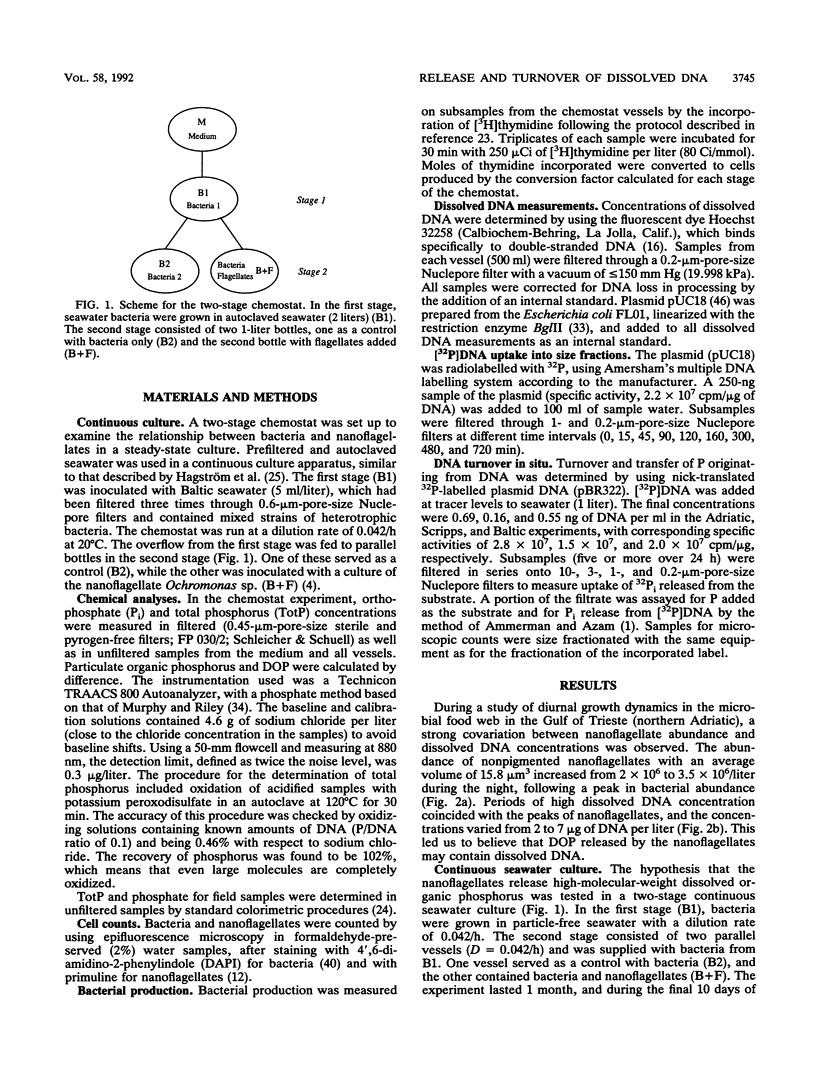
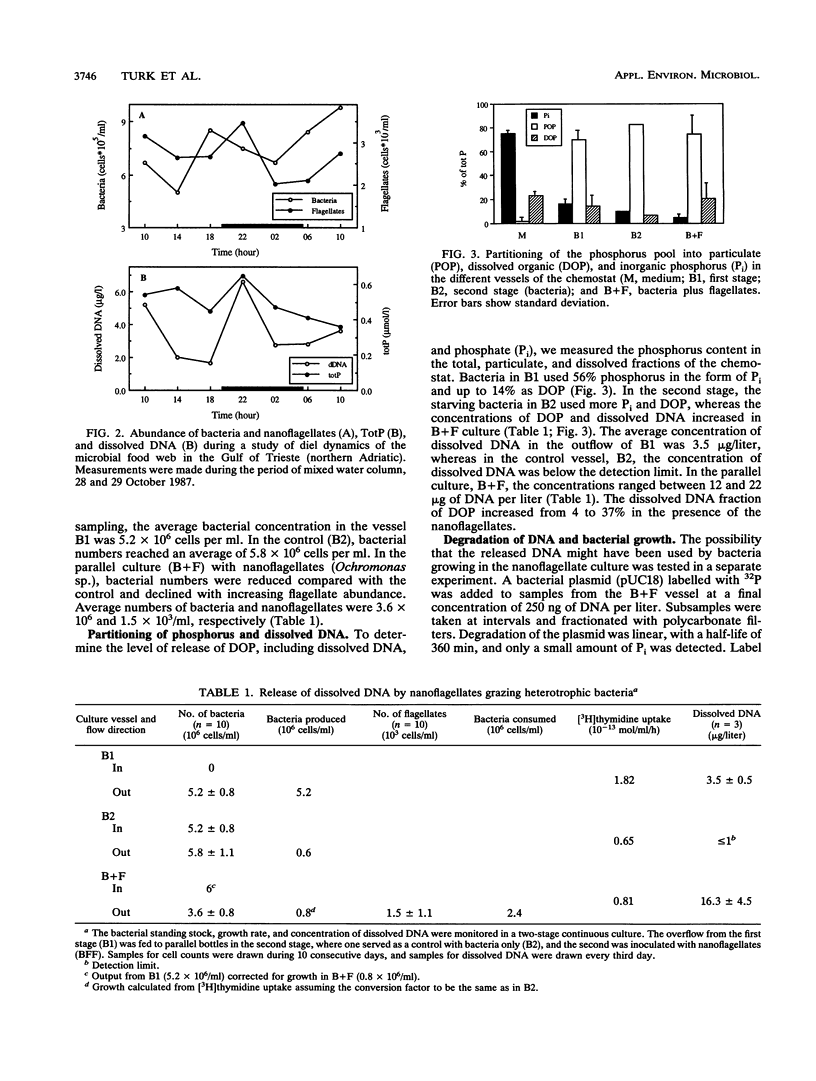
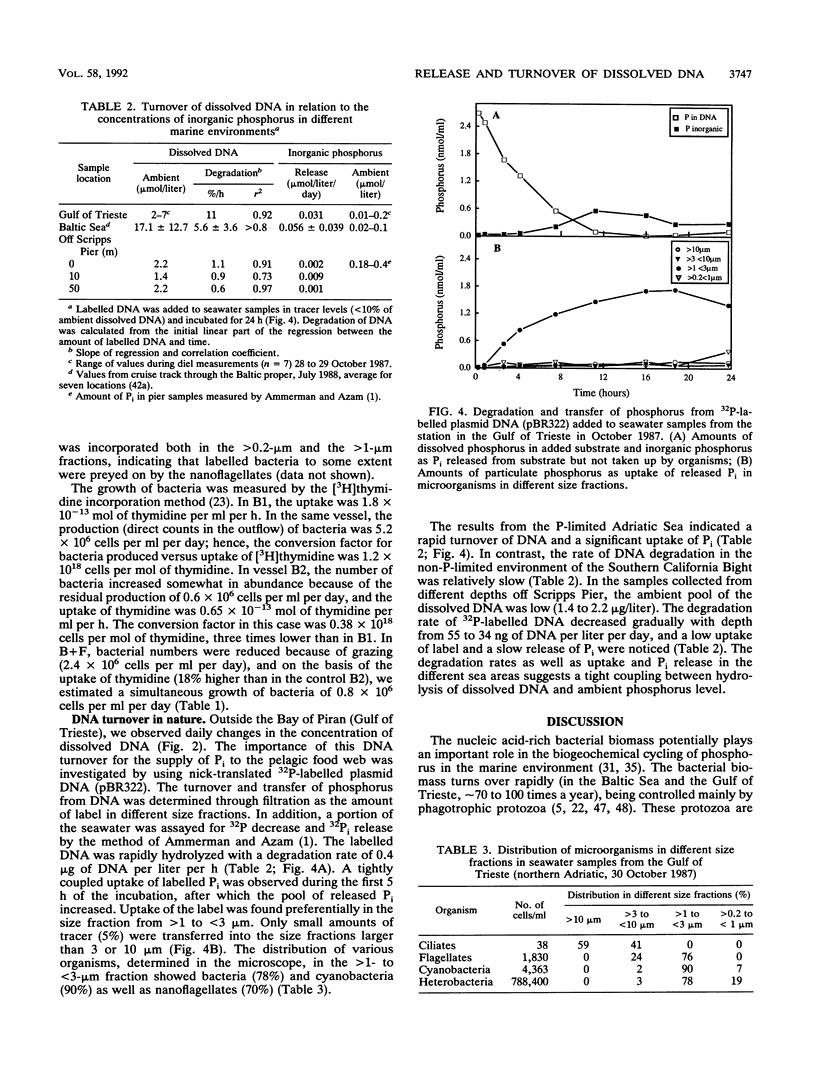
The concentrations of dissolved DNA and nanoflagellates were found to covary during a study of diel dynamics of the microbial food web in the Adriatic Sea. This observation was further investigated in a continuous seawater culture when nanoflagellates were fed bacteria grown in filtered seawater. Analysis of dissolved organic phosphorus and dissolved DNA showed a sixfold increase of dissolved DNA in the presence of the nanoflagellates (Ochromonas sp.). The amount of DNA released suggested that the majority of the consumed bacterial DNA was ejected. Phagotrophic nanoflagellates thus represent an important source of origin for dissolved DNA. The rate of breakdown of dissolved DNA and release of inorganic phosphorus in the pelagic ecosystem is suggested to be dependent on the ambient phosphate pool. In the P-limited northern Adriatic Sea, rapid degradation of the labelled DNA could be demonstrated, whereas the N-limited southern California bight water showed a much lower rate. Phosphorus originating from dissolved DNA was shown to be transferred mainly to organisms in the <3-μm-size fractions. On the basis of the C/P ratios, we suggest that a significant fraction of the phosphorus demand by the autotrophs may be sustained by the released DNA during stratified conditions. Thus, the nucleic acid-rich bacterial biomass grazed by protozoa plays an important role in the biogeochemical cycling of phosphorus in the marine environment.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ammerman J. W., Azam F. Bacterial 5-nucleotidase in aquatic ecosystems: a novel mechanism of phosphorus regeneration. Science. 1985 Mar 15;227(4692):1338–1340. doi: 10.1126/science.227.4692.1338. [DOI] [PubMed] [Google Scholar]
