Abstract
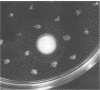
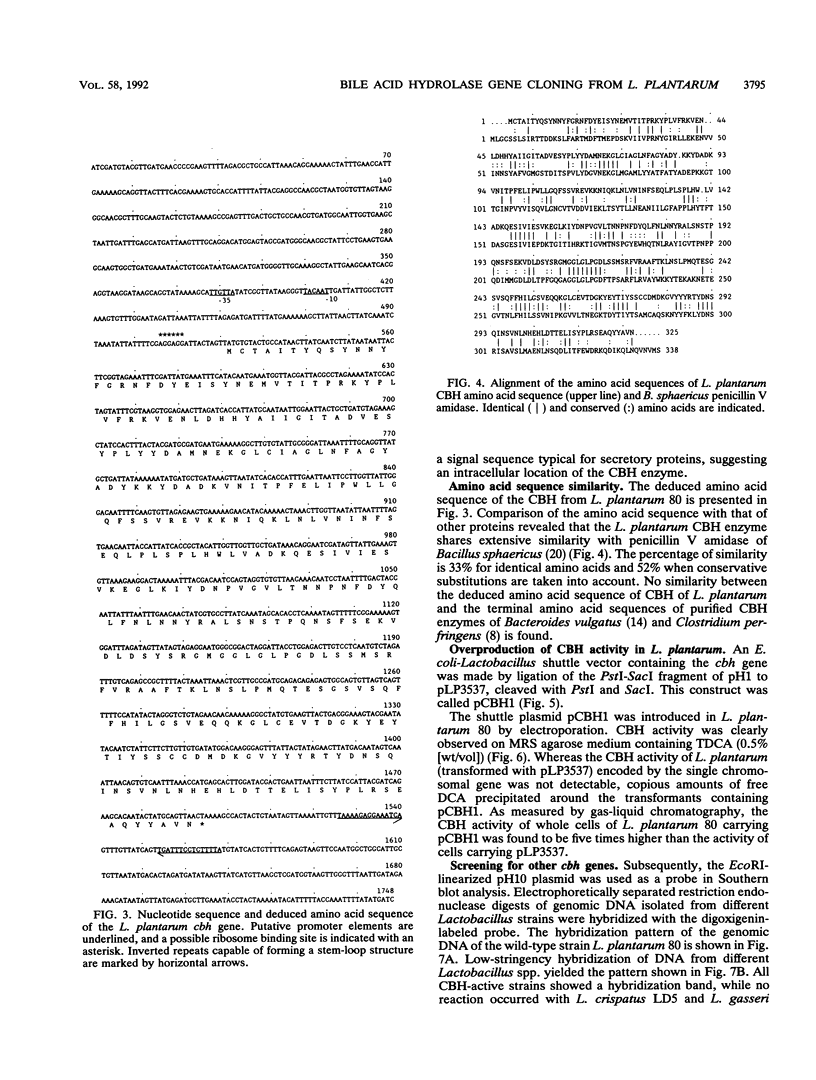
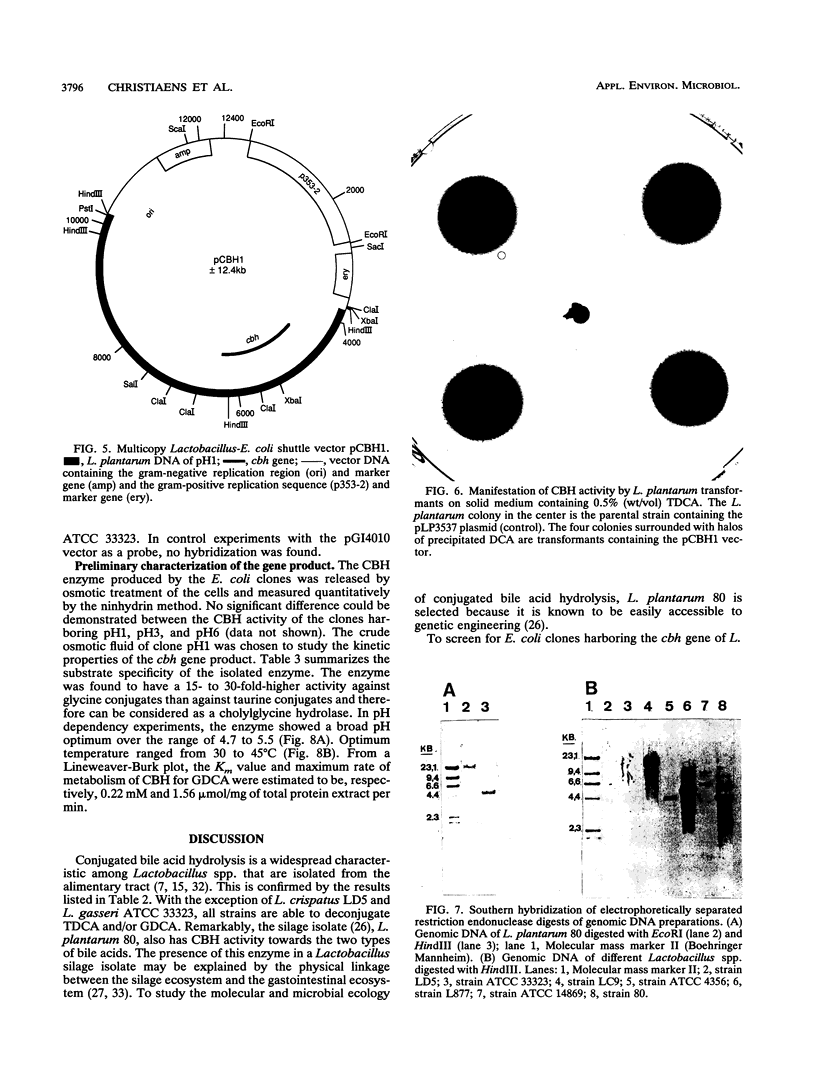
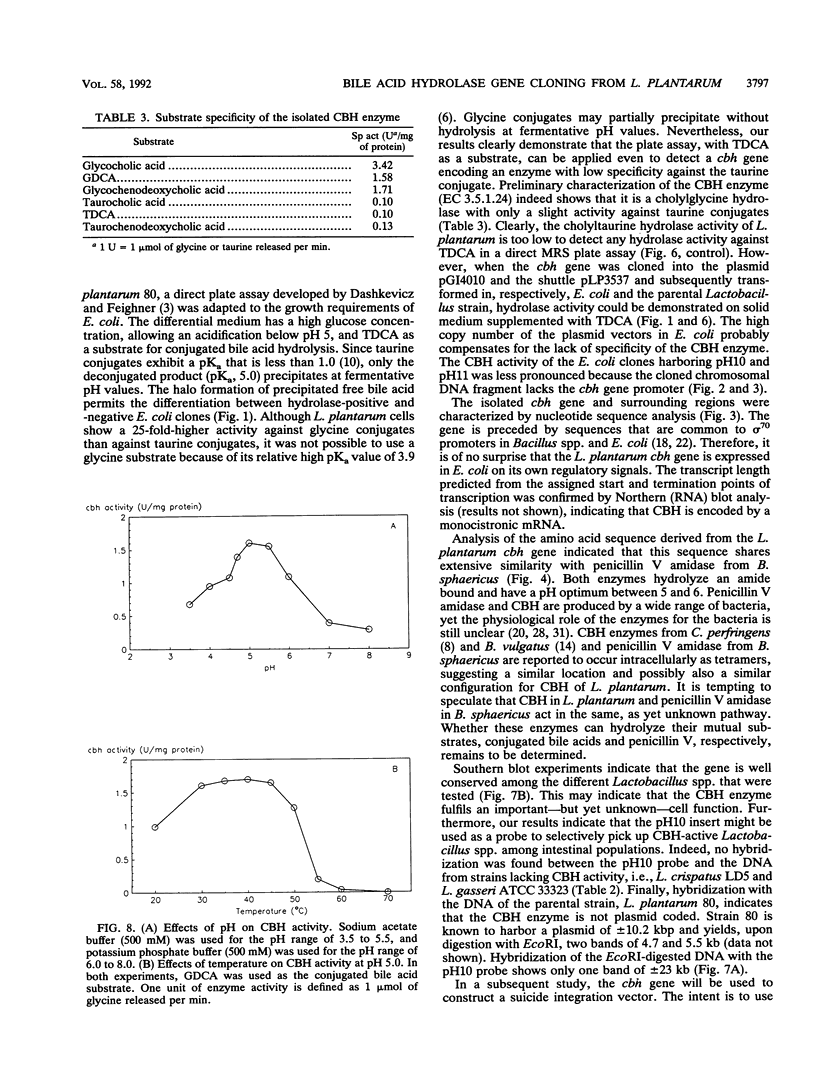
The conjugated bile acid hydrolase gene from the silage isolate Lactobacillus plantarum 80 was cloned and expressed in Escherichia coli MC1061. For the screening of this hydrolase gene within the gene bank, a direct plate assay developed by Dashkevicz and Feighner (M. P. Dashkevicz and S. D. Feighner, Appl. Environ. Microbiol. 53:331-336, 1989) was adapted to the growth requirements of E. coli. Because of hydrolysis and medium acidification, hydrolase-active colonies were surrounded with big halos of precipitated, free bile acids. This phenomenon was also obtained when the gene was cloned into a multicopy shuttle vector and subsequently reintroduced into the parental Lactobacillus strain. The cbh gene and surrounding regions were characterized by nucleotide sequence analysis. The deduced amino acid sequence was shown to have 52% similarity with a penicillin V amidase from Bacillus sphaericus. Preliminary characterization of the gene product showed that it is a cholylglycine hydrolase (EC 3.5.1.24) with only slight activity against taurine conjugates. The optimum pH was between 4.7 and 5.5. Optimum temperature ranged from 30 to 45 degrees C. Southern blot analysis indicated that the cloned gene has similarity with genomic DNA of bile acid hydrolase-active Lactobacillus spp. of intestinal origin.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980 Apr;138(2):179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- Dashkevicz M. P., Feighner S. D. Development of a differential medium for bile salt hydrolase-active Lactobacillus spp. Appl Environ Microbiol. 1989 Jan;55(1):11–16. doi: 10.1128/aem.55.1.11-16.