Abstract
The nucleotide-induced structural rearrangements in ATP binding cassette (ABC) transporters, leading to substrate translocation, are largely unknown. We have modeled nucleotide binding and release in the vitamin B12 importer BtuCD using perturbed elastic network calculations and biased molecular dynamics simulations. Both models predict that nucleotide release decreases the tilt between the two transmembrane domains and opens the cytoplasmic gate. Nucleotide binding has the opposite effect. The observed coupling may be relevant for all ABC transporters because of the conservation of nucleotide binding domains and the shared role of ATP in ABC transporters. The rearrangements in the cytoplasmic gate region do not provide enough space for B12 to diffuse from the transporter pore into the cytoplasm, which could suggest that peristaltic forces are needed to exclude B12 from the transporter pore.
INTRODUCTION
ABC transporters are membrane-spanning proteins containing the ATP binding cassette (ABC) motif (1). They use energy from ATP binding, hydrolysis, and ADP-Pi release to drive active transport of a large variety of substrates across biological membranes (2). There are 48 different transporters in humans and 80 in the gram-negative bacterium Escherichia coli, transferring different compounds such as anions, vitamins, lipids, and amino acids (3–5). (Mal)function of ABC transporters can have dramatic pathological consequences (6,7), such as cystic fibrosis, and also accounts for multidrug resistance (8), e.g., human tumor cell resistance to cytotoxic drugs used in chemotherapy (9,10). Thus, it is of great interest to understand in detail how ABC transporters use ATP to facilitate transport.
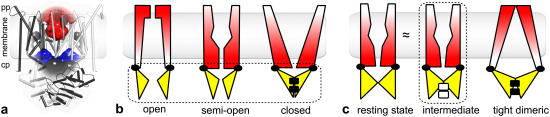
ABC transporters have two transmembrane domains (TMDs) and two nucleotide binding domains (NBDs), which can be organized into four, two, or one polypeptide chain. ATP hydrolysis takes place in an NBD dimer sandwich where highly conserved residues from both NBD monomers contribute to nucleotide binding and hydrolysis (4,11). Crystal structures of a number of NBDs, including four structures of MalK, the E. coli maltose transporter NBD (12,13), provide insight into their possible rearrangements during the transport cycle. Even though MalK contains an additional regulatory domain per monomer, it has been assumed that the nucleotide-dependent tweezers-like opening and closing motions of MalK are not an exclusive feature of this protein but rather reflect a general characteristic of all NBDs (12,13). In all ABC transporters, such NBD rearrangements are expected to propagate to the TMDs to enable transport. The structural basis of this propagation is not well understood yet (14). Three crystal structures of the complete lipid flippase MsbA have been solved (15), but the fold and orientation of the NBDs suggest that two of these structures may be subject to crystallization artifacts (4,11,16), and it is not clear how the lipid substrate enters and leaves the transporter. Besides MsbA, crystal structures have also been determined for two other full ABC transporters; namely the E. coli vitamin B12 importer BtuCD (17) and the bacterial multidrug transporter Sav1866 from Staphylococcus aureus (18). In this study we focus on BtuCD, whose structural organization is illustrated in Fig. 1 a. In the B12 uptake (Btu) system the substrate is delivered to the transporter by its associated periplasmic binding protein BtuF. Once docking has occurred and vitamin B12 has entered BtuC, it is most likely accommodated in a large cavity in the periplasmic half of the protein (17). The subsequent steps of the transport cycle are still unknown.
FIGURE 1.
(a) The BtuCD crystal structure (17) with a schematic illustration of the membrane. The BtuC-internal vitamin B12 uptake cavity is shown in red, the BtuC-BtuD interspace appears in blue, and clefts on the protein surface appear in gray. In the x-ray structure the B12 uptake cavity is accessible from the periplasm for any particle with a radius: 2.0 Å. (b) The MalK transport model (12). (c) The BtuCD model (17). The red to white color gradient of the TMDs emphasizes their gates' state of accessibility: a closed gate appears red; a white gate is open. NBDs are colored yellow. The black squares symbolize ATP; white squares symbolize vanadate. The dashed boxes mark known x-ray structures.
Two functional models have been proposed for the transport mechanism and the coupling between NBDs and TMDs. The MalK model (Fig. 1 b) is based on crystal structures of two ATP-free and one ATP-bound conformation of MalK (12). This model predicts that upon ATP binding the NBDs approach each other, forming a tight dimer (closed MalK configuration), which closes the TMDs toward the cytoplasm but induces an opening on the periplasmic side. Opening of the periplasmic gate allows the substrate to enter; subsequent release of hydrolysis products translocates the substrate and resets the transporter to the resting state (open MalK configuration). The crystal structure of the semiopen MalK is thought to represent an intermediate between the open and closed MalK configurations.
