Abstract
Integrin-mediated adhesion is regulated by multiple features of the adhesive surface, including its chemical composition, topography, and physical properties. In this study we investigated integrin lateral clustering, as a mechanism to control integrin functions, by characterizing the effect of nanoscale variations in the spacing between adhesive RGD ligands on cell spreading, migration, and focal adhesion dynamics. For this purpose, we used nanopatterned surfaces, containing RGD-biofunctionalized gold dots, surrounded by passivated gaps. By varying the spacing between the dots, we modulated the clustering of the associated integrins. We show that cell-surface attachment is not sensitive to pattern density, whereas the formation of stable focal adhesions and persistent spreading is. Thus cells plated on a 108-nm-spaced pattern exhibit delayed spreading with repeated protrusion-retraction cycles compared to cells growing on a 58-nm pattern. Cell motility on these surfaces is erratic and nonpersistent, leaving thin membrane tethers bound to the RGD pattern. Dynamic molecular profiling indicated that the adhesion sites formed with the 108-nm pattern undergo rapid turnover and contain reduced levels of zyxin. These findings indicate that a critical RGD density is essential for the establishment of mature and stable integrin adhesions, which, in turn, induce efficient cell spreading and formation of focal adhesions.
INTRODUCTION
The adhesion of a cell to its environment is mediated by transmembrane adhesion receptors and corresponding extracellular matrix (ECM) proteins. Integrins play a key role in such interactions, mediating the assembly of multimolecular complexes that bridge between the ECM and the actin cytoskeleton (1–3). It has been shown that after binding to specific adhesive ECM epitopes, such as RGD sequences (4), several intracellular anchor proteins are recruited to the adhesion site, ultimately inducing the assembly of focal complexes and focal adhesions (FA) and the associated cytoskeleton (5). The molecular composition and mechanical properties of the ECM affect these processes, yet the biochemical and biophysical basis for these cellular responses are largely unknown. In this article we explore the role of the ECM ligand density on the adhesive process.
During cell spreading and locomotion the assembly of early cell contacts to the ECM at the leading edge are driven by actin polymerization (6,7). In the nearby lamella, actomyosin contraction plays a major role in regulating FA structure and dynamics (3,8–10) as well as the position of the cell's front (11–14), consequently affecting the progression of the spreading or migration process. These protrusive and retractive events are tightly regulated by the small GTPases Rac1 and Rho, which trigger actin polymerization in the leading edge and actomyosin contractility, respectively (15–17). Local tension generated by actomyosin contractility and transduced via integrin adhesions, can affect these processes by modulating FA dynamics, membrane protrusion, and tail retraction (3,8–10). Thus, the focal complexes and FA that are formed during spreading serve as cytoskeletal organizing centers (18,19) as well as surface-sensing entities that control, locally and globally, adhesion-mediated signaling and coordinate the adhesive and migratory process (20,21).
It is currently widely appreciated that cells can sense and integrate multiple chemical and physical features of the underlying adhesive substrate. These include the chemical nature of the surface, the density and organization of the specific ECM proteins, the topography of the ECM, and its compliance (22–26). The understanding of how adhesion-mediated interactions are regulated by the spatial arrangement of the ECM has greatly benefited from advanced micro- and nanopatterning technologies (27,28). For example, assessment of the concentration dependence of RGD integrin interactions was achieved by varying the average surface density of the integrin ligands (29–31) or by nanoscale clustering of RGD peptides (32,33). However, only recently nanopatterned adhesive surfaces were engineered where accurate and regular RGD-to-RGD distances could be set. Using these surfaces as adhesion substrates indicated that a lateral spacing of 58 nm or less between nanogold-anchored RGD peptides is essential for FA formation (34). In this study we have addressed the mechanism involved in this “density sensing”, addressing the following questions: i), To what extent do cell spreading kinetics and adhesion stability depend on RGD ligand spacing? ii), Is there any critical ligand separation involved in the regulation of protrusion-retraction cycles during spreading and migration? iii), What is the relation between integrin lateral clustering and FA assembly and dynamics? To address these issues, rat embryo fibroblasts (REF52) were plated on surfaces presenting well-ordered nanopatterns consisting of 8-nm-sized gold particles, separated by either 58 or 108 nm. The area between the gold particles was passivated by polyethylene glycol and then a cyclic-RGD peptide, c(RGDfK)-thiol, was covalently linked to the gold particles. Given that each nanoparticle can accommodate only one integrin receptor we propose that by varying the spacing between the gold nanoparticles it is possible to control the lateral spacing between single ligand-bound integrins. The results presented here show that the spatial lateral spacing of ECM molecules regulates the dynamics of cell spreading, adhesion, and migration, suggesting that the local density of individual integrin receptors plays an essential role in controlling the dynamics and fate of the adhesive response.
MATERIALS AND METHODS
Preparation of RGD-nanopatterned surfaces
The method for the preparation of nanopatterned surfaces biofunctionalized with RGD peptides is based on the self-assembly of diblock copolymer micelles (34). Briefly, micelles of copolymers containing gold nanoparticles are deposited on glass coverslips; after treatment with hydrogen gas plasma, only the polymers are removed from the surface, leaving just gold nanoparticles of 8 nm diameter (35,36). These particles form a hexagonal lattice (as shown in the inserts in Fig. 1, B and C) and are separated by 28–110 nm, depending on the size of the copolymer used. The space between the gold nanoparticles is covered with polyethylene glycol ((PEG) molecular weight 2000) to prevent cell adhesion and protein adsorption. Each gold particle is biofunctionalized by binding the cyclic peptide c-RGDfK via a thiol group and an alkane spacer. Glass coverslips homogeneously coated with RGD peptides served as control; such surfaces were first coated with a 20-nm layer of gold (using a modular high vacuum coating unit from Bal-Tec Med 020, Bal-Tec AG, Balzers, Liechtenstein) and then incubated with the cyclic RGD peptides as described above.
FIGURE 1.
Phase contrast images of a cell spreading on surfaces presenting (A) homogeneously coated with RGD peptide, (B) 58-nm RGD-nanopattern, and (C) 108-nm RGD nanopattern. In panels B and C SEM micrographs of the nanopatterned surfaces are shown and the average spacing between gold nanoparticles, determined by measuring different fields of several samples, is indicated in the upper part (± SD). The kinetics of cell spreading are determined by measuring the cell area as a function of time (time 0 is the initial time of adhesion). Area values are normalized to the average of the maximum values of the control group. Each line in the plot is a measurement of a cell on that type of surface and the dotted black lines indicate the transition from initial spreading to stabilization phase. The lines corresponding to the cell shown in the image sequences are indicated in the lower right corner. The whole movie can be found in the online Supplementary Material.
