Abstract
The amelogenin gene is tightly regulated at the temporal and spatial level in accord with the developmental requirement for tooth formation. Previous studies have shown that CCAAT/enhancer-binding protein α (C/EBPα) is a transactivator of the mouse X-chromosomal amelogenin gene. C/EBPα contains four highly conserved regions (CR) named CR1, CR2, CR3 and CR4. Transient transfection assays showed that CR2 in isolation had an exceptional capacity to enhance transcription from the 2.3 kb mouse amelogenin promoter. The remaining conserved regions of C/EBPα, either in isolation or in selected combinations, were less effective in amelogenin transactivation than the full length C/EBPα. Msx2 has previously been shown to antagonize C/EBPα through protein-protein interactions with C/EBPα, and the carboxyl-terminus of Msx2 is required for protein-protein interactions. Co-immunoprecipitation analyses identified that the carboxyl-terminal domain (residues 218–359) of C/EBPα is required for the C/EBPα-Msx2 protein-protein interactions.
Keywords: Amelogenin, C/EBPα, Msx2, Gene regulation, Promoter, Tooth formation, Protein-protein interactions
Introduction
C/EBPα is the prototypic basic region/leucine zipper transcription factor consisting of a well-characterized carboxyl-terminal leucine zipper that exerts dimerization ability and has a neighboring DNA binding domain [1, 2]. The remaining amino-terminal amino acids (1–273) are organized into transactivation domains, which controls the transcriptional activity of C/EBPα in different sets of promoters [3–5]. In the search for the mechanism determining the ability of C/EBPα to activate the serum albumin promoter, a study identified three separable transactivation elements (TE-I, TE-II, and TE-III) located within the amino-terminal region of C/EBPα. Any two of these transactivation elements are able to activate the serum albumin promoter either in the context of the C/EBPα protein or when fused to the Gal4-DNA-binding domain. In addition, transactivation element III contains a negative regulatory domain, the function of which is abrogated upon C/EBPα binding to the albumin promoter [6]. Another study has mapped four conserved regions (CR1-4) within the C/EBPα transactivation domain based upon the ability of C/EBPα to induce spontaneous preadipocyte differentiation. The CR2 domain in isolation when fused to the DNA binding domain, is capable of stimulating spontaneous differentiation of 3T3-L1 preadipocytes. However, CR2 is not essential for adipogenesis because a combination of CR1 and CR3 is also able to reproduce this experimental end-point [5].
Here, we examine the role of these conserved regions (CR) of C/EBPα in the ability to activate the mouse amelogenin promoter. In addition, we identify that the carboxyl-terminus of C/EBPα is required for protein-protein interactions with Msx2, the transcriptional repressor of the amelogenin gene.
Materials and methods
Plasmids
The mouse C/EBPα expression plasmid (pSER28) was described previously [7]. The expression vectors containing different combinations of the four conserved regions (CR1-CR4) were prepared as previously described [5]. To generate C/EBPα ΔN (Δ1-217) expression vector, full length C/EBPα (pSER28) was digested with PstI, the 2.6 kb-PstI fragment was blunt-ended with T4 DNA polymerase (New England Biolabs, Ipswich, MA), digested with ApaI to release the 0.7 kb fragment, and inserted into the EcoRV-ApaI site of pcDNA3.1(+) (Invitrogen, Carlsbad, CA). To generate C/EBPα ΔC-V5 (Δ216-359) expression vector, pSER28 was digested with PstI, the 0.7 kb fragment was blunt-ended with T4 DNA polymerase (New England Biolabs, Ipswich, MA), digested with EcoRI, and inserted into the EcoRV-EcoRI site of pcDNA3.1/V5-HisA (Invitrogen, Carlsbad, CA).
Cell culture
A mouse ameloblast-like cell line (LS8) established from immortalizing primary cultures of enamel organ epithelial cells with SV40 large “T” antigen was used as previously described [8].
Transient transfection and luciferase assay
Transient transfection and luciferase assays were performed as described previously[9].