The BtuCD model (Fig. 1 c) assumes that the BtuCD crystal structure is close to the physiological resting state, with the cytoplasmic gate closed (17,19). In this state the transporter is open toward the periplasm, allowing the substrate to enter the B12 uptake cavity (Fig. 1 a) from the binding pocket of the periplasmic binding protein BtuF. Again, ATP binding brings the NBDs in a tight dimeric configuration. But in the BtuCD functional model this causes the transporter to close on the periplasmic side while simultaneously opening toward the cytoplasm to translocate vitamin B12. The cytoplasmic gate closes when the hydrolysis products are released and the transporter returns to its resting state.
Clearly, the MalK and BtuCD models predict opposite effects of NBD dimerization in the transport cycle. Irrespective of which transport model is correct, it is also unclear whether the structural rearrangements in the TMDs are large enough for the respective substrates to leave the transporter by diffusion or if peristaltic forces, generated by the TMDs on the substrate, are necessary for transport (17).
Computer simulations can provide insight into protein function and dynamics. Molecular dynamics (MD) simulations of ABC transporters have focused on rearrangements in the NBDs upon ATP binding and hydrolysis to ADP (20–22), and a recent simulation of the full BtuCD transporter found that ATP binding draws the NBDs closer to each other (23). At the same time a closing of the TMD section of the transporter was observed on the periplasmic side, but no significant opening of the cytoplasmic gate occurred.
In this study, we use a combination of elastic network normal mode analysis and MD simulation techniques to characterize the nucleotide-induced rearrangements in the TMDs of BtuCD to better understand the mechanism that drives substrate transport and gather evidence that can contribute to decide which one of the two proposed contradicting functional models is more likely. We assume that the three MalK crystal structures correspond to physiological states that also occur in BtuCD and derive from these structures two directions in conformational space that take MalK from the semiopen to closed and from semiopen to the open state. We then use these directions as bias in computer simulations of BtuCD to bring the BtuD NBDs into an orientation resembling the ones experimentally observed in the MalK structures and monitor the structural response of the TMDs. Our analysis is focused on the overall response of the TMDs and the detailed response of the cytoplasmic gate region.
METHODS
Perturbed anisotropic network model
In the anisotropic network model (ANM) BtuCD is described as a network of springs with a spring constant of 1 kcal/(mol Å2), connecting Cα atoms within a 13-Å cutoff (24,25). Normal mode analysis yields 3N − 6 nonzero eigenvalues with 3N − 6 corresponding eigenvectors for N Cαs (25,26). Each eigenvector describes a direction along which the protein vibrates with some frequency. Modes with low eigenvalues correspond to motions taking place on long timescales and in directions in which there is a low energy cost for deformation. In ANM calculations, only vibrations around an energy minimum are predicted; the effect of ligand binding cannot be accounted for directly. Further, it is not obvious how to determine the linear combination of low frequency modes that is functionally relevant (as reviewed in Ma (24)). Thus, we investigated the effect of ATP using a perturbational variant of the elastic network (PEN, perturbed elastic network) that resolves these issues (27). The effect of ligand binding is accounted for by introducing a small number of Cα-Cα distance constraints as a perturbation to the network energy. This perturbation gives a response vector that describes how all Cαs in the protein respond to changing a few Cα-Cα pair distances. Zheng and Brooks validated the method on 22 proteins for which two crystal structures, representing two conformational states of the same protein, are known (27). They chose 10 Cα pairs for the perturbation that cross the network cutoff and have the largest distance change between the two structures. Inherently, this is not possible for BtuCD since only one crystal structure is available. Therefore, we find the relevant Cα pairs from crystal structures of MalK. We map the perturbation sites from MalK to the equivalent sites in the BtuCD NBD, BtuD. The perturbations in BtuD then mimic nucleotide binding and release events and we monitor the response in the TMD BtuC. The perturbation sites in MalK were confined to regions conserved among the NBDs in ABC transporters, and therefore the same is true after mapping the sites onto BtuD. As a control, we ran simulations on MalK structures to verify the bias introduced from the perturbed normal mode indeed causes a transition between the two structures (details not shown). We implemented PEN in the ANM code from the Jernigan group (http://ribosome.bb.iastate.edu/software.html).
Molecular dynamics simulations
For the MD simulations, the crystal structure of BtuCD (1L7V) was placed in a preequilibrated palmitoyloleonyl-phosphatidyl-ethanolamine (POPE) bilayer consisting of 326 lipids using a new method (28) and solvated with 25,000 single point charge water molecules and 20 chloride ions (29). The full system contained ∼100 000 atoms. Titratable residues had default protonation states. The MD simulations were performed in GROMACS 3.2.1 using the ffgmx force field for the protein (30,31). Constant pressure was maintained by semiisotropic coupling to a Berendsen barostat with τ = 4 ps, a compressibility of 4.5 10−5 bar−1, and a reference pressure of 1 bar. Protein, bilayer, and solution were kept at 310 K by a Berendsen thermostat with τ = 0.1 ps. Bonds were constrained by LINCS (32) and SETTLE for water (33). The system was propagated with a 2-fs time step. Electrostatic interactions were evaluated using particle mesh Ewald (34,35), and Van der Waals interactions were evaluated directly with a twin-range cutoff of 0.9 and 1.4 nm. The system was equilibrated for 8 ns, when the Cα root mean square deviation (RMSD) of BtuCD converged at 0.23 nm relative to the crystal structure. Equilibrium data were collected during the next 4 ns.