Cell culture
REF 52 cells (a rat fibroblast line) were maintained in Dulbecco's modified Eagle's medium ((DMEM) Gibco Laboratories, Eggenstein, Germany) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine and 100 units/ml penicillin-streptomycin (all from Gibco Laboratories) at 37°C in 5% CO2. The cloning of the vectors and cell transfection with GFP-β3-integrin and YFP-paxillin have been described previously (15,37). For cell adhesion experiments on RGD surfaces, cells were plated at a density of 1–5 × 105/sample in DMEM containing 1% FBS. For live imaging a heated stage surrounded by a humidified chamber containing CO2 was used.
Critical point drying and scanning electron microscopy
Cells adhering to RGD-nanopatterned glass coverslips were fixed in 2% glutaraldehyde in phosphate buffered saline (PBS) for 15 min and then dehydrated in graded ethanol. Samples were then transferred into the chamber of the critical point dryer (CPD 030 critical point dryer; Bal-Tec) to replace the ethanol with liquid CO2 at a temperature of 10°C and a pressure of 50 bar. Then, a change in phase from liquid to gas CO2 was obtained at 40°C and at a pressure of 70 bar. Glass samples were sputter-coated with a carbon layer to be imaged with a field-emission scanning electron microscope (FE-SEM, LEO-1530, LEO, Oberkochen, Germany). To visualize the gold nanoparticles on the surface, an acceleration voltage of 3 kV was applied under a pressure of 5 × 10−6 mbar. The magnification used is 15,0000×. The geometrical order and nanoparticle distance were then analyzed using ImageJ software version 1.34s (38).
Cell spreading and motility measurements
Live monitoring of cell spreading on surfaces was performed in a heated and air humidified chamber built around a Zeiss Axiovert 135 microscope with a 20×/0.25 Ph2 objective (Carl Zeiss MicroImaging GmbH, Göttingen, Germany). The parameters were controlled by a LabVIEW (National Instruments, Austin, TX) environment. Phase-contrast time-lapse imaging of a field containing five to seven cells at a 30-s interval for 0–6 h after plating was performed with a charge-coupled device (CCD) camera (AxioCam HR, Carl Zeiss MicroImaging GmbH) controlled by the Axiovision software. Cell boundaries and centroids were identified using ImageJ. Three to five cells per field in 10 different samples per surface type were analyzed. The measurements of the cell area were analyzed at 1-min intervals and normalized to the average of the maximum area value obtained on homogenously coated RGD surfaces. Kymographs to examine lamellipodia velocity were determined at 30-s intervals. Cell motility was evaluated by analyzing the changes in the location of the center of mass at 10-min intervals.
Immunochemical reagents and indirect immunofluorescence
Primary antibodies that were used in this study include mouse anti-human vinculin monoclonal (Sigma-Aldrich Chemie GmbH, Munich, Germany) and mouse anti-zyxin monoclonal (gift from Daniel Louvard, Institut Curie, Paris, France). Phalloidin-TRITC and Cy5-conjugated goat anti-mouse were purchased from Molecular Probes (Eugene, OR). Cultured cells were fixed and permeabilized for 1 min in PBS containing 0.1% Triton X-100 (Sigma) and 3% paraformaldehyde, and then were fixed further with 3% paraformaldehyde in PBS for 30 min. The cells were incubated for 1 h at room temperature with the primary antibodies diluted in PBS, washed, and further incubated with the secondary antibodies for 1 h. After extensive washing with PBS, coverslips were mounted in Elvanol (Mowiol 4-88, Serafon, Ashdod, Israel) to inhibit photobleaching.
Digital microscopy
Immunofluorescence microscopy of fixed and living samples was carried out with an Olympus IX inverted microscope (Olympus, Hamburg. Germany). Images were acquired with a DeltaVision system (Applied Precision, Issaqua, WA) as previously described (30). In this study, cells were examined with a 60/1.3 NA plan-Neofluar objective (Olympus, Tokyo, Japan) resulting in a pixel length of 0.2206 μm. Cell imaging was carried out with a cooled CCD camera (Photometrics, Kew, Australia). Image acquisition and processing were performed with Resolve3D and Priism programs as described (37). In short, Cy5 and GFP or YFP images were subjected to high-pass filtration to remove background and low-intensity pixels. Ratio values were calculated pixel by pixel and displayed using a spectrum scale. For dynamic studies of paxillin clusters, cells were cultured in nanopatterned coverslip bottom dishes in DMEM containing 1% FBS for 3 h; the media was then exchanged for F12 medium (Invitrogen, Karlsruhe, Germany) before the samples were put in the microscope. The objective and stage were heated to maintain the cells at a stable temperature of 37°C and humidified with 5% CO2 throughout the experiment. Images were taken every minute for 120 min and processed in the same way as in the case of fixed samples. For the temporal fluorescence ratio imaging (FRIT) images acquired at two different time points were compared; the numeration and the denominator correspond to the later and earlier images, respectively. Thus, a blue color indicates a new structure and a red color indicates a structure that has disappeared. TIRF microscopy was conducted using an Olympus IX81 inverted microscope equipped with IX2-RFAEVA TIRF attachment (Olympus). Light from an argon ion laser model I77 (lines 488 nm, 514 nm) (Spectra-Physics, Mountain View, CA) was introduced into the sample through a high numerical aperture objective lens (63 OHR; NA 1.45, Olympus). Images were taken using a cooled CCD camera, Quantix 57, with EEV 57-10 back-illuminated CCD chip (Photometrics, Tucson, AZ). Temperature was kept at 37°C during the experiments, using the Cube&Box Temperature Control System from Life Imaging Services (Reinach, Switzerland).
