Abstract
Cardiomyopathy is a major cause of morbidity and mortality. Ventricular conduction delay, as shown by prolonged deflections in the electrocardiogram caused by delayed ventricular contraction (wide QRS complex), is a common feature of cardiomyopathy and is associated with a poor prognosis. Although the Gi-signaling pathway is up-regulated in certain cardiomyopathies, previous studies suggested this up-regulation was compensatory rather than a potential cause of the disease. Using the tetracycline transactivator system and a modified Gi-coupled receptor (Ro1), we provide evidence that increased Gi signaling in mice can result in a lethal cardiomyopathy associated with a wide QRS complex arrhythmia. Induced expression of Ro1 in adult mice resulted in a >90% mortality rate at 16 wk, whereas suppression of Ro1 expression after 8 wk protected mice from further mortality and allowed partial improvement in systolic function. Results of DNA-array analysis of over 6,000 genes from hearts expressing Ro1 are consistent with hyperactive Gi signaling. DNA-array analysis also identified known markers of cardiomyopathy and hundreds of previously unknown potential diagnostic markers and therapeutic targets for this syndrome. Our system allows cardiomyopathy to be induced and reversed in adult mice, providing an unprecedented opportunity to dissect the role of Gi signaling in causing cardiac pathology.
Keywords: G proteins, signal transduction, gene expression, genome, bioinformatics
Idiopathic dilated cardiomyopathy (IDC) is a major cause of heart failure characterized by cardiac dilation and reduced systolic function. In the United States, about half of the cases of dilated cardiomyopathy are associated with myocarditis or coronary artery disease, and half are considered idiopathic (1–4). Ventricular conduction delay, as shown by a prolonged depolarization in the electrocardiogram (wide QRS complex), is associated with up to 70% of IDC cases (5) and is an independent risk factor for death among IDC patients (6, 7). Several lines of evidence implicate altered Gi signaling in the development of cardiomyopathies such as IDC, but a direct relationship between Gi signaling and cardiomyopathy has not been demonstrated in vivo. We recently created a system that utilizes a specifically designed Gi-coupled receptor and inducible gene expression techniques to control Gi signaling in the adult mouse heart (8).
With over 2,000 members, G protein-coupled receptors (GPCRs) are the largest known class of cell-surface receptors (9, 10). The physiological effect of each receptor is primarily defined by the G protein pathways it activates. The best-studied G protein-signaling pathways are Gi, Gs, and Gq, which inhibit adenylyl cyclase, stimulate adenylyl cyclase, and stimulate phospholipase C, respectively (11, 12). In the heart, Gi-coupled receptors, such as the A1 adenosine and the M2 muscarinic receptors, decrease intracellular cAMP levels, contractility, and heart rate. Although each of these effects opposes the actions of Gs, Gi also has effects independent of Gs/cAMP regulation, such as controlling a potassium-selective ion channel (IKACh) and certain calcium channels (13).
Recently, several lines of evidence have implicated aberrant Gi signaling in the etiology of human IDC. First, in patients with IDC, Gi protein and mRNA levels are increased in ventricular tissues (2, 14–16). Second, these hearts have decreased adenylyl cyclase activity (14, 16), a known effect of Gi signaling. Third, autoantibodies that bind to and cause signaling through the Gi-coupled M2 muscarinic receptor are found in up to 40% of patients with IDC (17, 18). Despite the clinical association between increased Gi signaling and human IDC, many investigators have proposed that this signaling is a compensatory response to the “hyperadrenergic state” of heart failure (16, 19–21). Recent studies in transgenic and knockout mouse models have implicated cell-signaling molecules and myocyte structural proteins in the development (22) and potential treatment (23) of cardiomyopathy. Overexpression of signaling components of either the Gs or Gq pathway in mouse heart does not cause cardiomyopathy within the first months of life, but can eventually lead to cardiomyopathy if the mice are stressed or if expression is prolonged for more than 1 year (23–30). Similarly, expression of a dominant-active form of calcineurin leads to cardiac hypertrophy, which can eventually result in cardiomyopathy (23, 29). In contrast, overexpression of a dominant-negative cAMP response element-binding protein transcription factor (31) or targeted deletion of the gene encoding the muscle protein LIM (32) appears to cause a dilated cardiomyopathy without intervening hypertrophy. Although these mouse models provide insight into the molecular mechanisms of IDC, they do not address the potential role of Gi signaling in causing cardiomyopathy. Furthermore, none of these models allow the study of potentially cardiomyopathic signals in the adult mouse heart, free of developmental effects.
To determine the potential role of Gi signaling in causing cardiomyopathy, we used the tetracycline transactivator (tTA) system to express a modified Gi-coupled receptor in mouse heart (8, 33–35). This receptor is a modified human κ-opioid receptor that has a 200-fold decrease in binding and signaling in response to known endogenous agonists, but retains normal signaling in response to an exogenously administered small-molecule drug, spiradoline (36). We call this type of receptor a RASSL (receptor activated solely by a synthetic ligand). Ro1 (RASSL, opioid 1) was intended as a conditional signaling system that would allow drug-induced activation of Gi signaling in vivo. In tissue culture, activation of Ro1 increased Gi signaling (36). In the mouse heart, activation of Ro1 decreased heart rate by as much as 80%, the predicted result of an increase in Gi signaling (8).