Essential dynamics sampling
Long timescales of protein dynamics can be explored using essential dynamics sampling (EDS) (36,37). We used EDS to explore the directions that are relevant for nucleotide binding and release more efficiently. The input is a generalized direction in which the algorithm should increase the sampling. For this input direction we calculated the transition vectors from the semiopen to the open and from the semiopen to the closed structures of MalK. From these transition vectors, the (x, y, z)-elements for the Cαs close to functionally important and highly conserved regions, i.e., residues 30–44, 77–83, 124–138, 153–161, and 188–194 were used to bias both BtuD monomers. We ran 10 simulations biased in the direction corresponding to nucleotide binding and 10 simulations biased in the direction corresponding to nucleotide release, with different initial random velocities. The transitions to the tight dimeric structure took ∼75 ps, whereas the transitions to the loose dimeric structure took ∼150 ps in the biased simulation.
Both the perturbed network and the essential dynamics simulations only give the direction in which the protein deforms upon the simulated nucleotide binding and release but not the amplitude of the motion. Thus, we define the tight/loose dimeric structures of BtuCD as having the lowest RMSD from homologous parts of the closed/open MalK structures. The RMSD between the equilibrated BtuCD structure and homologous parts of the closed and open MalK crystal structures are 4.8 and 9.0 Å, respectively. In the biased MD simulations, the minimal RMSD between BtuCD and homolog parts of the closed MalK crystal structure, defining the tightly dimerized state of BtuCD, is 3.8 Å. The minimal RMSD between BtuCD and homologous parts of the open MalK crystal structure, defining the loose dimeric state, is 4.5 Å.
Cavity and pathway analysis
Cavity analysis on the all-atom BtuCD structures was performed by SURFNET (38), using a minimum probe sphere radius of 2.0 Å. To scan for possible vitamin B12 exit pathways we used HOLE (39).
Angle between inertia axes
The angle between the TMD inertia axes was calculated in VMD (40) using the additional plugins “orient” and “la101psx”.
X-shift plots
To quantify the structural responses in the transmembrane BtuC subunits and to detect possible opening or closing trends, we computed the shift in x-coordinates for corresponding Cα atoms with respect to a reference structure: the BtuCD x-ray structure in the case of the PEN analysis and the equilibrated crystal structure for the EDS simulation. The simulation x axis is almost the same as the e3 principal axis. We resolved the X-shift along the pore axis using windows of 2 Å.
RESULTS
Overall response of BtuCD
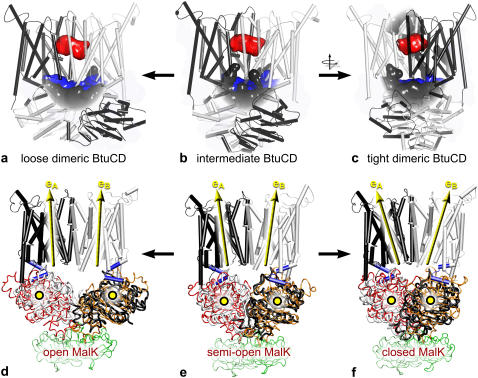
In Fig. 2 the start and end structures of BtuCD taken from one representative MD simulation for the loose and tight NBD dimerization transitions are shown, together with the corresponding MalK crystal structures used to model these transitions. As the structural overlay illustrates, on a global scale, EDS in MD simulations can be used to mimic the effect of nucleotide binding and release. During equilibration, without bias, the putative BtuC-internal vitamin B12 uptake cavity changes little, apart from losing its connection to the periplasm due to a closing motion of BtuC, consistent with previous simulations (23). During the EDS runs BtuC remains closed toward the periplasm for any particle with a radius >2 Å regardless of whether the NBDs are brought closer together or farther apart.
FIGURE 2.
Structural rearrangements in BtuCD (black and white). (a–c) BtuCD's main cavities: the putative B12 uptake cavity is shown in red; the BtuC-BtuD interdomain space appears blue, and clefts on the protein surface are colored gray. After 12 ns of equilibration the B12 uptake cavity in the crystal structure (b) is no longer connected to the periplasm. Bringing the BtuDs closer together (c) or farther apart (a) has no effect on the B12 uptake cavity. (d–f) Yellow circles represent the BtuD centers of mass; vectors eA and eB are the BtuC long axes of inertia. The MalK NBDs are shown in red and orange. MalK regulatory domains appear in green.
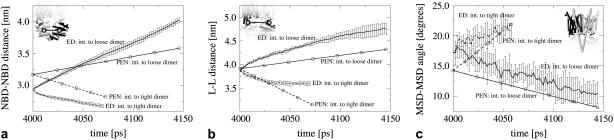
Fig. 3 a shows the center of mass distance between the two NBDs in BtuCD during the transition to the loose and tight dimeric state. As expected, both the elastic network and the MD simulations predict that the transition to the loose dimeric state of BtuCD increases the NBD-NBD distance and that this distance decreases upon transition to the tight dimeric state. The PEN calculations start from the BtuCD crystal structure, whereas the biased MD simulations start from an equilibrated BtuCD structure, which accounts for the different initial values. Thus, the initial values in the elastic network calculations correspond to the values in the crystal structure of BtuCD.