RESULTS
The lateral spacing between integrin ligands regulates cell spreading and adhesion stability
To evaluate the kinetics of cell spreading on dense and sparse nanopatterned substrates, we monitored temporal changes in the projected area of cells adhering to these surfaces. REF52 cells became attached to the 58-nm RGD-nanopatterned surface within 5–7 min after plating and underwent radial spreading, reaching a plateau in cell area within ∼70 min (Fig. 1 B), comparable to cells cultured on homogeneously modified RGD surfaces (Fig. 1 A). It is noteworthy that the average density of RGD molecules on homogeneous gold surface is estimated to be ∼40,000 molecules/μm2, assuming one RGD per 25 nm2. On the 58-nm patterned surface, the RGD density was ∼500 molecules/μm2. After longer incubation on the 58-nm RGD-nanopatterns the cells moderately retracted, and their projected area was reduced by 20–40%. Often, the cells assumed a polarized morphology, with advancing lamella at one end and retractions at the other end (Fig. 1 B and Supplementary Material, movie 1). The morphology and dynamics of the leading edge of cells adhering to the 58-nm patterned surfaces were different from those of the advancing lamella of cells spreading on homogeneously modified RGD surfaces. This is manifested by the formation of fine filopodia and local membrane retractions on the 58-nm pattern in comparison to a rather smooth and continuously advancing edge on the uniformly modified surface. REF52 cells cultured on the 108-nm surface displayed a delayed spreading and increased ruffling of the cell membrane (Fig. 1 C). After an initial period of nearly continuous spreading (∼120 min) these cells abruptly retracted, losing their contacts with the surface and showing major changes in shape and polarity (Fig. 1 C and Supplementary Material, movie 1). Time-lapse microscopy showed that cells cultured on the 108-nm RGD-nanopattern undergo repeated extension-retraction cycles, leading to major changes (up to 80% reduction) in the projected cell area. A quantitative analysis of the cell area as a function of time indicated that fibroblasts on the 58-nm RGD-nanopattern reach an average maximum area of 2712 ± 752 μm2, a value comparable to that of controls (2817 ± 794 μm2), whereas cells on the 108-nm RGD-nanopattern show an average maximum projected area of 1559 ± 527 μm2.
Measurement of the movements of the lamellipodium during the spreading stage, using kymograph analysis, revealed highly dynamic cycles of protrusions and retractions as indicated by positive and negative variations in edge velocities (Fig. 2). In cells adhering to the uniform surface the cycles of retraction and protrusion could be noticed during the first 50 min after plating, after which the edge became nearly stationary for ∼1 h (Fig. 2 A). During this period cells became polarized and started migrating, showing again cyclic movements. The protrusion velocity during early spreading and migration was typically 2 μm/min and the frequency of protrusion was 1.3 min−1. As indicated in the histogram on the right side of Fig. 2 A, the number of active protrusion and retraction events was similar, however, the number of stationary events, with minimal or null variation in velocity, was 7× higher than the active events. Kymograph analysis of cells plated on the 58-nm nanopatterned surfaces showed higher protrusive and retractive activity compared to the homogeneous RGD surface, with essentially no stationary phases but rather a continuous advancement of the lamellipodia (Fig. 2 B). After this period, cells became polarized and started migrating, showing again cyclic movements. The mean frequency was ∼1.5 min−1, similar to that measured on homogeneous RGD surfaces. The protrusion and retraction velocities were also similar, ∼2 μm/min. However, the halfwidth of the bar histogram in Fig. 2 B was broader due to fewer fast protrusion and retraction cycles, reaching velocities of 6–8 μm/min during the first 10 min after plating. The number of protrusive and retractive events, and the range of variations in velocity, were comparable to those observed on the homogenously modified RGD surfaces, except the decrease in the number of stationary events. Adhesion to the 108-nm pattern was significantly delayed. Spreading was first observed after 30 min, and was characterized by increased ruffling at the membrane edge (Fig. 2 C). Plating cells on the 108-nm pattern revealed frequent protrusion and retraction cycles that support an extensive and erratic migration as seen in Fig. 1 C. Protrusion and retraction cycles show a frequency of 3 min−1. Protrusion velocity was, typically, as high as 8 μm/min and retraction speed reached a value of −10 μm/min throughout the entire observation period, resulting in a rather broad halfwidth for the velocity distribution. The major effect of low ECM density was a delayed initiation of spreading and appearance of lamellipodia. During spreading and lamellipodia advancement, the number of stationary events was higher in cells adhering to homogenously modified surfaces and on 58-nm RGD nanopatterns whereas on 108-nm RGD-nanopatterns there was a broader variation in velocity (Fig. 2, bar histograms).
FIGURE 2.
Analysis of lamellipodial protrusion in REF52 cells, adhering to (A) homogenously modified RGD surface, (B) 58-nm, and (C) 108-nm RGD nanopattern. The initial time corresponds to the first appearance of the lamellipodia in the cell (for A and B, 5–7 min after attachment; for C, 30 min). The edge displacement along the three colored lines was monitored for 160 min at 30-s intervals using kymograms. The central plots show the rate of displacement over time. The plots on the right indicate the distribution of the number of protrusion and retraction events for each sample.
The turnover of focal adhesions increases in cells cultured on 108-nm RGD-nanopatterned surfaces
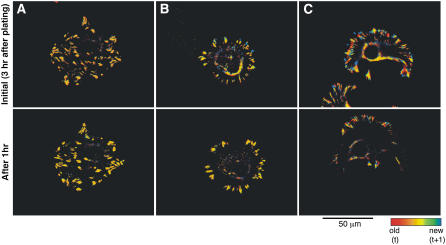
To explore the effect of RGD ligand spacing on the dynamics of focal adhesion formation, REF52 cells, expressing YFP-paxillin, were plated on homogeneously coated, 58-nm or 108-nm RGD-nanopatterns, and the organization of focal adhesion was examined by time-lapse fluorescence microscopy (Fig. 3 and Supplementary Material, movies 2 and 3). The adhesion sites of REF52 cells were identified by localization of YFP-paxillin; image background was reduced by high-pass filtration, and the intensity of individual substrate adhesion sites was analyzed after segmentation, using the “water” algorithm (37). To visualize changes in local fluorescence intensity we have calculated, based on these movies, temporal fluorescence ratio images (FRIT) where the ratio values between images taken at two time points (1-min interval) are represented by a spectral color scale. The color scale is such that structures that remain unchanged between the two time points appear yellow, new structures are blue, and those that disappear are depicted in red. (Fig. 3; upper panels show the initial time point and lower panels show the same region after 1 h). Analysis of the time-lapse movies of YFP-paxillin (Supplementary Material, movie 2) and the FRIT analysis (Supplementary Material, movie 3) of cells on the different nanopatterns shows that on the 58-nm RGD-nanopattern the paxillin-containing contacts move slowly along the cell margins; these contacts grow in size and density, as evident by the large yellow patches (Fig. 3 B, upper and lower panels), similar to those observed on the homogeneous RGD surface (Fig. 3 A, upper and lower panels). Cells cultured on the 108-nm RGD-nanopattern, on the other hand, displayed highly dynamic paxillin adhesions that were smaller and less dense than those formed on the 58-nm RGD-nanopatterns (Fig. 3 C, upper and lower panels). Please note the predominance of blue pixels (which represent new paxillin contacts) and red (which represent contacts that disappeared) in the FRIT images of cells cultured on the 108-nm RGD-nanopattern compared to the yellow pixels that are common in the cells cultured on the 58-nm-spaced nanopattern.
FIGURE 3.
