Abstract
The Rh system is one of the most important and complex blood group systems because of the large number of antigens and the serious complications for the fetus of a woman sensitized by transfusion or pregnancy. Major advances in our understanding of the Rh system have occurred with the cloning of the genes and with functional evidence that the Rh blood group proteins belong to an ancient family of membrane proteins involved in ammonia transport.
The arrangement and configuration of the genes at the RH locus promotes genetic exchange, generating new antigens. Importantly, RH genetic testing can now be applied to clinical transfusion medicine and prenatal practice. This includes testing for RHD zygosity, confirmation or resolution of D antigen status, and detection of altered RHD and RHCE genes in individuals at risk for producing antibodies to high incidence Rh antigens, particularly sickle cell disease patients. The Rh proteins form a core complex that is critical to the structure of the erythrocyte membrane, and may play a physiologically role in the sequestration of blood ammonia. The Rh family of proteins now includes non-erythroid Rh homologs present in many other tissues, and comparative genomics reveals Rh homologs in all domains of life.
Introduction
The history of the Rh system began in 1941 with its discovery as the cause of severe jaundice and fetal demise, i.e. erythroblastosis fetalis. The syndrome had been observed for many years to complicate pregnancies. The fact that it was due to an immune reaction to a paternal antigen was only realized with the delivery of a stillborn fetus, and an adverse reaction of the mother to a blood transfusion from the father.1 The syndrome, referred to as hemolytic disease of the newborn (HDN), was principally caused by maternal sensitization to Rh, specifically to the D antigen. HDN due to D incompatibility was prevalent in Caucasians, who have the highest incidence of the D-negative phenotype (15-17%), but was rare in other ethnic groups. The incidence of HDN was dramatically altered when it was realized that ABO incompatibility between a mother and the fetus had a partial protective effect against immunization to D. This suggested the rationale for the development of Rh immune globulin. By the 1960's, a mere 20 years after the discovery of Rh incompatibility, HDN due to anti-D could be effectively prevented. 2,3
The Rh system has long been known to be one of the most complex blood group systems. In addition to the presence or absence of the D antigen, other common Rh antigens include the allelic C or c, and E or e. However, over fifty different Rh antigens have been identified by investigating the specificity of antibodies produced after blood transfusion or pregnancy. Major advances in our understanding of the Rh system have occurred with the cloning of the genes. The arrangement and configuration of the RH locus result in gene conversion events that generate hybrid proteins, which represent additional Rh antigens. Importantly, genetic testing for RH can now be used in prenatal and clinical transfusion medicine to detect the presence of a recessive D-negative allele or to determine the inheritance of RH genes carrying mutations that encode altered Rh proteins. The latter is especially relevant for SCD patients who, because of genetic disparity with Caucasian donors, are at high risk for becoming alloimmunized.
The Rh complex is critical to the structure of the membrane. Rhnull erythrocytes, which lack Rh proteins, are stomatocytic and spherocytic, and affected individuals have hemolytic anemia.4 Recent findings indicate that Rh proteins mediate key interactions with the underlying cytoskeleton through protein 4.2 and ankyrin.5-7
The erythrocyte Rh blood group proteins are well known because of their importance in blood transfusion, but recent functional studies and structural modeling reveal that the Rh blood group proteins are members of an ancient family of proteins involved in ammonia transport.8-11 Non-erythroid Rh proteins have now been found in other tissues including the kidney, liver, brain, and skin 12-15, in locations where ammonia production and elimination occurs. The family of Rh proteins has now been expanded significantly through comparative genomics and structure-function studies that reveal the presence of Rh homologs in all domains of life. Current investigations can be compared to an “archeological dig” to uncover relationships between the Rh family members to gain insight into the mechanism of transport.
Terminology
Rh terminology distinguishes between the antigens, genes, and the proteins. The antigens are referred to by the letter designations, D, C, c, E, e, etc. The RH genes are designated by capital letters, with or without italics, and include erythroid RHD, RHCE, and RHAG, as well as the non-erythroid homologs expressed in other tissues, RHBG and RHCG. The different alleles of the RHCE gene are designated RHce, RHCe, RHcE, according to which antigens they encode. The proteins are indicated as RhD, RhCE (or according to the specific antigens they carry Rhce, RhCe, or RhcE), and include erythroid RhAG and those found in other tissues, RhBG and RhCG.