FIGURE 3.
Structural responses associated with nucleotide binding and release in BtuCD. (a) The distance between the two NBD centers of mass. (b) The distance between the centers of mass of the L-helix motifs in the two TMDs. (c) The tilt between the two TMDs defined as the angle between inertia axes of each TMD (eA and eB in Fig. 2, d–f). For the essential dynamics “intermediate to tight dimer” transitions (pushing NBDs closer), fewer data points could be collected as the NBDs begin detaching from the TMDs after 60 ps.The insets illustrate the measured quantities in the protein structure. Solid black curves correspond to the transition to the loose dimeric structure, dashed curves to the transition to the tight dimeric. Circles mark PEN results which, to facilitate the comparison, are adjusted to the same timescale as the MD simulations. Error bars are standard deviations obtained from 10 simulations carried out for each of the two transitions. There are no error bars on the PEN results since the method is deterministic and only the crystal structure was used. The PEN calculations start from the BtuCD crystal structure (int.), whereas the biased MD simulations start from an equilibrated BtuCD structure (int.). The x axes have the unit of time, which is somewhat artificial since the MD simulations are biased and the notion of time is absent in the PEN calculations.
The rearrangements in the NBDs are thought to propagate into the TMDs via the L-loop helices located in the NBD-TMD interface (17,19). The centers of mass distance between the L-loop helices for the two transitions in BtuCD are shown in Fig. 3 b. Comparing with Fig. 3 a, the distance between the L-helix motifs in opposing TMDs is strongly coupled to the NBD-NBD distance. Both the perturbed network and the MD simulations predict that a transition to the loose dimeric state, corresponding to nucleotide release, increases the L-helix distance. Nucleotide binding has the opposite effect. Fig. 3 c shows the response in the TMD-TMD tilt upon nucleotide binding and release. The TMD-TMD tilt, defined as the angle between the TMD inertia axes and illustrated in Fig. 2, d–f, increases upon tight NBD dimerization (nucleotide binding) and decreases upon transition to the loose dimeric state (nucleotide release).
Response in the transmembrane domains
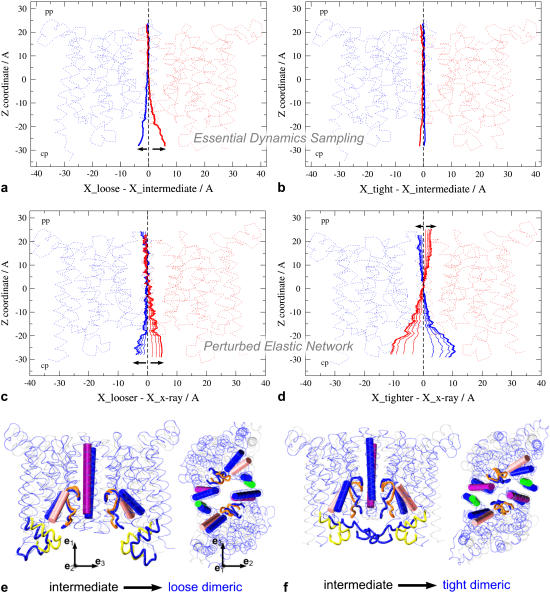
The structural response of BtuC quantified in terms of X-shifts is shown in Fig. 4. EDS MD induces a cytoplasmic opening trend upon driving the BtuDs apart—intermediate → loose dimeric (Fig. 4 a)—whereas bringing them closer together—intermediate → tight dimeric—has no detectable effect (Fig. 4 b). PEN analysis predicts an opening trend on the cytoplasmic side when the BtuD interdomain center of mass distance increases (Fig. 4, c and e) and a periplasmic opening trend when the BtuDs are brought closer together (Fig. 4, d and f). At the same time a clear closing trend is observed on the cytoplasmic side of the BtuCs.
FIGURE 4.
Structural response in BtuC subunits. (a–d) The structural responses as monitored by X-shift of the BtuC subunits in red and blue (see Methods) for four indicated cases. (e–f) Side view and top view of the PEN extreme conformations. The key structural elements in the cytoplasmic exit regions of the intermediate BtuC structure are rendered as a purple cylinder (TM5), green cylinder (C-terminal helix), pink cylinder (TM8), yellow loops (the L-loops), and orange tube (pore loop). The corresponding structural elements in the loose and tight dimeric structures are colored blue. For clarity not all structural elements are shown. The BtuC crystal structure is rendered in semitransparent gray. The amplitude of the responses is exaggerated to illustrate the motions of the different structural elements more clearly.