FRIT images (fluorescence ratio of images at different time points) of movies showing REF cells transfected with YFP-paxillin on (A) control surface, (B) 58-nm, and (C) 108-nm RGD-nanopattern. Upper panels show the initial time point after 3 h of plating cells on surfaces; lower panels show the same region after 1 h. Images were taken during the following 2 h of spreading at 1-min intervals. The images are temporal ratios of two consecutive frames: structures that appear only at the later image are shown in blue, whereas structures present at the earlier time point are red. Unchanged paxillin locations are represented in yellow. The whole movie (color and ratio) can be found in the online Supplementary Material.
Effect of nanopattern density on the organization and intensity of FA components
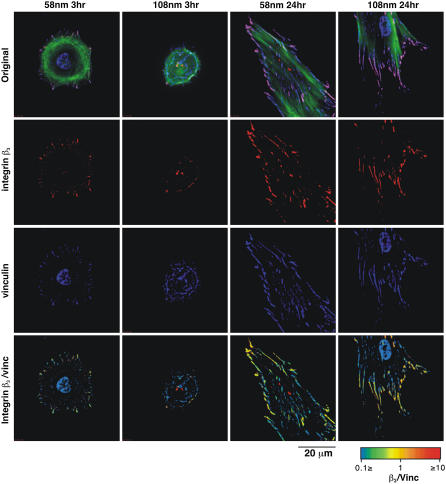
To compare the molecular composition and spatial distribution of FA formed on the dense (58 nm) and sparse (108 nm) nanopatterned surfaces, REF52 cells expressing GFP-integrin β3 were plated on these substrates for 3 or 24 h, then fixed and stained for vinculin and actin. Examination of the triple-labeled cells revealed more extensive spreading on the 58-nm RGD-nanopattern compared to the 108-nm nanopattern, with numerous vinculin- and integrin β3-rich peripheral FA (Fig. 4, left columns). After 24 h of incubation, the extent of spreading was similar on the two surfaces, yet there were striking differences in the distribution of the two FA proteins. Vinculin was associated with prominent FA in cells attached to the nanopatterned surfaces, however, its distribution was restricted mainly to the periphery of cells growing on the 108-nm nanopattern, whereas cells growing on the 58-nm pattern displayed numerous adhesions over their entire ventral surface. Interestingly, integrin β3 was primarily associated with the peripheral adhesions in both cases. This is apparent from the “yellow appearance” of the peripheral FA, and the abundance of “blue adhesions” in the central area on the cells growing on the 58-nm pattern.
FIGURE 4.
Immunostaining and fluorescence ratio images (FRI) of focal adhesion proteins. REF52 cells transfected with GFP-integrin β3 were fixed and immunostained with primary antibody against vinculin, followed by Cy5-conjugated secondary antibodies. Actin filaments were visualized with phalloidin-TRITC. Cells on 58-nm and 108-nm RGD-nanopatterns were observed at 3 h and 24 h after plating. The rows present the images with integrin β3 in red, vinculin in blue, and actin in green. The last row shows the ratio between integrin β3 and vinculin intensities. FRI are presented in a spectrum scale as indicated in the lookup table.
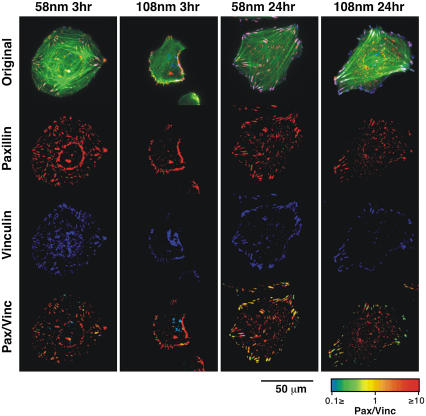
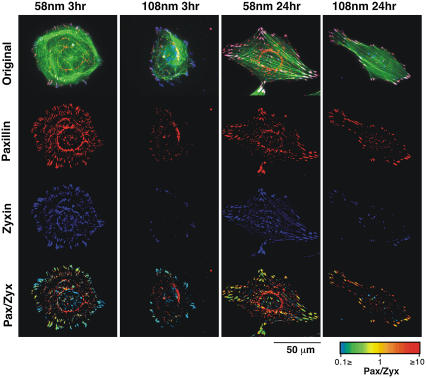
To test the effect of the spacing between RGD-peptides on the submembrane plaque components of FA, REF52 cells expressing YFP-paxillin were plated on the different substrates for 3 or 24 h and then fixed and immunolabeled for vinculin (Fig. 5) or zyxin (Fig. 6). Comparison of vinculin and paxillin revealed largely similar distributions, manifested by the largely yellow ratio images (Fig. 5, bottom row). In agreement with Fig. 4, cells growing for 3 h on the 58-nm pattern were considerably more spread than those plated on the 108-nm nanopattern. After longer incubation (24 h) vinculin- and paxillin-rich adhesions were detected throughout the ventral surface of cells growing on the 58-nm pattern, whereas those formed by cells growing on the 108-nm patterned surface were restricted to the cells periphery. Labeling of GFP-paxillin expressing REF52 cells for zyxin, revealed FA patterns largely similar to those described above, however, the levels of zyxin, relative to paxillin, varied considerably between FA, as evident from the heterogeneous color of FA in the ratio images (Fig. 6, bottom). Generally, the prominence of zyxin in FA formed with the 108-nm surface was considerably lower (∼40%) than that measured in FA formed with the 58-nm surface.
FIGURE 5.
Immunostaining and FRI of focal adhesion proteins. REF52 cells transfected with YFP-paxillin were fixed and immunostained with primary antibody against vinculin, followed by Cy5-conjugated secondary antibodies. Actin filaments were visualized with phalloidin-TRITC. Cells on 58-nm and 108-nm RGD-nanopatterns were observed at 3 h and 24 h after plating. The rows present the images with paxillin in red, vinculin in blue, and actin in green. The last row shows the ratio between paxillin and vinculin intensities. FRI are presented in a spectrum scale as indicated in the lookup table.
FIGURE 6.
Immunostaining and FRI of focal adhesion proteins. REF52 cells transfected with YFP-paxillin were fixed and immunostained with primary antibody against zyxin, followed by Cy5-conjugated secondary antibodies. Actin filaments were visualized with phalloidin-TRITC. Cells on 58-nm and 108-nm RGD-nanopatterns were observed at 3 h and 24 h after plating. The rows present the images with paxillin in red, zyxin in blue, and actin in green. The last row shows the ratio between paxillin and zyxin intensities. FRI are presented in a spectrum scale as indicated in the lookup table.