RH genes and Rh proteins
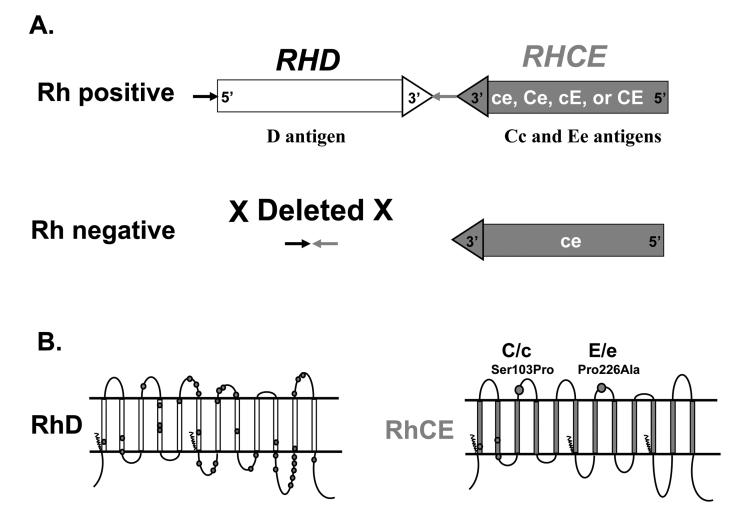
Two genes (RHD, RHCE) in close proximity on chromosome 1 encode the erythrocyte Rh proteins, RhD and RhCE; one carries the D antigen, and the other carries CE antigens in various combinations (ce, Ce, cE, or CE), (Fig.1A).16-19 The genes each have ten exons, are 97% identical, and arose via gene duplication. RhD and RhCE proteins differ by 32-35 of 416 amino acids (Fig. 1B shown as circles on RhD). This is in contrast to most blood group antigens that are encoded by single genes with alleles that differ by only one or a few amino acids. Individuals who lack RhD protein, “Rh or D negative”, most often have a complete deletion of the RHD gene (Fig. 1A). An important consideration in the immunogenicity of a protein is the degree of foreignness to the host. The large number of amino acid changes explains why exposure to RhD can result in a potent immune response in a D-negative individual.
Fig. 1.
A). Diagram of the RHD and RHCE locus. The two RH genes have opposite orientation, with the 3′ends facing each other. Rh negative Caucasian individuals have a complete deletion of RHD. B). Rh proteins in the RBC membrane. The RhD and RhCE proteins are predicted to have twelve transmembrane domains. Amino acid positions that differ between RhD and RhCE are shown as dark circles on RhD. The amino acid changes responsible for C/c and E/e polymorphisms are shown on RhCE.
RHCE, expressed in all but rare D- - individuals, encodes both C/c and E/e antigens on a single protein. C and c antigens differ by four amino acids, but only the amino acid change Ser103Pro is extracellular (Fig. 1B). The E and e antigens differ by one amino acid, Pro226Ala, located on the fourth extracellular loop of the protein.
The RH genes and proteins detailed in Fig.1 and Fig. 4A are typical for the majority of individuals, and commercial antibody reagents detect expression of these “conventional” D, C, c, E and e antigens shown. The proximity of the two RH genes, and their inverted orientation (Fig. 1A), augments opportunity for genetic exchange.20 Many RH genes carry point mutations, or have rearrangements and exchanges between RHD and RHCE that result from gene conversion events. The latter encode hybrid proteins that have RhCE-specific amino acids in RhD, or RhD-specific residues in RhCE. These can generate new antigens in the Rh blood group system, and alter or weaken expression of the conventional antigens.
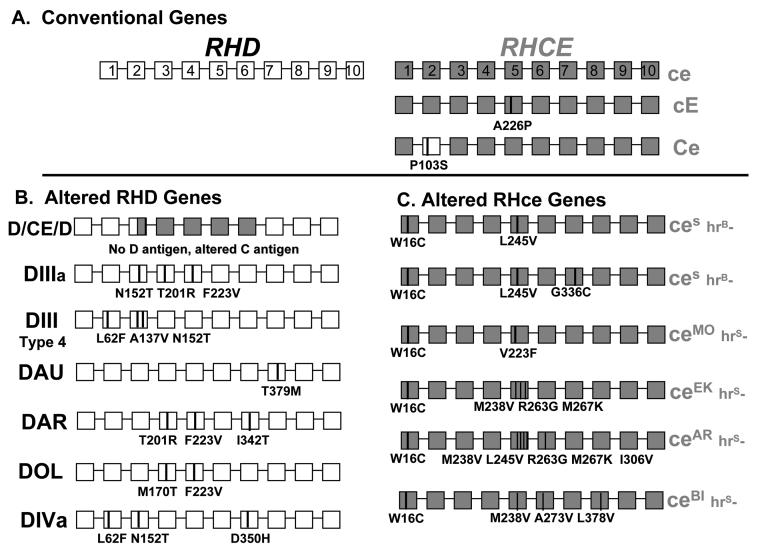
Fig. 4.