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feighner S. D., Dashkevicz M. P. Effect of dietary carbohydrates on bacterial cholyltaurine hydrolase in poultry intestinal homogenates. Appl Environ Microbiol. 1988 Feb;54(2):337–342. doi: 10.1128/aem.54.2.337-342.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feighner S. D., Dashkevicz M. P. Subtherapeutic levels of antibiotics in poultry feeds and their effects on weight gain, feed efficiency, and bacterial cholyltaurine hydrolase activity. Appl Environ Microbiol. 1987 Feb;53(2):331–336. doi: 10.1128/aem.53.2.331-336.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fini A., Roda A. Chemical properties of bile acids. IV. Acidity constants of glycine-conjugated bile acids. J Lipid Res. 1987 Jul;28(7):755–759. [PubMed] [Google Scholar]
- Gilliland S. E., Speck M. L. Deconjugation of bile acids by intestinal lactobacilli. Appl Environ Microbiol. 1977 Jan;33(1):15–18. doi: 10.1128/aem.33.1.15-18.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal-Srivastava R., Hylemon P. B. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J Lipid Res. 1988 Aug;29(8):1079–1085. [PubMed] [Google Scholar]
- Gorbach S. L., Tabaqchali S. Bacteria, bile, and the small bowel. Gut. 1969 Dec;10(12):963–972. doi: 10.1136/gut.10.12.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A. F., Roda A. Physicochemical properties of bile acids and their relationship to biological properties: an overview of the problem. J Lipid Res. 1984 Dec 15;25(13):1477–1489. [PubMed] [Google Scholar]