Formation of membrane microtethers and adhesion instability in migrating cells adhering to the RGD-nanopatterns
The data described above indicate that ligand spacing affects the protrusion-retraction cycles as well as the dynamics and molecular composition of matrix adhesions. Careful examination of these retractions by total internal reflection fluorescence (TIRF) microscopy, revealed a frequent formation of thin membrane tethers, rich in integrin β3 (Fig. 7 A). Examination of these structures by scanning electron microscopy showed the presence of membrane patches connected by a network of thin, long, and occasionally branching membrane cords (Fig. 7, B and C). These membrane tethers and patches were closely aligned along the gold nanodots present on the surface (Fig. 7 D).
FIGURE 7.
Fibroblasts on 108-nm RGD-nanopattern 3 h after adhesion. (A) TIRF image of REF cell expressing GFP-integrin β3. (B–D) Fibroblast adhering to nanopattern and its membrane structures have been visualized by SEM.
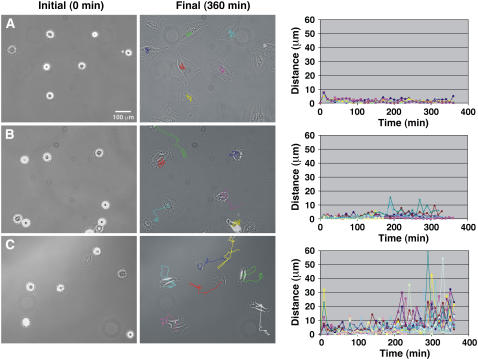
The apparent instability of FA in cells attached to the sparse RGD-nanopattern, greatly affected the migratory activity of the cells (Fig. 8 and Supplementary Material, movie 4). On the homogeneously coated RGD surfaces, REF52 cells were rather stationary during the first 6 h after plating (Fig. 8 A) and only occasionally acquired polarized morphology and started migrating. Cells cultured on the 58-nm RGD-nanopattern were similarly immobile, except for frequent peripheral protrusions and retractions (Fig. 8 B). On the 108-nm RGD-nanopattern rapid changes in cell position were evident, manifested by erratic cycles of protrusion and retraction, which dominated the dynamic behavior of the cells (Fig. 8 C).
FIGURE 8.
Cell motility on surfaces presenting (A) homogeneous coating of RGD peptides, (B) 58-nm RGD-nanopattern, and (C) 108-nm RGD nanopattern. The center of mass of cells adhering to the surfaces has been measured; plots show the velocity of several cells by evaluating the change in position of the center of mass (indicated as distance in μm) as a function of time (min). The initial time of measurement is the beginning of cell spreading. In the final image after 360 min, cell trajectories are indicated by colored lines. Each line in the plot corresponds to measurements of a single cell. The whole movie can be found in the online Supplementary Material.
DISCUSSION
The aim of this work was to elucidate the mechanisms underlying the differential effects of adhesive surfaces, with different ligand spacing at the nanoscale range, on cell adhesion and migration. For that purpose we compared the dynamic properties of cell adhesions and the migratory behavior of fibroblasts seeded on nanopatterned surfaces consisting of nanoparticles, with an interparticle spacing of 58 nm or 108 nm, conjugated to cRGDfK peptide. These particles have a diameter of 8 nm allowing each particle to interact with only one integrin receptor (39,40).
In a previous study we have shown that specific ligand functionalization and spacing on the surface is essential for the assembly of focal adhesions and stress fibers, since nonfunctionalized surfaces or surfaces with a spacing of ≥73 nm failed to induce the formation of such structures (34). It was concluded that nanoscale differences in ligand density are, indeed, sensed by living cells and affect the assembly of microns-scale matrix adhesion sites. However, the nature of the “molecular ruler” that is sensitive to the critical nanoscale spacing between ligand moieties is still unclear and, similarly, the cellular mechanisms responsible for the spacing-dependent cellular response were not defined in those early studies.
In this work we have focused on the long-range effects of adhesion to surfaces with different ligand spacing, namely, on global cell spreading, polarization, and migration. The dynamic analysis of cell spreading, migration, and FA formation shown here was performed, using two interligand spacing, namely 58 nm and 108 nm. These specific distances were selected based on previous work (34,41) and additional preliminary studies, indicating that cells cultured on the 58-nm nanopattern form normal FA, whereas those plated on the 108-nm nanopattern fail to develop FA. This study reveals major differences in dynamic cell behavior and focal adhesion composition and reorganization, which are regulated by ligand spacing. At the overall cellular level, these include delayed and considerably reduced spreading on the 108-nm-spaced pattern, and enhanced motile activity (Figs. 1 and 7). REF52 cells plated on homogeneously coated RGD surfaces or on the 58-nm RGD-nanopatterns reach maximum spreading within 1–2 h after plating, and are hardly migrating. On the 108-nm RGD-nanopattern, the apparent rate of spreading of the same cells was reduced by ∼50%. The kymograph analysis showed that this is not due to reduced lamellipodial protrusion but rather to excessive retractions due to an apparent instability of cell contacts formed on the 108-nm-spaced pattern.
Previous studies on the formation and reorganization of integrin adhesions during cell spreading and migration demonstrated that active membrane protrusion, induced by the small GTPase Rac1 (42) is followed by the establishment of many focal complexes, located close to the edge of the leading lamellae (43,44). These early adhesions were shown to be short lived (1–2 min range) and to be stationary relative to the matrix (45). They form under the advancing lamellae and fade or disappear; then the lamellipodium retracts or further advances. Some of the focal complexes “survive” after the retraction of the lamellipodium and are induced to grow and transform into a bona fide focal adhesion (15,45). This transformation is accompanied by an increase in the local concentration of different plaque proteins and recruitment of the plaque protein zyxin (21,46). It has been suggested that during spreading and migration cells produce initial adhesions with which they “sense” their environment by applying force to these new adhesions. This force application occurs regularly when the leading edge performs protrusion-retraction oscillations; local periodic contractions of lamellipodia that depend on matrix rigidity are due to an actin bound, contraction-activated signaling complex (10,47).
At the leading edge of cells spreading on homogeneously coated RGD surfaces, oscillations during advancement were observed until a plateau is reached and the leading edge becomes stable with only occasional oscillation when there is a change of direction. On the 58-nm pattern the protrusion-retraction cycles are quite similar, except that “stationary phases” are less prominent than on the homogeneous surface. On the 108-nm pattern there is erratic and poor spreading with protrusion-retraction cycles characterized by twofold higher frequency and 4–5 higher velocities, compared to the homogeneous surface and the 58-nm pattern. These results suggest a novel mechanism, regulating cell adhesion and protrusion, whereby nanoscale differences in adhesive ligand density (RGD) determine the rate of leading edge dynamics. Based on previous studies (34) and this work, it appears that there is a critical ligand (RGD) density (interligand spacing of <70 nm) that is essential for the formation of stable integrin-mediated adhesions. At larger spacing, integrin adhesion can still take place, yet the adhesions formed fail to develop into focal adhesions and to induce stress fiber assembly. We show here that these cryptic adhesions, formed under the protruding lamellae, are too weak to resist the mechanical forces applied by the leading lamellae during the “retractive phase” and thus fail to support spreading or develop persistent motility. Moreover, we show here that the erratic behavior of the leading edge of the cells plated on the 108-nm-spaced nanodots is attributable to lower resistance to retraction, rather than decreased protrusive activity since the frequency of protrusion and retraction cycles on the 108-nm pattern is considerably higher, compared to that on the 58-nm pattern or the homogeneous surfaces (Fig. 2).