Diagram of the RHD and RHCE genes. The ten RHD exons are shown as white boxes, and the RHCE as grey. A). RHD and RHCE genes responsible for the common D, C, c, E and e antigen polymorphisms. B). Altered RHD and C). Altered RHCE genes indicating the changes often found in African-Americans that complicate transfusion, especially for sickle cell patients.
Numerous mutations in RHD (greater than 100) are currently known. Most encode single amino acid changes, and many of these simply alter the quantity of RhD protein in the membrane, while others alter RhD membrane topology and D-epitopes. The latter are responsible for the enigmatic individual with D-positive RBCs who presents with anti-D following transfusion or pregnancy.
Mutations in both RHD and RHCE are found in Blacks, and other ethnically diverse groups, and alter D, C, or e antigen expression (detailed below). The incidence of variant RH genes in patients with sickle cell disease underlies the complex incompatibilities that can arise following transfusion in this patient population. Commercial typing reagents are not available to detect red cells that express variant Rh proteins, and individuals at risk go undetected until they produce antibodies reactive with all, or most, conventional Rh antigens. Finding compatible blood then becomes a serious and potentially life-threatening problem. Understanding of RH gene variation can now be applied to assess alloimmunization risk in SCD, because patients who inherit variant RH genes can now be identified at the genomic level.
D negative
D-negative is prevalent in Caucasians (15%-17%), but less common in Black Africans (5%) and Asians (3%).21 The D-negative phenotype has arisen numerous times, and multiple genetic events are responsible for loss of RhD expression in different populations. Caucasians have a deletion of the entire RHD gene,22 but Africans and Asians often have an inactive or silent RHD. Approximately 66% of South African, D-negative, Black persons have RHD with a 37-bp internal duplication that causes a premature stop codon and does not encode functional protein, while 15% carry a hybrid RHD-CE-D.23 This hybrid D-CE-D does not express D antigen and encodes an altered C antigen (Fig. 4B). In Asians, 10-30% of D-negative phenotypes are Del and have very low levels of D antigen not detectable by standard typing (see below).
RHD zygosity determination
Serologic testing for RBC expression of D, C/c, and E/e can only predict the likelihood that a sample is homozygous (D/D) or heterozygous (D/−). However, RHD zygosity can now be determined by assaying for the presence of a recessive D-negative allele. In the prenatal setting, paternal RHD zygosity testing is important to predict fetal D status when the mother has anti-D. The ethnic background of the parents is important to the design of the assay because different genetic events are responsible for D-negative phenotypes. In addition to testing for the absence of RHD, zygosity assays include detection of the region generated by deletion of RHD (arrows in Fig.1), the 37-bp insert RHD pseudogene, and the D-negative RHD-CE-D hybrid gene.23,24
Prediction of fetal risk for HDN
If the father is RHD homozygous, monitoring of the pregnancy will be required, but if the father is heterozygous, the D-status of the fetus should be determined. The goal is to prevent invasive and costly monitoring of the pregnancy when the infant is D-negative. Fetal DNA can be obtained by amniocentesis or villus sampling. However, cell-free, fetal-derived DNA is present in maternal plasma by approximately five weeks, which allows maternal plasma to be tested as a non-invasive sample source.25, 26 The assay detects the presence or absence of RHD, and because cell-free fetal DNA will be present in low amount, positive controls for isolation of sufficient fetal DNA is critical to validate negative results. Y chromosome markers are useful when the fetus is a male, but when the fetus is female, proper controls present a challenge and polymorphic paternal markers are then needed.27 Testing of samples from the parents also limits the possibility of misinterpretation due to inheritance of rare or familial RHD inactivating mutations or rearranged hybrid genes.
D positive
The majority of D-positive individuals inherit RHD as shown in Fig.1, but exceptions are encountered, and these can complicate determination of D status.