Co-immunoprecipitation assay and Western blotting
LS8 cells, ~90% confluent in a 100 mm cell culture plate, were washed twice in ice-cold phosphate-buffered saline. Ice-cold RIPA buffer (1 ml 1× PBS, 1% Nonidet P-40, 0.1 mg/ml phenylmethylsulfonyl fluoride (Sigma, St. Louis, MO), 30 μl/ml aprotinin (Sigma, St. Louis, MO), and 1 mM sodium orthovanadate) was added, and the cells were collected using a cell scraper (Corning, Acton, MA), and lysed by passing six times through a 22-gauge needle at 4°C. After centrifugation at 3,000 rpm for 15 min at 4°C, the protein concentration of the supernatant was measured using a Bio-Rad protein assay kit (Bio-Rad, Hercules, CA) with bovine serum albumin as standards. For immunoprecipitation, 500 μg of total protein was precleaned with 50 μl of protein G-agarose beads (Sigma, St. Louis, MO) prior to addition with 2 μg of primary antibody (Santa Cruz Biotechnology, Santa Cruz, CA) for overnight incubation at 4°C with rotation. Protein G-agarose beads (20 μl; IgG binding capacity at 10–20 μg per μl) was added and incubated for 2 h at 4°C with rotation. Immunoprecipitates were collected by centrifugation at 2,500 rpm for 5 min at 4°C. The pellets were washed three times with 1 ml of PBS. After the final wash, the pellets were resuspended in an equal volume of 2× SDS loading buffer, boiled for 5 min, and stored at −70°C. Samples were resolved by 12% SDS-polyacrylamide gel electrophoresis and transferred to Immobilon-P membrane (Millipore, Billerica, MA). The membranes were incubated in blocking buffer overnight at 4°C, followed by incubation with the primary antibody for 1 h and the appropriate horseradish peroxidase-conjugated secondary antibody (Amersham, Piscataway, NJ) for 1 h at room temperature. Protein-antibody complexes were visualized by enhanced chemiluminescence (ECL, Amersham, Piscataway, NJ).
Results
Physical dissection of the transactivation domain within C/EBPα responsible for activating the amelogenin promoter
It has previously been shown that C/EBPα is a transactivator of the mouse X-chromosomal amelogenin gene [8–10]. Four conserved regions of the C/EBPα transactivation domain, named CR1, CR2, CR3, and CR4, are identified among several species (Fig. 1A), and their functions contributing to adipogenesis have been previously analyzed [5]. To further investigate the region within the C/EBPα transactivation domain acting upon the amelogenin promoter, different combinations of the four conserved regions (CR1-CR4) in C/EBPα were used to examine their ability to alter amelogenin promoter activity using transient transfection assays performed in an ameloblast-like cell line termed LS8. Consistent with our previous findings, full length C/EBPα (CR1/2/3/4) activated amelogenin promoter by 4.4-fold (Fig. 1B). The C/EBPα isoform (CR3/4) had little effect on the promoter activity (1.1-fold, Fig. 1B). Interestingly, CR2 in isolation had exceptional capacity, 11.8-fold, to enhance transcription of the amelogenin promoter over baseline values (Fig. 1B). Moreover, CR2 in isolation was a stronger amelogenin transactivator than full length C/EBPα (11.8-fold compared to 4.4-fold). The remaining conserved regions of C/EBPα, either in isolation or in selected combinations, showed little effect on amelogenin transactivation, such as 0.9-fold for CR1, 1.2-fold for CR1/4, and 1.3-fold for CR1/3/4. Western blot analysis confirmed the expression of these various C/EBPα truncated isoforms when transfected into LS8 cells (Fig. 1C).
Fig. 1.

Effects of C/EBPα truncated isoforms on the p2207-luc mouse amelogenin promoter. (A) Schematic representation of various C/EBPα truncated isoforms. (B) Expression plasmids (200 ng) of various C/EBPα truncated isoforms were transiently cotransfected into LS8 cells with 250 ng of p2207-luc amelogenin promoter reporter construct. pCMV-lacZ was used as an internal control for transfection efficiency. The relative luciferase activity was the normalization of luciferase activity with β-galactosidase activity. The mean ± S.D. from at least three independent experiments was represented, and the level of p2207-luc in the absence of exogenous C/EBPα was set as 1. (C) LS8 cells were transfected with 1 μg of the expression vectors of various C/EBPα truncated isoforms, and lysed 24 h as described under “Materials and methods”. Equal amounts of protein were separated by 12% SDS-polyacrylamide gel electrophoresis and immunoblotted with the C/EBPα antibody and detected using Enhanced Chemiluminance assay (Amersham, Piscataway, NJ).