Filopodia are active extensions of membrane and cytoskeleton beyond the cell edge into previously unoccupied areas; therefore, these structures are frequently associated with the exploratory behavior of the leading edge of a motile cell (48). Erratic lateral motions of thin structures at the leading edge could not be observed at the chosen time interval of 30 s, however, the dynamic behavior of filopodia using real-time imaging is currently under investigation. Retraction fibers are due to retraction and condensation of membrane and cytoplasm around a residual cytoskeletal core. In migrating cells they represent the trailing edge and are thought to provide a specialized mechanism by which the trailing edge is released from the substratum to allow migration in the opposite direction (49). These trailing structures are thought to be less structurally rigid and more deformable than lamellipodial and filopodial structures at the leading edge of the cell (50). Our findings indicate that surface nanopatterning has an effect on the behavior at the trailing edge. In cells on homogeneously coated RGD surfaces, tail retraction leads to complete detachment or apparent loss of intact posterior FA (51–53), whereas cells migrating on the 58-nm or 108-nm patterns show the development of thin (15–20 nm) and long (30 μm), tube-like membrane tethers. Evidently, formation of such tethers was considerably enhanced on the sparse pattern, compared to the dense one. Density dependence of tether formation highlights some interesting features of the molecular cooperativity in the regulation of adhesion strength. Apparently, at high ligand density (at levels found on the homogeneously coated surface) individual molecular interactions are reinforced and mechanically interlinked, so that the entire “adhesive patch” (namely, focal complex or FA) behaves as one adhesive element. The resistance to contractile forces of adhesions formed with the nanodot surfaces (even with the 58-nm pattern) is dominated by individual, or just a few molecular interactions, which are sufficiently strong to produce thin tethers (54,55), yet insufficient to induce complete mechanical linkage between individual integrins in the same adhesion structure. The density dependence of this phenomenon is demonstrated by the much greater prominence of tethers formed on the 108-nm surface.
The failure to develop stable anterior protrusions with the sparse surface is shown here to correlate with the formation of immature and highly unstable integrin adhesions. This observation is corroborated here at two independent levels: the dynamic analysis of FA, based on time-lapse movies (Supplementary Material, movies 2 and 3) and the temporal ratio imaging (Figs. 3–6) indicate a considerably higher turnover of integrin adhesions, formed with the 108-nm-spaced nanopattern, compared to those formed with the uniform surface or the 58-nm pattern. The leading edge is a site where new adhesions form and paxillin recruitment to these contact sites is an early event in the formation of such adhesions likely serving as an adaptor protein with signaling roles (56). Upon recruitment of stabilizing structural proteins, like alpha-actinin and vinculin, these highly dynamic sites grow in size and molecular complexity and their turnover rate decreases (57). Cells cultured on the 108-nm RGD-nanopattern displayed highly dynamic paxillin adhesions that were smaller and less dense than those formed on the 58-nm RGD-nanopatterns. The smaller clusters move faster than the larger clusters, which instead remain intact for over 30 min as they translocate centripetally, indicating that these structures become stabilized on the 58-nm pattern.
Furthermore, the molecular properties of integrin adhesions, formed on the two surfaces, were distinctly different, displaying lower levels of some plaque components such as zyxin in adhesions formed on the 108-nm pattern (Fig. 6). It is noteworthy that zyxin is one of the focal adhesion plaque components that are absent from focal complexes (45) and recruited to adhesion sites after application of mechanical stress (21,46). Zaidel-Bar et al. (45) showed that zyxin is incorporated to adhesion sites upon local retraction, suggesting that this process is force dependent. Given the apparently lower retention force of integrin adhesions on the 108-nm pattern, and to some extent on the 58-nm pattern, one may propose that this force is insufficient for effective zyxin recruitment. It thus appears that the failure of adhesions, formed with the sparse surface, to mature and develop cytoskeletal interactions is correlated with and is most likely responsible for their failure to support normal spreading and persistent migration.
Taken together, the observations described in this article, shed new light on the importance of positioning of ECM epitopes, at a nanoscale resolution, on FA formation and maturation and on the regulation of cell migration.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biphysj.org.
Acknowledgments
We thank Dr. Jacques Blümmel for the synthesis of PEG, Dr. Philippe Girard and Mrs. Eva Kowalinski for their help with image analysis, and Mrs. Sabine Rinck-Janke for her assistance with SEM imaging.
E.A.C.A. is supported by a Marie Curie Intraeuropean Fellowship (EIF) Meif-ct-2003-501883. B.G. holds the E. Neter Chair in Cell and Tumor Biology. This study was supported by a grant from the Deutsche-Israeli Program (DIP), the Landesstiftung Baden-Württemberg, the Max Planck Society, and the STREP Nanocues (EU 6th framework program).
Abbreviations used: ECM, extracellular matrix; FA, focal adhesion; FBS, fetal bovine serum; FRI, fluorescence ratio imaging; FRIT, fluorescence ratio imaging in time; PBS, phosphate buffered saline; PEG, polyethylene glycol; REF, rat embryonic fibroblasts; RGD, arginine-glycine-aspartate; SEM, scanning electron microscope; TIRF, total internal reflection fluorescence.