- Hoskins L. C., Boulding E. T. Mucin degradation in human colon ecosystems. Evidence for the existence and role of bacterial subpopulations producing glycosidases as extracellular enzymes. J Clin Invest. 1981 Jan;67(1):163–172. doi: 10.1172/JCI110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josson K., Scheirlinck T., Michiels F., Platteeuw C., Stanssens P., Joos H., Dhaese P., Zabeau M., Mahillon J. Characterization of a gram-positive broad-host-range plasmid isolated from Lactobacillus hilgardii. Plasmid. 1989 Jan;21(1):9–20. doi: 10.1016/0147-619x(89)90082-6. [DOI] [PubMed] [Google Scholar]
- Kawamoto K., Horibe I., Uchida K. Purification and characterization of a new hydrolase for conjugated bile acids, chenodeoxycholyltaurine hydrolase, from Bacteroides vulgatus. J Biochem. 1989 Dec;106(6):1049–1053. doi: 10.1093/oxfordjournals.jbchem.a122962. [DOI] [PubMed] [Google Scholar]
- Lundeen S. G., Savage D. C. Characterization and purification of bile salt hydrolase from Lactobacillus sp. strain 100-100. J Bacteriol. 1990 Aug;172(8):4171–4177. doi: 10.1128/jb.172.8.4171-4177.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane G. T., Hay S., Gibson G. R. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J Appl Bacteriol. 1989 May;66(5):407–417. doi: 10.1111/j.1365-2672.1989.tb05110.x. [DOI] [PubMed] [Google Scholar]
- Masuda N. Deconjugation of bile salts by Bacteroids and Clostridium. Microbiol Immunol. 1981;25(1):1–11. doi: 10.1111/j.1348-0421.1981.tb00001.x. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Northfield T. C., Drasar B. S., Wright J. T. Value of small intestinal bile acid analysis in the diagnosis of the stagnant loop syndrome. Gut. 1973 May;14(5):341–347. [PMC free article] [PubMed] [Google Scholar]
- Olsson A., Uhlén M. Sequencing and heterologous expression of the gene encoding penicillin V amidase from Bacillus sphaericus. Gene. 1986;45(2):175–181. doi: 10.1016/0378-1119(86)90252-0. [DOI] [PubMed] [Google Scholar]