Using the tet system, we can specify the start, extent, and termination of Ro1 expression in the heart of an adult mouse. Here we report that prolonged high-level Ro1 expression alone caused hyperactive Gi signaling, a wide QRS complex arrhythmia, severe congestive heart failure, systolic dysfunction, and death, consistent with cardiomyopathy.
Methods
Transgenic Mice.
Mice generated with the cardiac-specific α myosin heavy chain (αMHC) promoter driving the tetracycline transactivator (αMHC-tTA) and tetO-Ro1 transgene constructs have been described (8, 35). Genotypes were determined by PCR (constructs and detailed protocols are available at http://gladstone.ucsf.edu/labs/conklin/). All αMHC-tTA/tetO-Ro1 mice were back-crossed for at least seven generations into an FVB/N background. All experiments were performed with littermate controls of either αMHC-tTA or αMHC-tTA/tetO-Ro1 genotype (as specified). Experimental and control mice were housed together during the described experiments. The transgenic mice used in these experiments have been donated to The Jackson Laboratory (Bar Harbor, ME) and are ready for distribution to nonprofit institutions.
Supplemental Data.
Detailed methods for doxycycline-mediated gene expression, ECG monitoring, pertussis toxin (PTX) treatment, spiradoline administration, histology, echocardiography, DNA-array analysis of gene expression with Affymetrix DNA arrays, statistical methods, QuickTime movie of mouse recovery, full DNA-array data sets, and interactive displays of DNA-array data may be found at www.pnas.org.
Results
Mortality in Adult Mice After Long-Term Cardiac Expression of Ro1.
To obtain cardiac expression of Ro1, we crossed mice harboring the αMHC-tTA (cardiac-specific) transgene (35) with mice harboring the tTA-responsive tetO-Ro1 transgene. In adult αMHC-tTA/tetO-Ro1 mice, we previously showed inducible and cardiac-restricted expression of Ro1 (8). To test the effects of chronic (>3 wk) Ro1 expression in adult mice, we weaned 15 αMHC-tTA/tetO-Ro1 mice from doxycycline at 8 wk of age (Fig. 1). All 15 of these adult mice survived the initial induction of receptor expression (10–14 days). However, 3 wk after doxycycline withdrawal, 1 mouse died; 8 wk after, 7 were dead; and 16 wk after, all but one had died. In another group of Ro1-expressing mice (n = 15), administration of doxycycline and reversal of Ro1 expression after 8 wk prevented further mortality (Fig. 1). None of the control mice (αMHC-tTA/tetO-Ro1, on doxycycline, n = 20) died during this experiment.
Figure 1.
Survival of mice expressing Ro1 in the heart: Kaplan–Meier survival plot of adult mice expressing Ro1. Doxycycline was withdrawn from 15 αMHC-tTA/tetO-Ro1 mice to induce Ro1 expression. All αMHC-tTA/tetO-Ro1 mice survived maximal induction of Ro1 (7–10 days). More than 40% of the mice died after 8 wk of Ro1 expression and 90% after 16 wk. Reinitiation (arrow) of doxycycline at 8 wk prevented further mortality in another group of αMHC-tTA/tetO-Ro1 mice (n = 15). None of the control mice died (αMHC-tTA/tetO-Ro1 on doxycycline for 16 wk, n = 20).
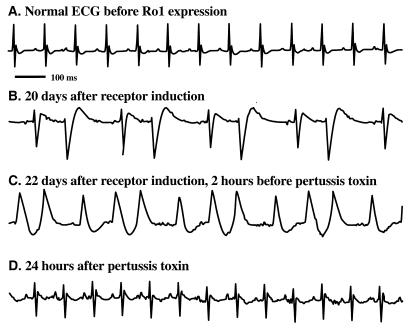
In a separate group of 20 mice expressing Ro1, we were able to monitor ECGs at regular intervals (5 min of each hour) in the days immediately before death. We noted a wide variety of cardiac arrhythmias, all characterized by wide QRS complexes, as shown in Fig. 7B in the supplemental material at www.pnas.org. In fact, 17 of the 20 mice showed signs of wide QRS complexes (>20 episodes of QRS ≥ 25 ms/250 beats) in the days before death. In over 100 h of recording in 10 normal mice, we never observed a wide QRS complex. Although most died shortly after developing wide QRS complexes, some mice survived for several days (Fig. 2), allowing us to directly test the role of Gi signaling in the wide QRS complex arrhythmia (see below).
Figure 2.
PTX reveals Gi-induced ventricular conduction delay and drug-induced signaling via expression of Ro1. Administration of PTX reduced ECG abnormalities in mice expressing Ro1. (A) Sinus rhythm (heart rate, 600 beats per min) with narrow QRS complex (<15 ms) before Ro1 expression. (B) Irregular rhythm with widened QRS complex (>25 ms) after 20 days of Ro1 expression. (C) Irregular rhythm with widened QRS maintained for 48 h before PTX injection. (D) Sinus rhythm (heart rate, 600 beats per min) with narrow QRS complex (<15 ms) 24 h after PTX injection.
PTX Blocks Gi-Induced Wide QRS Complexes in Ro1-Expressing Mice.