Carboxyl-terminus of C/EBPα is responsible for C/EBPα-Msx2 protein-protein interactions
It has been previously shown that there is functional antagonism between C/EBPα and Msx2 in regulating the mouse amelogenin gene transcription. Protein to protein interactions between the carboxyl-terminus of Msx2 (residues 184–267) and C/EBPα is required for this protein-protein interaction [11]. In an effort to map the domain within C/EBPα responsible for C/EBPα-Msx2 protein-protein interactions, various C/EBPα truncated isoforms and Msx2 expression vectors were cotransfected into LS8 cells with the amelogenin promoter report construct p2207-luc. In this experiment set, all tested C/EBPα truncated isoforms contained the CR2 domain because CR2 domain is indispensable for C/EBPα transactivation. Increasing amount of Msx2 potently attenuated either CR1/2/3/4 (full length C/EBPα), CR 2/3/4, CR2/4, CR1/2, or CR2-mediated transactivation of the amelogenin promoter in a dose dependent manner (Fig. 2). Deletion of CR1, CR2 and CR3 domains did not affect the inhibitory effect of Msx2 on CR2-mediated transactivation (Fig.2, lane 19–21), suggesting that CR1, CR3, and CR4 are not required for C/EBPα-Msx2 interactions. It is likely that either CR2 domain or the basic region/leucine zipper domain of C/EBPα accounts for Msx2-mediated inhibition. Co-immunoprecipitation assay was then performed to identify the region of C/EBPα required for productive interactions. Here, two C/EBPα constructs were generated to facilitate the study, C/EBPα ΔN (C/EBPα AA218-359) and C/EBPα ΔC (C/EBPα AA1-215). An amino-terminal FLAG epitope-tagged Msx2 expression plasmid was co-transfected into LS8 cells with the CR1/2/3/4, CR2/3/4, CR1/2, CR2/4, CR2, C/EBPα AA218-359, or C/EBPα AA1-215 expression plasmid, respectively. Comparable amounts of both Msx2 and various C/EBPα truncated isoforms were expressed in all transfected LS8 cell groups (Fig. 3A, lanes 1–7, 15–21). The Msx2 proteins could be readily detected when using an anti-FLAG antibody to pull down the desired target from total cell lysates (Fig. 3A, lanes 22–28). In addition to the Msx2 proteins, the light chain and heavy chain of anti-FLAG M2Ab antibody were also detected (Fig. 3A, lanes 22–28). For the co-immunoprecipitation portion, the various C/EBPα truncated isoforms (CR1/2/3/4, CR2/3/4, CR1/2, CR2/4, CR2), or the C/EBPα AA218-359 were co-immunoprecipitated efficiently with Msx2 (Fig. 3A, lanes 8–13). However, the C/EBPα AA1-215 protein was not detected in the immunocomplex (Fig. 3A, lane 14), suggesting that the amino-terminal domain (residues 1–215) are not required for C/EBPα-Msx2 protein-protein interactions. The reciprocal experimental strategy in which immunoprecipitation with a C/EBPα antibody or a V5 antibody was followed by immunoblotting using an anti-C/EBPα, anti-V5 or anti-FLAG antibody (Fig. 3B) was also performed and confirmed the finding that the carboxyl-terminal domain (residues 218–359) of C/EBPα are required for C/EBPα-Msx2 protein-protein interactions.
Fig. 2.