References
- 1.Hynes, R. O. 1987. Integrins: a family of cell surface receptors. Cell. 48:549–554. [DOI] [PubMed] [Google Scholar]
- 2.Pavalko, F. M., and C. A. Otey. 1994. Role of adhesion molecule cytoplasmic domains in mediating interactions with the cytoskeleton. Proc. Soc. Exp. Biol. Med. 205:282–293. [DOI] [PubMed] [Google Scholar]
- 3.Geiger, B., and A. D. Bershadsky. 2001. Transmembrane crosstalk between the extracellular matrix-cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2:793–805. [DOI] [PubMed] [Google Scholar]
- 4.Ruoslahti, E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697–715. [DOI] [PubMed] [Google Scholar]
- 5.Yamada, K. M., and B. Geiger. 1997. Molecular interactions in cell adhesion complexes. Curr. Opin. Cell Biol. 9:76–85. [DOI] [PubMed] [Google Scholar]
- 6.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 7.Ballestrem, C., B. Wehrle-Haller, and B. A. Imhof. 1998. Actin dynamics in living mammalian cells. J. Cell Sci. 111:1649–1658. [DOI] [PubMed] [Google Scholar]
- 8.Sheetz, M. P., D. Felsenfeld, C. G. Galbraith, and D. Choquet. 1999. Cell migration as a five-step cycle. Biochem. Soc. Symp. 65:233–243. [PubMed] [Google Scholar]
- 9.Bershadsky, A. D., N. Q. Balaban, and B. Geiger. 2003. Adhesion-dependent cell mechanosensitivity. Annu. Rev. Cell Dev. Biol. 19:677–695. [DOI] [PubMed] [Google Scholar]
- 10.Dobereiner, H. G., B. J. Dubin-Thaler, G. Giannone, and M. P. Sheetz. 2005. Force sensing and generation in cell phases: analyses of complex functions. J. Appl. Physiol. 98:1542–1546. [DOI] [PubMed] [Google Scholar]
- 11.Ponti, A., M. Machacek, S. L. Gupton, C. M. Waterman-Storer, and G. Danuser. 2004. Two distinct actin networks drive the protrusion of migrating cells. Science. 305:1782–1786. [DOI] [PubMed] [Google Scholar]
- 12.Ponti, A., A. Matov, M. Adams, S. Gupton, C. M. Waterman-Storer, and G. Danuser. 2005. Periodic patterns of actin turnover in lamellipodia and lamellae of migrating epithelial cells analyzed by quantitative fluorescent speckle microscopy. Biophys. J. 89:3456–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobereiner, H. G., B. J. Dubin-Thaler, G. Giannone, and M. P. Sheetz. 2005. Force sensing and generation in cell phases: analyses of complex functions. J. Appl. Physiol. 98:1542–1546. [DOI] [PubMed] [Google Scholar]
- 14.Dubin-Thaler, B. J., G. Giannone, H. G. Dobereiner, and M. P. Sheetz. 2004. Nanometer analysis of cell spreading on matrix-coated surfaces reveals two distinct cell states and STEPs. Biophys. J. 86:1794–1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballestrem, C., B. Hinz, B. A. Imhof, and B. Wehrle-Haller. 2001. Marching at the front and dragging behind: differential alphaVbeta3-integrin turnover regulates focal adhesion behavior. J. Cell Biol. 155:1319–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Rooij, J., A. Kerstens, G. Danuser, M. A. Schwartz, and C. M. Waterman-Storer. 2005. Integrin-dependent actomyosin contraction regulates epithelial cell scattering. J. Cell Biol. 171:153–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wittmann, T., G. M. Bokoch, and C. M. Waterman-Storer. 2003. Regulation of leading edge microtubule and actin dynamics downstream of Rac1. J. Cell Biol. 161:845–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borisy, G. G., and T. M. Svitkina. 2000. Actin machinery: pushing the envelope. Curr. Opin. Cell Biol. 12:104–112. [DOI] [PubMed] [Google Scholar]
- 19.Burridge, K., and M. Chrzanowska-Wodnicka. 1996. Focal adhesions, contractility, and signaling. Annu. Rev. Cell Dev. Biol. 12:463–518. [DOI] [PubMed] [Google Scholar]
- 20.Geiger, B., and A. D. Bershadsky. 2001. Assembly and mechanosensory function of focal contacts. Curr. Opin. Cell Biol. 13:584–592. [DOI] [PubMed] [Google Scholar]
- 21.Zaidel-Bar, R., M. Cohen, L. Addadi, and B. Geiger. 2004. Hierarchical assembly of cell-matrix adhesion complexes. Biochem. Soc. Trans. 32:416–420. [DOI] [PubMed] [Google Scholar]
- 22.Asthagiri, A. R., C. M. Nelson, A. F. Horwitz, and D. A. Lauffenburger. 1999. Quantitative relationship among integrin-ligand binding, adhesion, and signaling via focal adhesion kinase and extracellular signal-regulated kinase 2. J. Biol. Chem. 274:27119–27127. [DOI] [PubMed] [Google Scholar]
- 23.Balaban, N. Q., U. S. Schwarz, D. Riveline, P. Goichberg, G. Tzur, I. Sabanay, D. Mahalu, S. Safran, A. Bershadsky, L. Addadi, and B. Geiger. 2001. Force and focal adhesion assembly: a close relationship studied using elastic micropatterned substrates. Nat. Cell Biol. 3:466–472. [DOI] [PubMed] [Google Scholar]
- 24.Choquet, D., D. P. Felsenfeld, and M. P. Sheetz. 1997. Extracellular matrix rigidity causes strengthening of integrin-cytoskeleton linkages. Cell. 88:39–48. [DOI] [PubMed] [Google Scholar]
- 25.Katz, B. Z., E. Zamir, A. Bershadsky, Z. Kam, K. M. Yamada, and B. Geiger. 2000. Physical state of the extracellular matrix regulates the structure and molecular composition of cell-matrix adhesions. Mol. Biol. Cell. 11:1047–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loftis, M. J., D. Sexton, and W. Carver. 2003. Effects of collagen density on cardiac fibroblast behavior and gene expression. J. Cell. Physiol. 196:504–511. [DOI] [PubMed] [Google Scholar]
- 27.Lehnert, D., B. Wehrle-Haller, C. David, U. Weiland, C. Ballestrem, B. A. Imhof, and M. Bastmeyer. 2004. Cell behaviour on micropatterned substrata: limits of extracellular matrix geometry for spreading and adhesion. J. Cell Sci. 117:41–52. [DOI] [PubMed] [Google Scholar]
- 28.Chen, C. S., M. Mrksich, S. Huang, G. M. Whitesides, and D. E. Ingber. 1997. Geometric control of cell life and death. Science. 276:1425–1428. [DOI] [PubMed] [Google Scholar]
- 29.Pizzo, A. M., K. Kokini, L. C. Vaughn, B. Z. Waisner, and S. L. Voytik-Harbin. 2005. Extracellular matrix (ECM) microstructural composition regulates local cell-ECM biomechanics and fundamental fibroblast behavior: a multidimensional perspective. J. Appl. Physiol. 98:1909–1921. [DOI] [PubMed] [Google Scholar]