PTX specifically blocks Gi signaling by ADP-ribosylation of all members of the Gi family found in the heart (Gαi1, Gαi2, Gα0). In each of four mice that had developed wide QRS complexes after Ro1 expression, we administered a single injection of PTX (30 ng/g of body weight) and monitored QRS width and rhythm abnormalities. All four mice had a 90% reduction in the number of prolonged QRS events (from 92% ± 14% to 5% ± 10% wide QRS events in the first 250 beats in each hour, P < 0.01) and reverted to a sinus rhythm for more than 3 days, consistent with the prolonged actions of PTX in vivo (Fig. 2, with the full time course in Fig. 7B in the supplemental material). Thus, maximal expression of Ro1 causes increased Gi signaling in the heart that results in ventricular conduction delay (as reflected by wide QRS complexes) and may also contribute to the pathophysiology of the cardiomyopathy (see below). We have further evidence that this basal Gi signaling is due to overexpression of Ro1, since it does not occur at more moderate expression levels of Ro1 (see Discussion and Fig. 7H in the supplemental material).
Chronic Expression of Ro1 Causes Pathologic and Physiologic Changes Characteristic of Cardiomyopathy.
All 14 mice that died after long-term Ro1 expression (Fig. 1) had clinical or necropsy evidence of a cardiomyopathy. Just before death, 8 of the 14 mice developed signs of clinical distress characterized by labored breathing, rough hair coat, complete inactivity, failure to eat or drink, and fluid retention (Fig. 3B). At necropsy, these 8 mice had edema, ascites, and pleural effusions. Other mice showed signs of dehydration caused by lack of fluid intake (see QuickTime movie 1 in the supplemental material). Cross sections of hearts from all 14 mice demonstrated enlarged ventricular chamber size and decreased ventricular wall thickness compared with controls (Fig. 3 C and D).
Figure 3.
Expression of Ro1 in mouse heart causes a cardiomyopathy. (A and B) Control mouse (A, αMHC-tTA) and Ro1-expressing mouse with anasarca (B). (C and D) Photomicrographs (×5) of hematoxylin/eosin-stained cross sections of mouse heart. The ventricular chambers were larger and the ventricular walls thinner in the mouse expressing Ro1 (D) than in the control mouse (C).
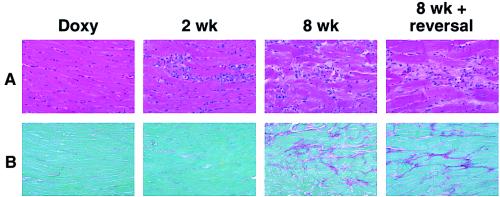
To assess the pathologic and physiologic changes associated with Ro1 expression, we analyzed mice expressing Ro1 and littermate controls at multiple time points. Heart weight to body weight ratios were unreliable because the sick mice exhibited extreme weight fluctuations caused by severe edema (Fig. 3) or cachexia (see QuickTime movie 1 in the supplemental material). We compared heart weight to weight of control mice (αMHC-tTA littermates) and found no significant differences at 8 wk of expression (controls 146 ± 33 mg, Ro1 175 ± 29 mg), indicating that hypertrophy is not a prominent feature of this cardiomyopathy. After 2 wk of Ro1 expression, histologic analysis showed a cellular infiltrate of the myocardium (Fig. 4A). After 4 wk, there was evidence of patchy myocyte disarray and collagen deposition in the myocardium. This collagen replacement was maximal after 8 wk (Fig. 4), when the mean amount of collagen deposition was 8 times greater in Ro1-expressing mice than in control mice (αMHC-tTA/tetO-Ro1, on doxycycline) (see Fig.7D in the supplemental material at www.pnas.org). These results demonstrate inducible pathologic changes that follow a predictable pattern of cellular infiltration followed by myocyte disarray and collagen replacement of the myocardium.
Figure 4.
Progression of inducible cardiac pathology in mice expressing Ro1. (A) Photomicrographs (×40) of hematoxylin/eosin-stained sections of myocardium from mice expressing Ro1. Cellular infiltration begins at 2 wk after doxycycline (Doxy) removal and is followed by myofibril disarray at 8 wk. Note the lack of improvement in myocardial histology in mice that had Ro1 expression for 8 wk followed by Ro1 suppression for 4 wk (8 wk + reversal). (B) Photomicrographs (×50) of Sirius red-stained sections of myocardium from mice expressing Ro1. Collagen deposition begins at 4 wk (not shown) and is most prominent at 8 wk. Note lack of improvement in the 8 wk + reversal group.
Myocardial force was measured in right ventricular papillary muscle from mice expressing Ro1 (8-wk time point). Absolute force (g/mm2) was 50% lower in mice expressing Ro1 than in control mice (αMHC-tTA). Furthermore, the rates of muscle contraction and relaxation were prolonged in Ro1-expressing mice (see Fig. 7 E and F in the supplemental material at www.pnas.org). These results suggest that expression of a Gi-coupled receptor for 8 wk in mouse myocardium impairs both the absolute force generated by the heart and the time course of contraction and relaxation. These physiologic findings are consistent with the observed phenotype of cardiomyopathy.