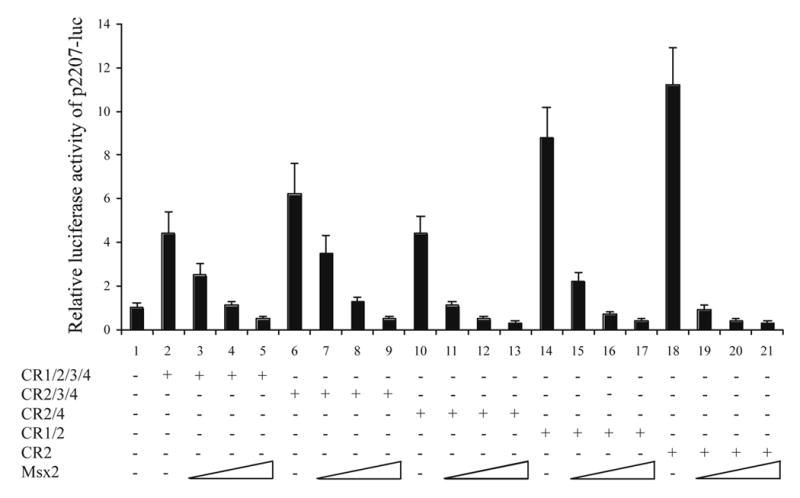
Antagonism of C/EBPα truncated isoforms mediated transactivation on the amelogenin promoter by Msx2. In the presence of 250 ng of p2207-luc reporter construct and 75 ng of pCMV-lacZ, various amounts (100, 200, and 400 ng) of the Msx2 expression plasmid were cotransfected into LS8 cells in the presence (+) or in the absence (−) of 200 ng of the expression plasmid encoding CR1/2/3/4, CR2/3/4, CR2/4, CR1/2, or CR2, respectively. The relative luciferase activity is the normalization of luciferase activity with β-galactosidase activity. The mean ± S.D. from at least three independent experiments is represented, and the basal level of p2207-luc was set as 1.
Fig. 3.
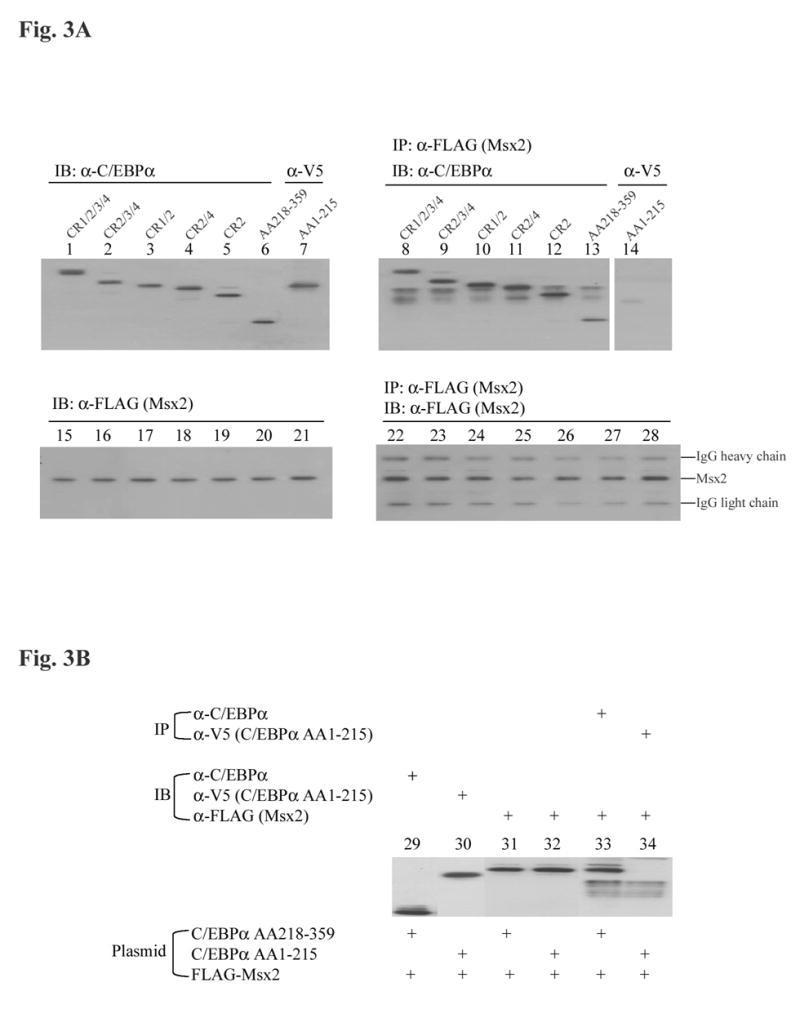
Co-immunoprecipitation of various C/EBPα truncated isoforms and Msx2 from LS8 cells. (A) In a 100-mm tissue culture dish, 2 μg of amino-terminal FLAG tagged Msx2 expression plasmid was cotransfected into LS8 cells with 2 μg of various C/EBPα truncated isoforms, CR1/2/3/4 (lane 1, 8, 15 and 22), CR2/3/4 (lane 2, 9, 16 and 23), CR1/2 (lane 3, 10, 17 and 24), CR2/4 (lane 4, 11, 18, and 25), CR2 (lane 5, 12, 19 and 26), C/EBPα AA218-359 (lane 6, 13, 20, and 27), C/EBPα AA1-215 (lane 7, 14, 21 and 28). After 24 h incubation, whole cell lysates were prepared as described under “Materials and methods”. For immunoblot (IB), 10 μg of cell lysates were electrophoresed, transferred to Immobilon-P membrane (Millipore, Billerica, MA), and immunoblotted with a C/EBPα antibody (Santa Cruz Biotechnology, Santa Cruz, CA, lane 1 to 6), a V5 antibody (Invitrogen, Carlsbad, CA, lane 7), or an anti-FLAG antibody (M2Ab, Sigma, St. Louis, MO, lane 15 to 21). 