- Posno M., Leer R. J., van Luijk N., van Giezen M. J. F., Heuvelmans P. T. H. M., Lokman B. C., Pouwels P. H. Incompatibility of Lactobacillus Vectors with Replicons Derived from Small Cryptic Lactobacillus Plasmids and Segregational Instability of the Introduced Vectors. Appl Environ Microbiol. 1991 Jun;57(6):1822–1828. doi: 10.1128/aem.57.6.1822-1828.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage D. C. Microbial ecology of the gastrointestinal tract. Annu Rev Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- Scheirlinck T., Mahillon J., Joos H., Dhaese P., Michiels F. Integration and expression of alpha-amylase and endoglucanase genes in the Lactobacillus plantarum chromosome. Appl Environ Microbiol. 1989 Sep;55(9):2130–2137. doi: 10.1128/aem.55.9.2130-2137.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TROLL W., CANNAN R. K. A modified photometric ninhydrin method for the analysis of amino and imino acids. J Biol Chem. 1953 Feb;200(2):803–811. [PubMed] [Google Scholar]
- Tannock G. W., Dashkevicz M. P., Feighner S. D. Lactobacilli and bile salt hydrolase in the murine intestinal tract. Appl Environ Microbiol. 1989 Jul;55(7):1848–1851. doi: 10.1128/aem.55.7.1848-1851.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Van Renterghem B., Huysman F., Rygole R., Verstraete W. Detection and prevalence of Listeria monocytogenes in the agricultural ecosystem. J Appl Bacteriol. 1991 Sep;71(3):211–217. doi: 10.1111/j.1365-2672.1991.tb04450.x. [DOI] [PubMed] [Google Scholar]
- Vandamme E. J., Voets J. P. Microbial penicillin acylases. Adv Appl Microbiol. 1974;17(0):311–369. doi: 10.1016/s0065-2164(08)70563-x. [DOI] [PubMed] [Google Scholar]
- Vandevoorde L., Christiaens H., Verstraete W. Prevalence of coaggregation reactions among chicken lactobacilli. J Appl Bacteriol. 1992 Mar;72(3):214–219. doi: 10.1111/j.1365-2672.1992.tb01826.x. [DOI] [PubMed] [Google Scholar]