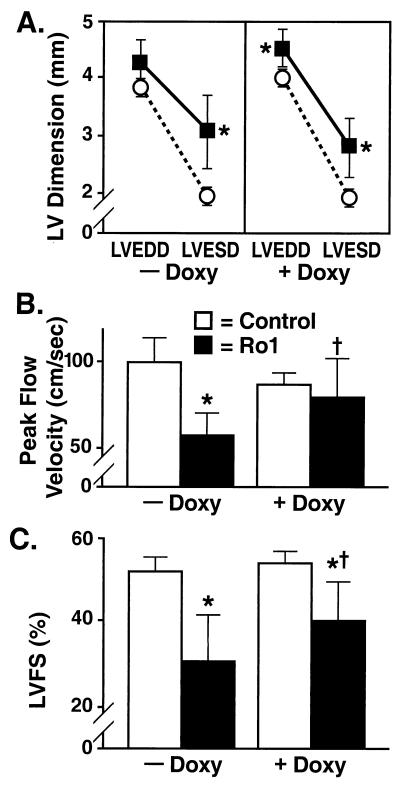
We next compared the in vivo cardiac physiology of Ro1-expressing mice and controls (αMHC-tTA). In mice expressing Ro1 (8-wk time point), transthoracic echocardiography demonstrated increased left ventricular (LV) systolic chamber size and impaired left ventricular fractional shortening (LVFS) and aortic peak flow velocity. The end-systolic diameter was 50% greater in mice expressing Ro1 than in control mice (Fig. 5A; −Doxy), and aortic peak flow velocity and LVFS were nearly 40% lower than in controls (Fig. 5 B and C; −Doxy). Interestingly, the LV end-diastolic diameter was not increased in these mice (which otherwise appeared healthy). However, when we selectively performed echocardiography on mice that showed signs of edema, there were clear signs of dilated cardiomyopathy in systole and diastole (see Fig. 7G in the supplemental material at www.pnas.org). Therefore, systolic dysfunction is an early marker of this cardiomyopathy.
Figure 5.
Echocardiographic evidence of cardiomyopathy and recovery in mice expressing Ro1. (A) LV dimension. Mice expressing Ro1 for 8 wk showed an increase in LV end-systolic diameter (LVESD) (■, Ro1, αMHC-tTA/tetO-Ro1, n = 9; ○, control, αMHC-tTA, n = 9). After administration of doxycycline (Doxy) to these same mice for 4 wk, there was a persistent increase in LVESD and a new increase in LV end-diastolic diameter (LVEDD). (B) Peak flow velocity. After 8 wk of Ro1 expression, aortic peak flow velocity decreased by nearly 40%. After 4 wk of doxycycline administration, peak flow velocity increased significantly. (C) LVFS. After 8 wk of Ro1 expression, LVFS decreased by nearly 40%. LVFS increased significantly after suppression of Ro1 expression but remained less than LVFS in control mice. *, P < 0.05 vs. controls; **, P < 0.001 vs. controls; †, P < 0.05 vs. same mice at the previous time point.
Suppression of Ro1 Expression Partially Reverses Cardiomyopathy.
We predicted from our survival curve data (Fig. 1) that suppressing Ro1 expression after 8 wk would reverse the cardiac pathologic and physiologic changes. To test this hypothesis, we suppressed Ro1 expression for 4 wk in the same nine mice that underwent echocardiographic examination after 8 wk of Ro1 expression. In mice with signs of clinical distress, suppression of Ro1 caused rapid improvement (<48 h) in breathing, activity, and fluid balance (see QuickTime movie 1, supplemental material at www.pnas.org). Pathologic examination of the hearts of these mice revealed no reduction in collagen deposition as compared with the 8-wk time point (Fig. 4 and Fig. 7C in the supplemental material). By echocardiography, suppression of Ro1 expression significantly improved aortic peak flow velocity and LVFS (Fig. 5 B and C; +Doxy). These results demonstrate that suppressing Ro1 expression prevents further mortality and allows partial recovery of cardiac function, but does not reverse some of the structural changes, such as chamber dilation (Fig. 5A; +Doxy) and collagen deposition (Fig. 4B).
Gene-Expression Changes in Ro1-Induced Cardiomyopathy.
To obtain gene-expression profiles in this syndrome, we used Affymetrix “murine 6500” DNA arrays to assess the expression levels of over 6,000 genes in the ventricles of eight control mice and nine mice expressing Ro1 for 8 wk (a separate set of DNA arrays was collected for each mouse). Examination of the genes immediately downstream of all G protein pathways showed a remarkable regulation of the Gi pathway and relative sparing of the proximal portions of the Gq pathway (see Fig. 6 and Fig. 8 A and B in the supplemental material at www.pnas.org). As part of an expected fetal gene-expression program (25, 29, 31, 32, 37, 38), these mice showed increases in the expression of genes encoding atrial natriuretic factor, brain natriuretic peptide, skeletal muscle actin, and β-myosin heavy chain and a decrease in the expression of the gene encoding αMHC. To identify genes not previously associated with cardiomyopathy, we ranked all of the genes and expressed sequence tags by their fold up-regulation (for the 8-wk time point vs. control). A full list is available in Table 1 in the supplemental material at www.pnas.org, and selected findings are highlighted in Discussion.
Figure 6.