500 μg of cell lysates were subjected to immunoprecipitation (IP) with an anti-FLAG antibody (lane 8 to 14). The immunoprecipitates were then electrophoresed, transferred to Immobilon-P membrane, and immunoblotted with an anti-C/EBPα antibody (lane 8 to 13), or an anti-V5 antibody (lane 14), or an anti-FLAG antibody (lane 22 to 28). (B) Co-immunoprecipitation analysis was performed as in (A). Total cell lysates of 10 μg were immunoblotted with an antibody against C/EBPα (lane 29), V5 (lane 30), or FLAG (lane 31 and 32). Cell lysates were immunoprecipitated with an anti-C/EBPα antibody, followed by immunoblotting with an anti-FLAG antibody (lane 33); or with an anti-V5 antibody, followed by immunoblotting with an anti-FLAG antibody (lane 34).
Discussion
Here we report that the CR2 (conserved region 2) of the C/EBPα transactivation domain was best able to stimulate transcription from the mouse amelogenin promoter. The remainders of the conserved regions in select combinations had little effect on the mouse amelogenin promoter. This finding in the regulation of the mouse amelogenin gene is different from the observations for the C/EBPα functions during adipogenesis, in which a combination of CR1 and CR3 is also able to induce adipogenesis [5]. The difference may result from the ability of C/EBPα to interact with selected co-activators in the context of different physiologic requirements.
Several studies have shown that CR2 interacts with basal transcription apparatus and certain co-activators. TBP and TFIIB, two essential components of the RNA polymerase II basal transcriptional apparatus, have been identified to co-operatively interact with the region covering CR2 domain [12]. The retinoblastoma (Rb) protein complex has been shown to interact with C/EBPα and to activate C/EBPα-mediated transcription [13, 14], while the Rb-binding motifs of C/EBPα (residues 67–82) are located within the CR2 domain (residues 55–108) [15]. Furthermore, p300 is reported to co-activate C/EBPα-mediated transactivation, which is mediated, in part, by functional interaction with CR2 domain of C/EBPα [5].