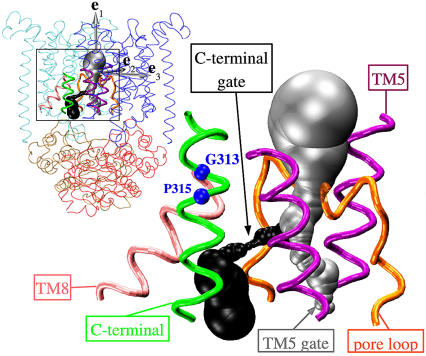
To monitor the response in the cytoplasmic gate region in greater detail, we first carried out a HOLE analysis to identify putative translocation pathways through the transmembrane part of BtuCD in the crystal structure. We find two possible cytoplasmic exit paths. Fig. 5 shows the two pathways in the BtuCD structure. The gray-colored pathway, referred to as the TM5 exit, essentially follows the membrane normal, in between the transmembrane helices number 5 (TM5) of each of the two TMD monomers. The other exit path, referred to as the C-terminal exit, departs from the TM5 exit path ∼1 nm below the vitamin B12 uptake cavity and reaches the cytoplasm through a passage in between the structural elements entitled the pore loop (residues 83–92), lower transmembrane helix 5 (TM5, residues 142–155), transmembrane helix 8 (TM8, residues 256–267), and the lower C-terminal helix (residues 313–325). Fig. 6 shows the pore radius profiles for these paths. Note that due to the rotational symmetry of the BtuC dimer, there is a second C-terminal exit in the other BtuC monomer, thus providing a total of two equivalent C-terminal exits in BtuC. Descriptions of the C-terminal exit region below refer to both of these exits.
FIGURE 5.
Two possible cytoplasmic exits in BtuCD. The gray surface is the TM5 exit path; the black surface is the C-terminal exit path. The protein is rendered as TM5 helix (purple tube), TM8 (pink tube), pore loop (orange tube), and C-terminal helix (green tube). The residues G313 and P315 are shown as blue spheres. The inset shows the two pores in the full BtuCD structure. The gray arrows show the principal components of the inertia tensor of the TMDs.
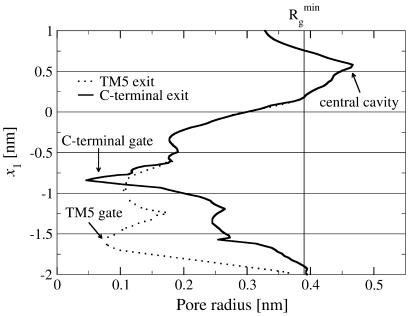
FIGURE 6.
Pore radii as functions of the position projected on the pore axis e1. The graph shows the radii for two possible pores through the crystal structure of BtuCD: the TM5 exit path (dotted) and the C-terminal exit path (solid). We have indicated the dimensions of B12 by the vertical line at 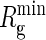 , which is the minimal radius of gyration of B12.
, which is the minimal radius of gyration of B12.
The response of the structural elements comprising the two putative exits is shown in Fig. 4, e and f. The TM5 helices change their tilt relative to the pore axis upon nucleotide binding and release. The change in the tilt primarily takes place in the e3-direction but not in the e2-direction. The transition to the loose dimeric structure causes the TM8 helices and the pore loop to retract from the pore center line. Transition to the tight dimeric structure has the opposite effect. The response of the C-terminal helix upon transition to the loose dimeric configuration does not affect the size of the C-terminal exit (see Fig. 4 e). However, upon transition to the tight dimeric configuration of BtuCD, the C-terminal helix moves out of the C-terminal exit path.
We analyzed these structural rearrangements further by monitoring changes in Cα-Cα pair distances in the cytoplasmic gate region. The Cα-Cα pairs used in the analysis all line the TM5 and C-terminal pores, and the lines connecting each Cα-Cα pair approximately cross the pore centerline. To allow for comparison, the same analysis was carried out on the BtuCD structures predicted by PEN. The distance analyses of both the MD simulations and the perturbed network (data not shown) show a significant increase in the Cα-Cα pair distances in the C-terminal exit region, which indicates that this gate opens upon transition to the loose dimeric state of BtuCD. Nucleotide binding has the opposite effect. In the TM5 exit region, the magnitudes of the structural rearrangements are smaller and primarily take place in the e2-direction. Although the rearrangements in the C-terminal exit region are larger than the ones found in the TM5 exit region, the largest Cα-Cα pair distance increase in the C-terminal exit is merely ∼2 Å.
DISCUSSION
It is commonly thought that ATP binding induces tight dimerization of the NBDs in ABC transporters, whereas the transition to a loose dimeric state may be coupled to the release of hydrolysis products (reviewed in Davidson and Chen (4); 12,17). These transitions are thought to cause structural rearrangements in the TMDs that facilitate transport, but the exact nature of this coupling is not well understood (14). Due to the long timescales involved, global rearrangements are difficult to capture in direct MD simulations. Therefore, instead of including the nucleotides explicitly in direct MD simulations (23), we modeled the effects of nucleotide binding and release in a PEN model and in biased MD simulations—using experimentally observed transitions in MalK as templates for perturbing and biasing the NBDs in BtuCD—and then monitored the response in the TMDs. Thus our simulations are restricted to investigating the effect of conversion of the NBDs from open to closed on the TMDs, where the closed state corresponds to the ATP-bound conformation and the open state corresponds to a nucleotide configuration we do not identify. Additional NBD conformations may exist in the entire cycle of ATP binding, hydrolysis, and Pi/ADP release, with possible corresponding structural changes in the TMDs.