- Bloem J., Starink M., Bär-Gilissen M. J., Cappenberg T. E. Protozoan grazing, bacterial activity, and mineralization in two-stage continuous cultures. Appl Environ Microbiol. 1988 Dec;54(12):3113–3121. doi: 10.1128/aem.54.12.3113-3121.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratbak G., Heldal M., Norland S., Thingstad T. F. Viruses as partners in spring bloom microbial trophodynamics. Appl Environ Microbiol. 1990 May;56(5):1400–1405. doi: 10.1128/aem.56.5.1400-1405.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Børsheim K. Y., Bratbak G., Heldal M. Enumeration and biomass estimation of planktonic bacteria and viruses by transmission electron microscopy. Appl Environ Microbiol. 1990 Feb;56(2):352–356. doi: 10.1128/aem.56.2.352-356.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron D. A. Technique for enumeration of heterotrophic and phototrophic nanoplankton, using epifluorescence microscopy, and comparison with other procedures. Appl Environ Microbiol. 1983 Aug;46(2):491–498. doi: 10.1128/aem.46.2.491-498.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concino M. F., Goodgal S. H. DNA-binding vesicles released from the surface of a competence-deficient mutant of Haemophilus influenzae. J Bacteriol. 1982 Oct;152(1):441–450. doi: 10.1128/jb.152.1.441-450.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deflaun M. F., Paul J. H., Davis D. Simplified method for dissolved DNA determination in aquatic environments. Appl Environ Microbiol. 