Ro1-induced gene-expression changes in the G protein signaling cascade. All genes known to be downstream of G proteins were mapped, and gene-expression changes were indicated by using GenMAPP (www.GenMAPP.org). Significant changes (P < 0.05) are indicated by colors. The fold change is noted at the right of each box. Genes that are absent on the DNA array are in clear boxes. Nonsignificant (P > 0.05) changes are indicated by yellow. The abbreviation definitions, GenBank numbers, and expression values are all available in an interactive version of this figure and another figure focusing on information in Fig. 8 A and B in the supplemental material at www.pnas.org. Abbreviations include CaL, L-type calcium channel; AC, adenylyl cyclase; PLC, phospholipase C; Rho GEF, rho guanine nucleotide exchange factor; IP3, inositol trisphosphate; DAG, diacylglycerol; PDE, phosphodiesterase; PKA, protein kinase A; PKC, protein kinase C; CalM, calmodulin; Calc, calcineurin; CREB, cAMP response element binding protein; cAMP-dependent transcription factor; NFAT, nuclear factor of activated cells; NHE, Na+/H+ exchanger.
Discussion
A Mouse Model of Inducible Cardiomyopathy.
The Ro1 model of cardiomyopathy provides several insights that may advance our understanding of the etiology of cardiomyopathy. First, it demonstrates that the expression of a Gi-coupled receptor in mouse heart can cause a lethal cardiomyopathy with systolic dysfunction and LV chamber dilation. In contrast to previous data suggesting that up-regulation of the Gi pathway in cardiomyopathy is a compensatory response to hyperactive Gs signaling, our findings suggest a causal role for the Gi pathway. Second, our PTX experiments prove that Gi signaling causes ventricular conduction delay, a cardinal feature of dilated cardiomyopathy associated with a poor prognosis (6, 7). The cause of ventricular conduction delay in human cardiomyopathy is unknown. Our findings raise the possibility that Gi signaling plays a similar role in humans. Third, the Ro1 model allows precise timing of both gene induction and gene suppression in an adult mouse, which should allow dissection of the steps leading to this cardiomyopathy and those leading to recovery.
The role of Gi signaling in causing ventricular conduction delay as revealed by the PTX experiments is somewhat surprising. The simplest explanation is that PTX blocks Ro1 signaling via Gi; however, PTX could also have other nonspecific effects on the heart. Further evidence that the cardiomyopathy is due to Ro1 signaling has been obtained in a recent experiment using the κ opioid receptor antagonist nor-binaltorphimine (nor-BNI). Because Ro1 is based on the κ opioid receptor, we hypothesized that nor-BNI injected three times a week (10 μmol/kg) during 8 wk of expression would block the cardiomyopathy. We found that the nor-BNI blocked the fibrosis response (N.C., T.N., and B.R.C., unpublished results). The results obtained with nor-BNI and PTX indicate that Ro1 causes the cardiomyopathy via Gi signaling. Better control of the delivery of PTX to the heart, as might be provided by a tTA-controlled version of PTX, would allow us to assess the role of Gi signaling at specific times in the heart.
An important aspect of this mouse model is that the tet system permits precise timing of gene induction and suppression. Therefore, we can avoid potential deleterious effects of Ro1 expression during cardiac development and study the effects of Ro1 at precise time points in the adult mouse heart. Because cardiomyopathy is primarily a disease of adults, the adult mouse heart is a physiologically relevant model system. Our findings in this study suggest several pathological steps in the cardiomyopathy phenotype: initial cellular infiltration of the myocardium followed by myocyte disarray and eventual collagen deposition that correlate with a significant decrease in cardiac contractility and mouse survival. Although suppressing Ro1 expression allowed partial functional recovery, as shown by echocardiography (Fig. 7G and QuickTime movie 1 in the supplemental material at www.pnas.org), and protected against further mortality, collagen deposition was not reversed (see Fig. 4 and Fig. 7D in the supplemental material). The Ro1 model of cardiomyopathy allows us to examine tissues at early time points in the disease process that are rarely seen clinically. Thus, our control over Ro1 expression will allow investigation of pathologic, physiologic, and gene-expression steps leading to heart failure and to recovery from heart failure.
Basal Signaling by Ro1 in the Heart.
Our original intent in making Ro1 was to create a conditional signaling system that would allow drug-induced, high-level activation of Gi signaling in vivo. Activation of Ro1 by the small-molecule drug spiradoline causes a profound and reversible bradycardia, but expression of Ro1 alone is not associated with a decrease in heart rate (8). However, prolonged Ro1 expression in adult mice causes significant pathologic and physiologic abnormalities, suggesting some basal activity.
Likely mechanisms for basal Ro1 signaling include high-level expression of the receptor alone in cardiac myocytes or activation of the receptor by endogenous agonists at high local concentrations. Basal activity of a highly overexpressed GPCR in the heart has been characterized for the β2-adrenergic receptor (22). Basal signaling caused by high-level receptor expression is supported by the finding that a reduced level of receptor expression blocks the development of pathologic changes, while still maintaining ligand-activated control of heart rate (see Fig. 7 D and H in the supplemental material at www.pnas.org). These data suggest that ideal regulation of G protein signaling in vivo is achieved with markedly lower receptor expression levels than those we have achieved. Knowledge of appropriate receptor expression levels for various tissues will be important in building future RASSL-based conditional signaling systems.
DNA-Array Analysis of Cardiomyopathy Models.
An important goal of our combined physiologic and genomic studies is to understand on a genomic scale the changes in gene expression underlying the progression to cardiomyopathy. Knowledge of these changes should allow a more precise description of cardiomyopathy.