Which transport model?
When the NBDs of BtuCD dissociate into the loose dimeric configuration, the distance between the L-loop helices, which connect the two TMDs to the NBDs, increases and the tilt angle between the two TMDs decreases (Fig. 3). The transition to the tight dimeric state has the opposite effect. Our data indicate that transition to the loose dimeric configuration induces an opening trend of the TMDs toward the cytoplasm and a closing trend on the periplasmic side, whereas the transition to the tight dimeric state has the opposite effect. These results are consistent with the MalK transport model (4,12) and disagree with the BtuCD functional model (17; reviewed in Locher (19)). Instead the adaptation of the MalK model for the BtuCD transport cycle as presented in the schematics in the review article by Van der Does and Tampé appears to be valid (16). The results of the PEN and the biased MD simulations are qualitatively similar, which shows some universality in our prediction.
Recently Vergani et al. (41) reported that the tightly dimerized NBD conformation of the CFTR, an ABC protein with TMDs comprising an ion channel, corresponds to the so-called open-burst state. In ABC exporters and importers, this state is thought to be equivalent to a TMD configuration where the periplasmic gate is open and the cytoplasmic gate is closed. In MsbA, site-directed spin labeling and electron paramagnetic resonance spectroscopy suggest that ATP binding closes the cytoplasmic gate (42). If there is a common transport mechanism among ABC importers and exporters, these experimental results also support the MalK transport model. Other ways to experimentally test if BtuCD follows the MalK model—and is indeed open to the periplasm and closed to the cytoplasm in the absence of nucleotides—could be to appropriately mark for example the upper half of the translocation channel and measure the corresponding signals in absence and presence of ATP, possibly by spectroscopic methods.
A valid concern is to what extend the transitions we find in BtuCD are biased by the MalK-like input. To this end it is important to note that MalK and the NBDs of BtuCD have almost complete sequence identity in the regions that are involved in ATP binding and hydrolysis and an almost identical structure at the binding sites. The structural changes in the above mentioned regions, induced by nucleotide binding and release, are therefore expected to be the same in the two transporters (4,11,17). We assume that the absence of regulatory domains in BtuCD compared to MalK does not cause a substantial difference in the mechanisms of these two importers.
Cytoplasmic gate opening
Our analyses of the putative transport pore suggest that there may exist two types of cytoplasmic exit paths, which we refer to as the TM5 exit and the C-terminal exit, respectively (Fig. 5). If nucleotide binding or release were responsible for opening the TM5 exit, one would expect the response in the cytoplasmic ends of the TM5 helices to go toward the e2-direction (Fig. 5). Such a motion is, however, observed in neither the perturbed network nor the biased MD simulations, which is consistent with previous MD simulations (23). The structural elements that make up the C-terminal exits, on the other hand, are found to retract from the pore centerline upon transition to the loose dimeric state of BtuCD in both the PEN model and the biased MD simulations. Therefore, release of hydrolysis products, which could reset the transporter in the resting state, seems to be responsible for opening the C-terminal exit. However, the observed gate opening is not large enough to accommodate vitamin B12. Thus, it is interesting that the C-terminal helix, which lines the C-terminal exit, contains a molecular hinge, i.e., GXP motif (43) (see Fig. 5). This hinge region (G313-P315), located just ∼0.2 nm below the central cavity, could provide considerable structural flexibility in the C-terminal exit region. Indeed, the crystallographic B-factors do increase down through the C-terminal helix, although this is a common characteristic in protein crystal structures. Therefore, it is not possible to associate this increase in B-factors with the GXP motifs or with the function of BtuCD without further investigations—especially since at 3.2 Å the resolution of the BtuCD x-ray structure does not permit a quantitative interpretation of B-factors.
Two transport scenarios
Two contradicting models have been proposed to explain the ABC transporter functional mechanism. Our data indicate that of these two the MalK model appears to be more likely. However, compared to the cartoon transport models presented in the literature (4,12,16,17), the magnitude of the cytoplasmic gate opening we find is strikingly small. Even the structural changes in the C-terminal exit are not large enough to allow B12 to diffuse from the central cavity and out into the cytoplasm. This finding points to one of the following two transport scenarios:
The structural response from nucleotide binding and release are in fact as small as predicted here. In this case, the modest opening indicates that peristaltic forces from the TMDs pushing B12 toward the cytoplasm are important. The changes in the TMD tilt upon nucleotide release drive the periplasmic ends of the TMDs closer together, which could generate a force pushing B12 toward the cytoplasm via the C-terminal exit. In line with these speculations, the C-terminal helix could act as a passive hatch, which moves to the side as B12 propagates through the C-terminal exit. The term “passive hatch” refers to the fact that such a motion of the C-terminal helix is not seen in this study and is therefore expected to be independent of nucleotide binding and release. If this is true, and the C-terminal helix is simply pushed to the side by B12, it is important that the C-terminal helix is moved easily, which could indicate the role of the GXP motifs in the C-terminal helices.