1986 Oct;52(4):654–659. doi: 10.1128/aem.52.4.654-659.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward D. W., Garon C. F., Judd R. C. Export and intercellular transfer of DNA via membrane blebs of Neisseria gonorrhoeae. J Bacteriol. 1989 May;171(5):2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., Cannon J. P. Production of dissolved DNA, RNA, and protein by microbial populations in a Florida reservoir. Appl Environ Microbiol. 1990 Oct;56(10):2957–2962. doi: 10.1128/aem.56.10.2957-2962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jeffrey W. H., David A. W., Deflaun M. F., Cazares L. H. Turnover of extracellular DNA in eutrophic and oligotrophic freshwater environments of southwest Florida. Appl Environ Microbiol. 1989 Jul;55(7):1823–1828. doi: 10.1128/aem.55.7.1823-1828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H., Jiang S. C., Rose J. B. Concentration of viruses and dissolved DNA from aquatic environments by vortex flow filtration. Appl Environ Microbiol. 1991 Aug;57(8):2197–2204. doi: 10.1128/aem.57.8.2197-2204.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul John H., Deflaun Mary F., Jeffrey Wade H., David Andrew W. Seasonal and Diel Variability in Dissolved DNA and in Microbial Biomass and Activity in a Subtropical Estuary. Appl Environ Microbiol. 1988 Mar;54(3):718–727. doi: 10.1128/aem.54.3.718-727.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr B. F., Sherr E. B., Rassoulzadegan F. Rates of digestion of bacteria by marine phagotrophic protozoa: temperature dependence. Appl Environ Microbiol. 1988 May;54(5):1091–1095. doi: 10.1128/aem.54.5.1091-1095.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]