DNA arrays are our primary tool in this analysis. Because they yield overwhelming amounts of data, we developed a bioinformatics tool, Gene MicroArray Pathway Profiler (GenMAPP), which maps the data to known biochemical pathways (Fig. 6; Fig. 8 A and B in the supplemental material at www.pnas.org). GenMAPP will be freely available and can be viewed on the Internet (www.GenMAPP.org).
Using GenMAPP, we found a particularly striking pattern of changes in the expression of genes downstream of G protein signaling. Most changes are downstream of the Gi signaling pathway; relatively few occur in the proximal portions of the Gq pathway (Fig. 6). The direct effectors of Gi are modulated in the expected way. Gi activates GIRK-1 (IKACh subunit; Fig. 6) (13), which is down-regulated by 50%. Gi inhibits adenylyl cyclase, and therefore it is expected that adenylyl cyclase would be up-regulated, as shown for adenylyl cyclase type VII (39). These combined effects should make the heart less sensitive to the effects of Gi signaling at the level of the potassium channel as well as adenylyl cyclase. Several genes common to the Gi and Gs pathways (ATF3, protein kinase A) are actively regulated in a complex pattern of feedback regulation, which may help explain some of the alterations in the cAMP pathway that have been noted in cardiomyopathy (40). These data support the hypothesis that each G protein-signaling pathway has a unique gene-expression signature.
The fibrotic response that is characteristic of this cardiomyopathy is well represented in DNA-array data, since fibronectin, laminin (types α5, β2c, and γ2), and collagen (types I-α1 I-α2, and III-Iα2) are all up-regulated (see Fig. 8B in the supplemental material at www.pnas.org). The specificity of the fibrosis gene response is also indicated by the fact that the genes associated with the acute immune response are not significantly changed in these same mice (see Fig. 8B in the supplemental material at www.pnas.org).
We confirmed the cardiomyopathy phenotype by demonstrating a fetal gene-expression program in mice expressing Ro1 for 8 wk. Interestingly, 3 of the 10 most up-regulated genes, βMHC, fibulin, and atrial natriuretic factor, are known markers of cardiomyopathy. However, the 7 others have not been previously examined in cardiomyopathy. These genes are fibroblast secreted protein-12, an early-response gene and possibly a secreted protein (41); NAD-dependent methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, a mitochondrial enzyme (42); osteoblast-specific factor 2, an early-response gene, transcription factor, and determinant of bone differentiation (43); the α2-macroglobulin receptor/low density lipoprotein receptor-related protein, a membrane-bound or secreted receptor involved in lipid metabolism, cell migration, and tissue invasion (44, 45); FK506 binding protein 51; signal recognition particle receptor; and the phosphorylase kinase catalytic subunit. The identification of genes not previously known to be associated with cardiomyopathy should aid the development of diagnostic and therapeutic targets. A full listing of these data (Table 1 in the supplemental material at www.pnas.org) should supply a rich source of information for future analysis.
Although ours is among the first DNA-array analyses of a G protein-signaling pathway that we are aware of, other models are likely to emerge and provide powerful new tools for biomedical research. DNA arrays would be expected to lead to several other improvements for studying the Ro1 model and other heart failure models. First, DNA-array analysis should connect a specific signaling event to potential therapeutic targets. Second, it could be applied to human cardiomyopathy specimens. Human cardiomyopathy is likely a heterogeneous disease with multiple causes, but matching gene-expression patterns in human and mouse tissues should allow for the analogous mouse models to be identified. Third, the identification of genes involved in the progression of cardiomyopathy should provide new diagnostic markers, such as secreted proteins, that could be measured in blood samples.
Does Increased Gi Signaling Cause Cardiomyopathy in Humans?
Gi-coupled receptors in the heart include receptors for opiates (46), endothelin, somatostatin, and acetylcholine (M2-muscarinic), as well as many “orphan” receptors identified by the Human Genome Project. These Gi-coupled receptors could cause cardiomyopathy by several mechanisms. First, activating mutations of GPCRs could increase basal signaling levels enough to cause cardiac disease. Similar mutations have been shown to cause human thyroid hyperplasia, premature testicular maturation, and other endocrinopathies (47). Second, increased levels of ligands acting on any of these receptors could activate Gi signaling and contribute to cardiomyopathy. Third, autoantibodies that bind to and cause signaling through endogenous Gi-coupled receptors could cause increased Gi signaling in the heart (17), as occurs in inflammatory insults to myocardium, such as Chagas cardiomyopathy. In each of these disease models, chronic activation of the Gi pathway would result in a phenotype similar to that in the Ro1 model. By showing that the Gi pathway has a possible causative role in prolonged ventricular conduction and cardiomyopathy, we implicate an entire class of GPCRs and provide insights into a wide variety of potential therapeutic targets for this syndrome.
Supplementary Material
Acknowledgments
We thank Dale Newland for histochemistry assistance, Gary Howard and Stephen Ordway for editorial assistance, and Jane Peredo and Bethany Taylor for manuscript preparation. We thank Robert Mahley, Morris Schambelan, and members of the Conklin lab for their advice and constant support. This work was supported by an American Heart Association Clinician Investigator Award (K.D.), the San Francisco General Hospital General Clinical Research Center MO1RR00083 (B.R.C.), and National Institutes of Health Grants CA71779 (C.H.R.), HL60664 and HL61689 (B.R.C.), and SBIR 1R44DK53325–01 (J.A.W. and B.R.C.).