The structural rearrangements we find are too small, and larger rearrangements, coupled to events that are not accounted for in this study, control transport. For example, the structural responses to nucleotide binding and release could be altered by the presence of the periplasmic binding protein BtuF. The presence of binding proteins stimulate ATPase activity (4) and in the schematic transport mechanism presented by Locher and Borths (14), BtuF sends a “signal” from the periplasmic side of BtuCD to the hydrolysis sites, which trigger ATP hydrolysis. The signaling between BtuF and the hydrolysis sites could involve changes in the transporter structure that in turn could change the response to nucleotide binding and release. This hypothesis could be tested by the same techniques as employed in this study once the structure of the BtuF-BtuCD complex is solved experimentally or accurately modeled.
Acknowledgments
This research was supported by the Canadian Institutes of Health Research (CIHR) and WestGrid. D.P.T. is an AHFMR Senior Scholar and CIHR New Investigator. C.K. is an Alberta Heritage Foundation for Medical Research Postdoctoral Fellow. This work was also supported by the Danish National Research Foundation via a grant to MEMPHYS - Center for Biomembrane Physics.
References
- 1.Higgins, C. F. 1992. ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8:67–113. [DOI] [PubMed] [Google Scholar]
- 2.Linton, K. J., and C. F. Higgins. 2007. Structure and function of ABC transporters: the ATP switch provides flexible control. Pflugers Arch. 453:555–567. [DOI] [PubMed] [Google Scholar]
- 3.Biemans-Oldehinkel, E., M. K. Doeven, and B. Poolman. 2006. ABC transporter architecture and regulatory roles of accessory domains. FEBS Lett. 580:1023–1035. [DOI] [PubMed] [Google Scholar]
- 4.Davidson, A. L., and J. Chen. 2004. ATP-binding cassette transporters in bacteria. Annu. Rev. Biochem. 73:241–268. [DOI] [PubMed] [Google Scholar]
- 5.Holland, I. B., and M. A. Blight. 1999. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J. Mol. Biol. 293:381–399. [DOI] [PubMed] [Google Scholar]
- 6.Dean, M., A. Rzhetsky, and R. Allikmets. 2001. The human ATP-binding cassette (ABC). Transporter Superfamily. Genome Res. 11:1156–1166. [DOI] [PubMed] [Google Scholar]
- 7.Riordan, J. R., J. M. Rommens, B. Kerem, N. Alon, R. Rozmahel, Z. Grzelczak, J. Zielenski, S. Lok, N. Plavsic, J.-L. Chou, M. L. Drumm, M. C. Iannuzzi, F. S. Collins, and L.-C. Tsui. 1989. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 245:1066–1073. [DOI] [PubMed] [Google Scholar]
- 8.Borst, P., and R. O. Elferink. 2002. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 71:537–592. [DOI] [PubMed] [Google Scholar]
- 9.Gottesman, M. M., and S. V. Ambudkar. 2001. ABC transporters and human disease. J. Bioenerg. Biomembr. 33:453–458. [DOI] [PubMed] [Google Scholar]
- 10.Horio, M., M. M. Gottesman, and I. Pastan. 1988. ATP-dependent transport of vinblastine in vesicles from human multidrug-resistant cells. P. Natl. Acad. Sci. USA. 85:3580–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones, P. M., and A. M. George. 2004. The ABC transporter structure and mechanism: perspectives on recent research. Cell. Mol. Life Sci. 61:682–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J., G. Lu, J. Lin, A. L. Davidson, and F. A. Quiocho. 2003. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol. Cell. 12:651–662. [DOI] [PubMed] [Google Scholar]
- 13.Lu, G., J. M. Westbrooks, A. L. Davidson, and J. Chen. 2005. ATP hydrolysis is required to reset the ATP-binding cassette dimer into the resting-state conformation. P. Natl. Acad. Sci. USA. 102:17969–17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Locher, K. P., and E. Borths. 2004. ABC transporter architecture and mechanism: implications from the crystal structures of BtuCD and BtuF. FEBS Lett. 564:264–268. [DOI] [PubMed] [Google Scholar]
- 15.Reyes, C. L., A. Ward, J. Yu, and G. Chang. 2006. The structures of MsbA: insight into ABC transporter-mediated multidrug efflux. FEBS Lett. 580:1042–1048. [DOI] [PubMed] [Google Scholar]
- 16.van der Does, C., and R. Tampe. 2004. How do ABC transporters drive transport? Biol. Chem. 385:927–934. [DOI] [PubMed] [Google Scholar]
- 17.Locher, K. P., A. T. Lee, and D. C. Rees. 2002. The E. coli BtuCD structure: a framework for ABC transporter architecture and mechanism. Science. 296:1091–1098. [DOI] [PubMed] [Google Scholar]
- 18.Dawson, R. J., and K. P. Locher. 2006. Structure of a bacterial multidrug ABC transporter. Nature. 443:180–185. [DOI] [PubMed] [Google Scholar]