Weak D
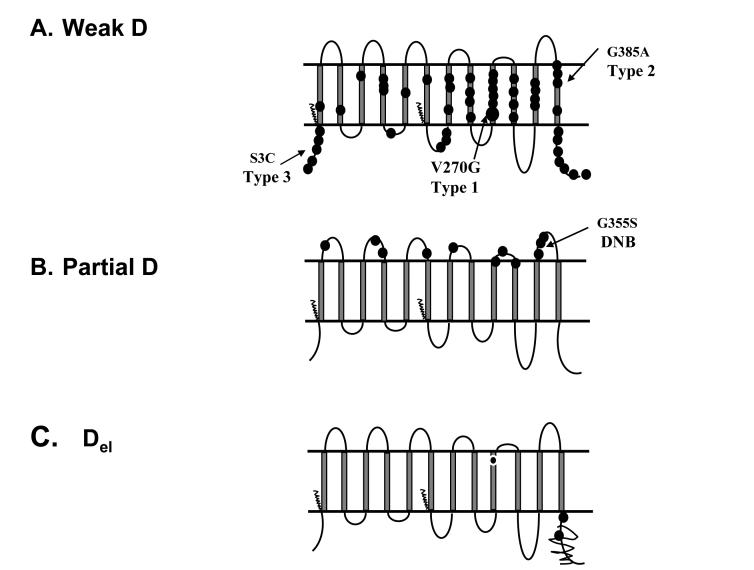
Reduced expression of D antigen occurs in an estimated 0.2%-1% of Caucasians.28 Historically, RBCs that react with anti-D only after extended testing with the indirect antiglobulin test (IAT) are called weak D. However, the number of samples classified as weak D depends on the characteristics of the typing reagent. Weak D expression primarily results from single point mutations in RHD that encode amino acid changes predicted to be intracellular or in the transmembrane regions of RhD (Fig. 2A).29 These affect the efficiency of insertion, and, therefore, the quantity of RhD protein in the membrane, reflected in the reduced number of D antigen sites on the RBCs. Over 50 different mutations, the most common being a Val270Gly designated Type 1, cause weak D expression (Fig. 2A). Mutations are catalogued on the RhesusBase and blood group mutation websites and are updated regularly.30,31
Fig. 2.
Diagram of amino acid changes in RhD proteins shown as circles.
A). Weak D phenotypes. Amino acid changes that cause weak D expression, shown as circles, are located predominantly in transmembrane and cytoplasmic regions. Weak D Type 1 (V270G) predominates in Europeans, as well as Type 2 and Type 3, which together represent the majority of weak D, are indicated. B). Partial D phenotypes. Amino acid changes that cause some partial D phenotyes are predicted to be located in the extracellular loops. The DNB mutation is frequent in Europeans. C). Del phenotypes. Amino acid changes that severely reduce the quantity of RhD resulting in RBCs that type serologically D-negative are shown. The scribble line indicates loss of the 3′ region, characteristic of Asian mutations, and the European M295I mutation is shown in transmembrane nine.
Del
Del red cells express very low levels of D antigen that cannot be detected by routine testing but these adsorb and elute anti-D, hence the name. Del are often found in Asian ethnic groups (10-30% of D-negative) and result from a mutation causing reduced synthesis of the 3′ region of RhD (Fig. 2C). Del are less frequently in Europeans (0.027%) and result from an amino acid change, M295I.29,32,33
Partial D
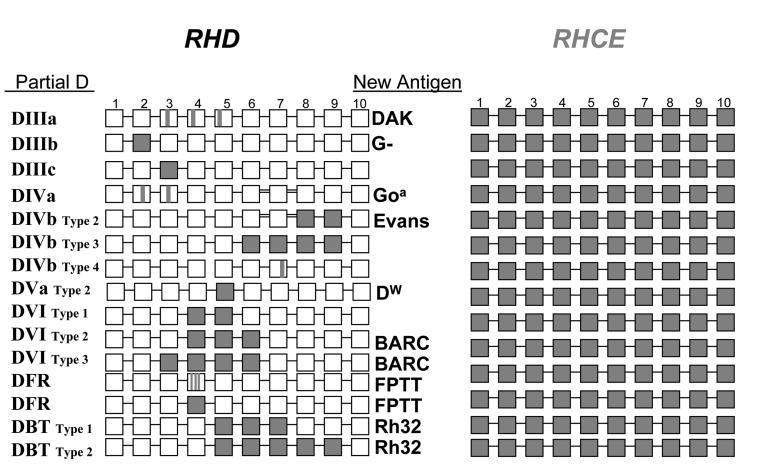
RBCs with partial D antigen type as D-positive (some in direct tests, and others by IAT), but individuals often produce anti-D when stimulated by transfusion or pregnancy. Some partial D, similar to weak D, result from point mutations in RHD that cause single amino acid changes. But, in contrast to weak D, these changes are located on the extracellular regions and alter or create new epitopes (Fig. 2B). Many partial D result from hybrid genes that have regions of RHD replaced by the corresponding regions of RHCE. Some examples are shown in Fig. 3. These replacements can involve short regions encompassing several codons, entire exons, or large regions of the gene, and the novel sequence of amino acids that result from RhD joined with RhCE can generate new Rh antigens (e.g., DAK, Goa, Evans, Dw, BARC, FPTT, Rh32 etc) (Fig. 3).