- 30.Massia, S. P., and J. A. Hubbell. 1990. Covalently attached GRGD on polymer surfaces promotes biospecific adhesion of mammalian cells. Ann. N. Y. Acad. Sci. 589:261–270. [DOI] [PubMed] [Google Scholar]
- 31.Massia, S. P., and J. A. Hubbell. 1991. An RGD spacing of 440 nm is sufficient for integrin alpha V beta 3-mediated fibroblast spreading and 140 nm for focal contact and stress fiber formation. J. Cell Biol. 114:1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maheshwari, G., G. Brown, D. A. Lauffenburger, A. Wells, and L. G. Griffith. 2000. Cell adhesion and motility depend on nanoscale RGD clustering. J. Cell Sci. 113:1677–1686. [DOI] [PubMed] [Google Scholar]
- 33.Koo, L. Y., D. J. Irvine, A. M. Mayes, D. A. Lauffenburger, and L. G. Griffith. 2002. Co-regulation of cell adhesion by nanoscale RGD organization and mechanical stimulus. J. Cell Sci. 115:1423–1433. [DOI] [PubMed] [Google Scholar]
- 34.Arnold, M., E. A. Cavalcanti-Adam, R. Glass, J. Blummel, W. Eck, M. Kantlehner, H. Kessler, and J. P. Spatz. 2004. Activation of integrin function by nanopatterned adhesive interfaces. ChemPhysChem. 5:383–388. [DOI] [PubMed] [Google Scholar]
- 35.Spatz, J.P., S. Mossmer, C. Hartmann, M. Möller, T. Herzog, M. Krieger, H.G. Boyen, P. Ziemann, and B. Kabius. 2000. Ordered deposition of inorganic clusters from micellar block copolymer films. Langmuir. 16:407–415. [Google Scholar]
- 36.Glass, R., M. Möller, J. P. Spatz. 2003. Block copolymer micelle nanolithography. Nanotechnology. 14:1153–1160. [Google Scholar]
- 37.Zamir, E., B. Z. Katz, S. Aota, K. M. Yamada, B. Geiger, and Z. Kam. 1999. Molecular diversity of cell-matrix adhesions. J. Cell Sci. 112:1655–1669. [DOI] [PubMed] [Google Scholar]
- 38.Rasband, W. S. 2005. ImageJ. U.S. National Institutes of Health, Bethesda, MD. http://rsb.info.nih.gov/ij/ [Online].
- 39.Xiong, J. P., T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D. L. Scott, A. Joachimiak, S. L. Goodman, and M. A. Arnaout. 2001. Crystal structure of the extracellular segment of integrin alpha Vbeta3. Science. 294:339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiong, J. P., T. Stehle, R. Zhang, A. Joachimiak, M. Frech, S. L. Goodman, and M. A. Arnaout. 2002. Crystal structure of the extracellular segment of integrin alpha Vbeta3 in complex with an Arg-Gly-Asp ligand. Science. 296:151–155. [DOI] [PubMed] [Google Scholar]
- 41.Cavalcanti-Adam, E. A., T. Volberg, A. Micoulet, J. Blümmel, J. Auernheimer, H. Kessler, and J. P. Spatz. 2006. Lateral spacing of integrin ligands influences cell spreading and focal adhesion assembly. Eur. J. Cell Biol. 85:219–224. [DOI] [PubMed] [Google Scholar]
- 42.Hall, A. 1998. Rho GTPases and the actin cytoskeleton. Science. 279:509–514. [DOI] [PubMed] [Google Scholar]
- 43.Nobes, C. D., and A. Hall. 1995. Rho, rac, and cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell. 81:53–62. [DOI] [PubMed] [Google Scholar]
- 44.Rottner, K., A. Hall, and J. V. Small. 1999. Interplay between Rac and Rho in the control of substrate contact dynamics. Curr. Biol. 9:640–648. [DOI] [PubMed] [Google Scholar]
- 45.Zaidel-Bar, R., C. Ballestrem, Z. Kam, and B. Geiger. 2003. Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116:4605–4613. [DOI] [PubMed] [Google Scholar]
- 46.Lele, T. P., J. Pendse, S. Kumar, M. Salanga, J. Karavitis, and D. E. Ingber. 2006. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J. Cell. Physiol. 207:187–194. [DOI] [PubMed] [Google Scholar]
- 47.Giannone, G., B. J. Dubin-Thaler, H. G. Dobereiner, N. Kieffer, A. R. Bresnick, and M. P. Sheetz. 2004. Periodic lamellipodial contractions correlate with rearward actin waves. Cell. 116:431–443. [DOI] [PubMed] [Google Scholar]
- 48.Lauffenburger, D. A., and A. F. Horwitz. 1996. Cell migration: a physically integrated molecular process. Cell. 84:359–369. [DOI] [PubMed] [Google Scholar]
- 49.Sheetz, M. P. 1994. Cell migration by graded attachment to substrates and contraction. Semin. Cell Biol. 5:149–155. [DOI] [PubMed] [Google Scholar]
- 50.Schmidt, C. E., A. F. Horwitz, D. A. Lauffenburger, and M. P. Sheetz. 1993. Integrin-cytoskeletal interactions in migrating fibroblasts are dynamic, asymmetric, and regulated. J. Cell Biol. 123:977–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abercrombie, M., J. E. Heaysman, and S. M. Pegrum. 1970. The locomotion of fibroblasts in culture. 3. Movements of particles on the dorsal surface of the leading lamella. Exp. Cell Res. 62:389–398. [DOI] [PubMed] [Google Scholar]
- 52.Abercrombie, M., J. E. Heaysman, and S. M. Pegrum. 1970. The locomotion of fibroblasts in culture. I. Movements of the leading edge. Exp. Cell Res. 59:393–398. [DOI] [PubMed] [Google Scholar]
- 53.Abercrombie, M., G. A. Dunn, and J. P. Heath. 1977. The shape and movement of fibroblasts in culture. Soc. Gen. Physiol. Ser. 32:57–70. [PubMed] [Google Scholar]
- 54.Roux, A., G. Cappello, J. Cartaud, J. Prost, B. Goud, and P. Bassereau. 2002. A minimal system allowing tubulation with molecular motors pulling on giant liposomes. Proc. Natl. Acad. Sci. USA. 99:5394–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cuvelier, D., P. Chiaruttini, P. Bassereau, and P. Nassoy. 2005. Pulling long tubes from firmly adhered vesicles. Europhys. Lett. 71:1015–1021. [Google Scholar]
- 56.Schaller, M. D., and J. T. Parsons. 1995. pp125FAK-dependent tyrosine phosphorylation of paxillin creates a high-affinity binding site for Crk. Mol. Cell. Biol. 15:2635–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luakaitis, C. M., D. J. Webb, K. Donais, and A. F. Horwitz. 2001. Differential dynamics of alpha 5 integrin, paxillin, and alpha-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153:1427–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]