Abbreviations
- IDC
idiopathic dilated cardiomyopathy
- RASSL
receptor activated solely by a synthetic ligand
- Ro1
RASSL, opioid 1
- tTA
tetracycline transactivator
- αMHC
α myosin heavy chain
- GPCRs
G protein-coupled receptors
- IKACh
potassium-selective ion channel
- LV
left ventricle or ventricular
- LVFS
LV fractional shortening
- PTX
pertussis toxin
- GenMAPP
Gene MicroArray Pathway Profiler
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Deceased February 20, 2000.
References
- 1.Olson T M, Keating M T. J Clin Invest. 1996;97:528–532. doi: 10.1172/JCI118445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwartz K, Mercadier J-J. Curr Opin Cardiol. 1996;11:227–236. doi: 10.1097/00001573-199605000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Cohn J N, Bristow M R, Chien K R, Colucci W S, Frazier O H, Leinwand L A, Lorell B H, Moss A J, Sonnenblick E H, Walsh R A, et al. Circulation. 1997;95:766–770. doi: 10.1161/01.cir.95.4.766. [DOI] [PubMed] [Google Scholar]
- 4.Manolio T A, Baughman K L, Rodeheffer R, Pearson T A, Bristow J D, Michels V V, Abelmann W H, Harlan W R. Am J Cardiol. 1992;69:1458–1466. doi: 10.1016/0002-9149(92)90901-a. [DOI] [PubMed] [Google Scholar]
- 5.Xiao H B, Roy C, Gibson D G. Br Heart J. 1994;72:167–174. doi: 10.1136/hrt.72.2.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cianfrocca C, Pelliccia F, Nigri A, Critelli G. J Electrocardiol. 1992;25:295–303. doi: 10.1016/0022-0736(92)90035-x. [DOI] [PubMed] [Google Scholar]
- 7.Olshausen K V, Stienen U, Schwarz F, Kübler W, Meyer J. Am J Cardiol. 1988;61:146–151. doi: 10.1016/0002-9149(88)91321-5. [DOI] [PubMed] [Google Scholar]
- 8.Redfern C H, Coward P, Degtyarev M Y, Lee E K, Kwa A T, Hennighausen L, Bujard H, Fishman G I, Conklin B R. Nat Biotechnol. 1999;17:165–169. doi: 10.1038/6165. [DOI] [PubMed] [Google Scholar]
- 9.Strader C D, Fong T M, Tota M R, Underwood D. Annu Rev Biochem. 1994;63:101–132. doi: 10.1146/annurev.bi.63.070194.000533. [DOI] [PubMed] [Google Scholar]
- 10.Lefkowitz R J. J Biol Chem. 1998;273:18677–18680. doi: 10.1074/jbc.273.30.18677. [DOI] [PubMed] [Google Scholar]
- 11.Gilman A G. Biosci Rep. 1995;15:65–97. doi: 10.1007/BF01200143. [DOI] [PubMed] [Google Scholar]
- 12.Neer E J. Cell. 1995;80:249–257. doi: 10.1016/0092-8674(95)90407-7. [DOI] [PubMed] [Google Scholar]
- 13.Wickman K, Clapham D E. Physiol Rev. 1995;75:865–885. doi: 10.1152/physrev.1995.75.4.865. [DOI] [PubMed] [Google Scholar]
- 14.Holmer S R, Homcy C J. Circulation. 1991;84:1891–1902. doi: 10.1161/01.cir.84.5.1891. [DOI] [PubMed] [Google Scholar]
- 15.Fu L-X, Liang Q-M, Waagstein F, Hoebeke J, Sylvén C, Jansson E, Sotonyi P, Hjalmarson Å. Cardiovasc Res. 1992;26:950–955. doi: 10.1093/cvr/26.10.950. [DOI] [PubMed] [Google Scholar]
- 16.Schnabel P, Böhm M. Cell Signal. 1996;8:413–423. doi: 10.1016/s0898-6568(96)00087-3. [DOI] [PubMed] [Google Scholar]
- 17.Fu L-X, Magnusson Y, Bergh C-H, Liljeqvist J Å, Waagstein F, Hjalmarson Å, Hoebeke J. J Clin Invest. 1993;91:1964–1968. doi: 10.1172/JCI116416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goin J C, Leiros C P, Borda E, Sterin-Borda L. Neuroimmunomodulation. 1994;1:284–291. doi: 10.1159/000097178. [DOI] [PubMed] [Google Scholar]
- 19.Bristow M R. Lancet. 1998;352, Suppl. I:8–14. doi: 10.1016/s0140-6736(98)90311-7. [DOI] [PubMed] [Google Scholar]
- 20.Feldman A M, Cates A E, Veazey W B, Hershberger R E, Bristow M R, Baughman K L, Baumgartner W A, Van Dop C. J Clin Invest. 1988;82:189–197. doi: 10.1172/JCI113569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Böhm M, Kirchmayr R, Erdmann E. Cardiovasc Res. 1995;30:611–618. [PubMed] [Google Scholar]
- 22.Rockman H A, Koch W J, Lefkowitz R J. Am J Physiol. 1997;272:H1553–H1559. doi: 10.1152/ajpheart.1997.272.4.H1553. [DOI] [PubMed] [Google Scholar]
- 23.Sussman M A, Lim H W, Gude N, Taigen T, Olson E N, Robbins J, Colbert M C, Gualberto A, Wieczorek D F, Molkentin J D. Science. 1998;281:1690–1693. doi: 10.1126/science.281.5383.1690. [DOI] [PubMed] [Google Scholar]