Fig 3.
Most Parital D phenotypes result from gene conversion. The ten RHD exons are shown as white boxes, and the RHCE as grey. Some examples of new antigens that result from hybrid proteins encoded by regions of RHCE inserted in RHD are indicated. For a comprehensive summary, see the Facts Book.72
Clinical Considerations for Weak D, Partial D, and Del
Of clinical concern, particularly when determining the D status of a woman of child-bearing age, is the distinction between a partial D and weak D phenotypes. The former may make anti-D, whereas the latter are unlikely to do so. It is impossible to distinguish between these with commercial serologic reagents. The extensive history of transfusing patients who have weak D RBCs with D-positive blood strongly suggests that weak D Types 1, 2, and 3 (Fig. 2A), which comprise 90 percent of weak D individuals, do not make anti-D and can safely receive D-positive blood. In contrast, patients with partial D RBCs often make anti-D and should receive D-negative blood and be considered candidates for RhIG. This is a consideration for only performing a direct test to determine the D type in prenatal and transfusion practice. It is important to note that the current monoclonal FDA-licensed anti-D typing reagents are formulated to not react in direct tests with partial DVI RBCs. DVI is the most common partial D in Caucasians,34 and anti-D produced by partial DVI has resulted in fatal HDN. 35 Elimination of the IAT will better serve women with DVI RBCs by classifying them as D-negative for transfusion and RhIG prophylaxis.
As donors, weak D and partial D RBCs can stimulate the production of anti-D in a D-negative recipient. Therefore donor centers are required to use extended methods that detect and label these as D-positive. This can result in an individual who is appropriately labeled D-positive as a donor, but classified D-negative as a transfusion recipient. Communication may be required to avoid confusion for the patient or donor, the physician, or the nursing staff. In the age of genomics, with the potential for future health care and treatment options based on genetic polymorphisms, RHD polymorphisms that result in altered D antigen expression should be part of a patient's medical history.
Lastly, since there are no serologic reagents that detect Del RBCs, these will go undetected, type as D-negative, and be labeled D-negative as donors. Whether it is important to use genetic screening to eliminate donors with Del red cells from the D-negative donor pool has been debated. 36-38 Although Del RBCs have stimulated anti-D in two cases39,40, more data are needed to assess the potential risk in communities with large Asian donor populations. Of relevance, in approximately 10-13% of cases in Asia, D-negative individuals have been transfused with Del blood.
D typing discrepancies
Multiple factors can complicate the determination of the D status. These include the different methods used in various laboratories, the different monoclonal antibodies in FDA-licensed reagents that can react differently with variant D antigens, and the large number of different RHD genes, which can affect both the level of expression and, potentially, the structure of the molecule and D-epitopes.
D epitopes expressed on Rhce proteins (DHAR, ceCF, ceRT, ceSL)
The discovery that some Rhce proteins carry D-specific amino acids or express a D-like epitope that reacts with some monoclonal anti-D reagents further complicates serologic determination of D status. Two examples, DHAR, found in individuals of German ancestry, and Crawford, ceCF , found in African-Americans, are often responsible for serologic D typing discrepancies in the U.S. 41. These two are notable because of their strong reactivity (3+/4+) with some FDA-licensed monoclonal reagents and complete lack of reactivity with others.
Less dramatic are discrepancies caused by amino acid changes that mimic a D-epitope structure (epD6) that cause RBCs to be weakly reactive with some monoclonal anti-D, designated ceRT and ceSL. 42,43
In conclusion, because all of these lack RhD, they can readily be sensitized44(and our observations) if not appropriately classified as D-negative.