The expression pattern of Msx2 has been identified in murine teeth during odontogenesis. Msx2 is expressed in undifferentiated inner enamel epithelial cells but is down-regulated in differentiated ameloblast cells [16]. The role of Msx2 in tooth formation has also been elucidated in Msx2-deficient mice. Msx2-deficient molar tooth germs developed normally up to the cap stage. The defect starts to be observed at E16.5 as a modest reduction in enamel organ volume. By the late bell stage, the amount of the epithelial derived stellate reticulum (SR) decreases, and by the day 1 postnatal stage, highly duplicated epithelial cells reside in the intercuspal regions. Adult Msx2-deficient mice show brittle and misaligned incisors as well as degenerated molars [17]. Functional assays have demonstrated that Msx2 represses mouse amelogenin gene expression through direct protein-protein interactions with C/EBPα, a strong transactivator of amelogenin gene [11]. Msx2-C/EBPα physical interactions have also been identified to participate in regulating osteogenic versus adipogenic differentiation of aortic myofibroblasts [18].
In this study, we identify that the CR2 domain of C/EBPα is the only transactivation domain responsible for activation of the mouse amelogenin gene. In addition, the carboxyl-terminus of C/EBPα (residues 216–359) is required for protein-protein interactions with Msx2. Although Msx2 is reported as a general transcription repressor [19], this mechanism is not likely to play a significant role in our model system since previous data has only shown a modest inhibitory effect on the amelogenin promoter construct p51-luc in the presence of Msx2 [11].
Since the carboxyl-terminal stretch of amino acids contains basic region leucine zipper domain that is responsible for the DNA binding and dimer formation of C/EBPα, there are several possibilities to account for repression of Msx2 on C/EBPα-mediated transactivation. Msx2 may interfere with C/EBPα binding to its cis-element upon the formation of Msx2-C/EBPα protein complexes. Alternatively, Msx2 may perturb C/EBPα dimer formation, since dimerization is a prerequisite for C/EBPα binding [20, 21]. These data, together with our previous findings [8, 11], help to better understand the molecular mechanism for C/EBPα and Msx2 to orchestrate amelogenin production during tooth formation.
Supplementary Material
Acknowledgments
This work was funded by grant DE-06988 from the National Institute of Dental and Craniofacial Research, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Landschulz WH, Johnson PF, Adashi EY, Graves BJ, McKnight SL. Isolation of a recombinant copy of the gene encoding C/EBP. Genes Dev. 1988;2:786–800. doi: 10.1101/gad.2.7.786. [DOI] [PubMed] [Google Scholar]
- 2.Landschulz WH, Johnson PF, McKnight SL. The DNA binding domain of the rat liver nuclear protein C/EBP is bipartite. Science. 1989;243:1681–1688. doi: 10.1126/science.2494700. [DOI] [PubMed] [Google Scholar]
- 3.Friedman AD, McKnight SL. Identification of two polypeptide segments of CCAAT/enhancer-binding protein required for transcriptional activation of the serum albumin gene. Genes Dev. 1990;4:1416–1426. doi: 10.1101/gad.4.8.1416. [DOI] [PubMed] [Google Scholar]
- 4.Pei DQ, Shih CH. An “attenuator domain” is sandwiched by two distinct transactivation domains in the transcription factor C/EBP. Mol Cell Biol. 1991;11:1480–1487. doi: 10.1128/mcb.11.3.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erickson RL, Hemati N, Ross SE, MacDougald OA. p300 coactivates the adipogenic transcription factor CCAAT/enhancer- binding protein alpha. J Biol Chem. 2001;276:16348–16355. doi: 10.1074/jbc.m100128200. [DOI] [PubMed] [Google Scholar]
- 6.Nerlov C, Ziff EB. Three levels of functional interaction determine the activity of CCAAT/enhancer binding protein-alpha on the serum albumin promoter. Genes Dev. 1994;8:350–362. doi: 10.1101/gad.8.3.350. [DOI] [PubMed] [Google Scholar]