- 24.Akhter S A, Luttrell L M, Rockman H A, Iaccarino G, Lefkowitz R J, Koch W J. Science. 1998;280:574–577. doi: 10.1126/science.280.5363.574. [DOI] [PubMed] [Google Scholar]
- 25.Wakasaki H, Koya D, Schoen F J, Jirousek M R, Ways D K, Hoit B D, Walsh R A, King G L. Proc Natl Acad Sci USA. 1997;94:9320–9325. doi: 10.1073/pnas.94.17.9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hein L, Stevens M E, Barsh G S, Pratt R E, Kobilka B K, Dzau V J. Proc Natl Acad Sci USA. 1997;94:6391–6396. doi: 10.1073/pnas.94.12.6391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwase M, Uechi M, Vatner D E, Asai K, Shannon R P, Kudej R K, Wagner T E, Wight D C, Patrick T A, Ishikawa Y, et al. Am J Physiol. 1997;272:H585–H589. doi: 10.1152/ajpheart.1997.272.1.H585. [DOI] [PubMed] [Google Scholar]
- 28.D'Angelo D D, Sakata Y, Lorenz J N, Boivin G P, Walsh R A, Liggett S B, Dorn G W, II. Proc Natl Acad Sci USA. 1997;94:8121–8126. doi: 10.1073/pnas.94.15.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Molkentin J D, Lu J-R, Antos C L, Markham B, Richardson J, Robbins J, Grant S R, Olson E N. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakata Y, Hoit B D, Liggett S B, Walsh R A, Dorn G W, II. Circulation. 1998;97:1488–1495. doi: 10.1161/01.cir.97.15.1488. [DOI] [PubMed] [Google Scholar]
- 31.Fentzke R C, Korcarz C E, Lang R M, Lin H, Leiden J M. J Clin Invest. 1998;101:2415–2426. doi: 10.1172/JCI2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arber S, Hunter J J, Ross J, Jr, Hongo M, Sansig G, Borg J, Perriard J-C, Chien K R, Caroni P. Cell. 1997;88:393–403. doi: 10.1016/s0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M, Bujard H. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. Science. 1995;268:1766–1769. doi: 10.1126/science.7792603. [DOI] [PubMed] [Google Scholar]
- 35.Yu Z, Redfern C S, Fishman G I. Circ Res. 1996;79:691–697. doi: 10.1161/01.res.79.4.691. [DOI] [PubMed] [Google Scholar]
- 36.Coward P, Wada H G, Falk M S, Chan S D H, Meng F, Akil H, Conklin B R. Proc Natl Acad Sci USA. 1998;95:352–357. doi: 10.1073/pnas.95.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akhter S A, Milano C A, Shotwell K F, Cho M-C, Rockman H A, Lefkowitz R J, Koch W J. J Biol Chem. 1997;272:21253–21259. doi: 10.1074/jbc.272.34.21253. [DOI] [PubMed] [Google Scholar]
- 38.Gottshall K R, Hunter J J, Tanaka N, Dalton N, Becker K D, Ross J, Jr, Chien K R. Proc Natl Acad Sci USA. 1997;94:4710–4715. doi: 10.1073/pnas.94.9.4710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang W-J, Gilman A G. Cell. 1992;70:869–872. doi: 10.1016/0092-8674(92)90236-6. [DOI] [PubMed] [Google Scholar]
- 40.Serruys P W, Emanuelsson H, van der Giessen W, Lunn A C, Kiemeney F, Macaya C, Rutsch W, Heyndrickx G, Suryapranata H, Legrand V, et al. Circulation. 1996;93:412–422. doi: 10.1161/01.cir.93.3.412. [DOI] [PubMed] [Google Scholar]
- 41.Kireeva M L, Latinkic B V, Kolesnikova T V, Chen C-C, Yang G P, Abler A S, Lau L F. Exp Cell Res. 1997;233:63–77. doi: 10.1006/excr.1997.3548. [DOI] [PubMed] [Google Scholar]
- 42.Bélanger C, MacKenzie R E. J Biol Chem. 1989;264:4837–4843. [PubMed] [Google Scholar]
- 43.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 44.Quinn K A, Grimsley P G, Dai Y-P, Tapner M, Chesterman C N, Owensby D A. J Biol Chem. 1997;272:23946–23951. doi: 10.1074/jbc.272.38.23946. [DOI] [PubMed] [Google Scholar]
- 45.Stefansson S, Muhammad S, Cheng X-F, Battey F D, Strickland D K, Lawrence D A. J Biol Chem. 1998;273:6358–6366. doi: 10.1074/jbc.273.11.6358. [DOI] [PubMed] [Google Scholar]
- 46.Pepe S, Xiao R-P, Hohl C, Altschuld R, Lakatta E G. Circulation. 1997;95:2122–2129. doi: 10.1161/01.cir.95.8.2122. [DOI] [PubMed] [Google Scholar]
- 47.Spiegel A M. J Inherit Metab Dis. 1997;20:113–121. doi: 10.1023/a:1005393501786. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.