- 19.Locher, K. P. 2004. Structure and mechanism of ABC transporters. Curr. Opin. Struct. Biol. 14:426–431. [DOI] [PubMed] [Google Scholar]
- 20.Campbell, J. D., S. S. Deol, F. M. Ashcroft, I. D. Kerr, and M. S. P. Sansom. 2004. Nucleotide-dependent conformational changes in HisP: molecular dynamics simulations of an ABC transporter nucleotide-binding domain. Biophys. J. 87:3703–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell, J. D., and M. S. P. Sansom. 2005. Nucleotide binding to the homodimeric MJ0796 protein: a computational study of a prokaryotic ABC transporter NBD dimer. FEBS Lett. 579:4193–4199. [DOI] [PubMed] [Google Scholar]
- 22.Jones, P. M., and A. M. George. 2002. Mechanism of ABC transporters: a molecular dynamics simulation of a well characterized nucleotide-binding subunit. P. Natl. Acad. Sci. USA. 99:12639–12644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oloo, E. O., and D. P. Tieleman. 2004. Conformational transitions induced by the binding of MgATP to the vitamin B-12 ATP-binding cassette (ABC) transporter BtuCD. J. Biol. Chem. 279:45013–45019. [DOI] [PubMed] [Google Scholar]
- 24.Ma, J. 2005. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 13:373–380. [DOI] [PubMed] [Google Scholar]
- 25.Atilgan, A. R., S. R. Durell, R. L. Jernigan, M. C. Demirel, O. Keskin, and I. Bahar. 2001. Anisotropy of fluctuation dynamics of proteins with an elastic network model. Biophys. J. 80:505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doruker, P., A. R. Atilgan, and I. Bahar. 2000. Dynamics of proteins predicted by molecular dynamics simulations and analytical approaches: application to α-amylase inhibitor. Proteins. 40:512–524. [PubMed] [Google Scholar]
- 27.Zheng, W., and B. R. Brooks. 2005. Normal-modes-based prediction of protein conformational changes guided by distance constraints. Biophys. J. 88:3109–3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kandt, C., W. L. Ash, and D. P. Tieleman. 2007. Setting up and running MD simulations of membrane proteins. Methods. In press. [DOI] [PubMed]
- 29.Hermans, J., H. J. C. Berendsen, W. F. v. Gunsteren, and J. P. M. Postma. 1984. A consistent empirical potential for water-protein interactions. Biopolymers. 23:1513–1518. [Google Scholar]
- 30.Berendsen, H. J. C., D. v. d. Spoel, and R. v. Drunen. 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 91:43–56. [Google Scholar]
- 31.Lindahl, E., B. Hess, and D. v. d. Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. (Online). 7:306–317. [Google Scholar]
- 32.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 18:1463–1472. [Google Scholar]
- 33.Miyamoto, S., and P. A. Kollman. 1992. SETTLE: an analytical version of the SHAKE and RATTLE algorithm for rigid water models. J. Comput. Chem. 13:952–962. [Google Scholar]
- 34.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an N.log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 35.Essmann, U., L. Perera, M. L. Berkowitz, T. Darden, H. Lee, and L. G. Pedersen. 1995. A smooth particle mesh Ewald method. J. Chem. Phys. 103:8577–8593. [Google Scholar]
- 36.de Groot, B. L., A. Amadei, R. M. Scheek, N. A. J. van Nuland, and H. J. Berendsen. 1996. An extended sampling of the configurational space of HPr from E. coli. Proteins. 26:314–322. [DOI] [PubMed] [Google Scholar]
- 37.de Groot, B. L., A. Amadei, D. M. van Aalten, and H. J. Berendsen. 1996. Toward an exhaustive sampling of the configurational spaces of the two forms of the peptide hormone guanylin. J. Biomol. Struct. Dyn. 13:741–751. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski, R. A. 1995. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 13:323–330, 307–328. [DOI] [PubMed] [Google Scholar]
- 39.Smart, O. S., J. M. Goodfellow, and B. A. Wallace. 1993. The pore dimensions of gramicidin A. Biophys. J. 65:2455–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Humphrey, W., A. Dalke, and K. Schulten. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–28. [DOI] [PubMed] [Google Scholar]
- 41.Vergani, P., S. W. Lockless, A. C. Nairn, and D. C. Gadsby. 2005. CFTR channel opening by ATP-driven tight dimerization of its nucleotide-binding domains. Nature. 433:876–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong, J., G. Yang, and H. S. Mchaourab. 2005. Structural basis of energy transduction in the transport cycle of MsbA. Science. 308:1023–1028. [DOI] [PubMed] [Google Scholar]
- 43.Sansom, M. S., and H. Weinstein. 2000. Hinges, swivels and switches: the role of prolines in signalling via transmembrane alpha-helices. Trends Pharmacol. Sci. 21:445–451. [DOI] [PubMed] [Google Scholar]