- 7.Ross SE, Erickson RL, Hemati N, MacDougald OA. Glycogen synthase kinase 3 is an insulin-regulated C/EBPalpha kinase. Mol Cell Biol. 1999;19:8433–8441. doi: 10.1128/mcb.19.12.8433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou YL, Snead ML. Identification of CCAAT/enhancer-binding protein alpha as a transactivator of the mouse amelogenin gene. J Biol Chem. 2000;275:12273–12280. doi: 10.1074/jbc.275.16.12273. [DOI] [PubMed] [Google Scholar]
- 9.Xu Y, Zhou YL, Luo W, Zhu QS, Levy D, MacDougald OA, Snead ML. NF-Y and CCAAT/enhancer-binding protein alpha synergistically activate the mouse amelogenin gene. J Biol Chem. 2006;281:16090–16098. doi: 10.1074/jbc.M510514200. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Zhou YL, Ann DK, MacDougald OA, Shum L, Snead ML. Transcription factor sumoylation and factor YY1 serve to modulate mouse amelogenin gene expression. Eur J Oral Sci. 2006;114(Suppl 1):169–177. doi: 10.1111/j.1600-0722.2006.00319.x. discussion 201-162, 381. [DOI] [PubMed] [Google Scholar]
- 11.Zhou YL, Lei Y, Snead ML. Functional antagonism between Msx2 and CCAAT/enhancer-binding protein alpha in regulating the mouse amelogenin gene expression is mediated by protein-protein interaction. J Biol Chem. 2000;275:29066–29075. doi: 10.1074/jbc.M002031200. [DOI] [PubMed] [Google Scholar]
- 12.Nerlov C, Ziff EB. CCAAT/enhancer binding protein-alpha amino acid motifs with dual TBP and TFIIB binding ability co-operate to activate transcription in both yeast and mammalian cells. Embo J. 1995;14:4318–4328. doi: 10.1002/j.1460-2075.1995.tb00106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Classon M, Kennedy BK, Mulloy R, Harlow E. Opposing roles of pRB and p107 in adipocyte differentiation. Proc Natl Acad Sci U S A. 2000;97:10826–10831. doi: 10.1073/pnas.190343597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charles A, Tang X, Crouch E, Brody JS, Xiao ZX. Retinoblastoma protein complexes with C/EBP proteins and activates C/EBP-mediated transcription. J Cell Biochem. 2001;83:414–425. doi: 10.1002/jcb.1239. [DOI] [PubMed] [Google Scholar]
- 15.Chen PL, Riley DJ, Chen Y, Lee WH. Retinoblastoma protein positively regulates terminal adipocyte differentiation through direct interaction with C/EBPs. Genes Dev. 1996;10:2794–2804. doi: 10.1101/gad.10.21.2794. [DOI] [PubMed] [Google Scholar]
- 16.MacKenzie A, Ferguson MW, Sharpe PT. Expression patterns of the homeobox gene, Hox-8, in the mouse embryo suggest a role in specifying tooth initiation and shape. Development. 1992;115:403–420. doi: 10.1242/dev.115.2.403. [DOI] [PubMed] [Google Scholar]
- 17.Satokata I, Ma L, Ohshima H, Bei M, Woo I, Nishizawa K, Maeda T, Takano Y, Uchiyama M, Heaney S, Peters H, Tang Z, Maxson R, Maas R. Msx2 deficiency in mice causes pleiotropic defects in bone growth and ectodermal organ formation. Nat Genet. 2000;24:391–395. doi: 10.1038/74231. [DOI] [PubMed] [Google Scholar]
- 18.Cheng SL, Shao JS, Charlton-Kachigian N, Loewy AP, Towler DA. MSX2 promotes osteogenesis and suppresses adipogenic differentiation of multipotent mesenchymal progenitors. J Biol Chem. 2003;278:45969–45977. doi: 10.1074/jbc.M306972200. [DOI] [PubMed] [Google Scholar]
- 19.Newberry EP, Latifi T, Battaile JT, Towler DA. Structure-function analysis of Msx2-mediated transcriptional suppression. Biochemistry. 1997;36:10451–10462. doi: 10.1021/bi971008x. [DOI] [PubMed] [Google Scholar]
- 20.Vinson CR, Sigler PB, McKnight SL. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science. 1989;246:911–916. doi: 10.1126/science.2683088. [DOI] [PubMed] [Google Scholar]
- 21.Hurst HC. Transcription factors 1: bZIP proteins. Protein Profile. 1995;2:101–168. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