RhCE
Altered RHce genes are prevalent in Blacks and mixed ethnic backgrounds. Individuals with these variant alleles type as e-positive, but they often make alloantibodies with e-like specificities. Many altered RHce genes have now been characterized (Fig. 4C). The amino acid changes from conventional RHce are shown, and most carry a Tryptophan.. Cysteine at amino acid position 16 (W16C) encoded in exon 1, as well as additional changes, primarily in exon 5. Individuals homozygous for these alleles lack high incidence Rh antigens (hrB and hrS). The ceS alleles are associated with RBCs that are hrB-, and numerous alleles (ceMO, ceEK, etc) are associated with RBCs that are hrS- (Fig. 4C). Importantly, because of the multiple and varied changes responsible for hrS- and hrB- phenotypes, the antibodies produced by these individuals are not all compatible with RBCs from the other variants.
As an additional complication, these variant RHce are often linked to the D-negative hybrid RHD-CE-D (Fig. 4B) that encodes an altered C antigen, or are linked to a variant RHD (DIII, DAU, DAR etc.). Patients with these alleles are at risk of producing alloantie, −C, and also anti-D, in spite of having RBCs that are positive for these Rh antigens by serologic typing. This is not a rare occurrence; for example, the hybrid RHD-CE-D allele encoding altered C has a frequency of 25% in African-American Blacks.45 Genetic testing for RH can now be used to identify patients homozygous for these altered alleles. It is very difficult to find compatible donors for these patients after they become alloimmunized, and the challenge is to develop a registry of donors that are genotyped for these variants.
Transfusion Management in sickle cell disease (SCD)
Transfusion of patients with sickle cell disease (SCD) represents a significant challenge in clinical transfusion medicine. SCD may be the single disease for which transfusion therapy may increase in the next decade as a result of the stroke prevention trial in sickle cell anemia (STOP). This clinical trial was halted before its scheduled closure because of the significant benefit of chronic red cell transfusion in reducing the risk of stroke.46 STOP II further confirmed the markedly lower risk of stroke in participants receiving blood transfusions. 47 Nevertheless, serious complications of chronic transfusion include iron overload and alloimmunization, and these risks must be considered in medical decision-making regarding treatment. The recent availability of oral iron chelation agents is predicted to make transfusion a more acceptable treatment option 48, and genetic testing for RH has the potential to reduce alloimmunization.
The approach to management of alloimmunization in SCD varies, and although the goal is to provide blood with maximal survival, how best to accomplish this has been the subject of debate.49,50 Some programs aim to prevent or reduce the risk and incidence of alloantibody production by transfusing RBCs that are phenotype-matched, i.e. antigen-matched for D, C, E and Kell. Less often, Duffy (Fya/b) and Kidd (Jka/b) blood groups are also included. In addition to antigen-matching, some programs actively recruit and supply RBCs from African-American donors to SCD patients whenever possible. These approaches clearly reduce the risk and incidence of alloantibody production. Others provide antigen-negative blood only after a patient has made an antibody, arguing that it is logistically difficult and expensive to provide these units prophylactically, and not all patients will become sensitized and require antigen-matched products. Both approaches have strong proponents, and the community resources and donor center support are often the deciding factor in implementation of a specific approach in the absence of consensus.
Knowledge of RH gene variation sheds light on immunization in SCD patients. Although transfusion of antigen-matched units reduces the incidence of alloantibody production, some SCD patients still become sensitized. Many of the antibodies present as autoantibodies, and most represent complex, high incidence specificities within the Rh system. The prevalence in the sickle cell population of RH alleles that encode variant or altered e, C, or D antigens explains why these patients become immunized despite conventional antigen-matching, and why these incompatibilities are complex to resolve and compatible blood is difficult to obtain. Importantly, RH genotyping can identify SCD patients who are homozygous for variant alleles and at risk for production of “apparent auto” and alloantibodies to high incidence Rh antigens. RH genotyping of SCD patients, partnered with RH genotyping of donors, would have a positive impact to reduce alloimmunization in SCD and would optimize the use of minority donations, as not all SCD patients require blood from minority donors.
Rh glycoproteins (RhAG, RhBG, RhCG)
The Rh blood group proteins are well known because of their importance in blood transfusion. However, the mammalian family of Rh proteins has been expanded with the discovery of Rh-associated glycoprotein, RhAG, in erythrocytes, and the related proteins, RhBG and RhCG, in other tissues. Erythrocyte RhAG is not polymorphic, and while it is not associated with any blood group antigen, it is important for targeting RhCE and RhD to the membrane, as mutations in RHAG are responsible for loss of Rh antigen expression (Rhnull).51,52
Protein sequences with similarities to the mammalian Rh proteins were first found in C. elegans, and these homologs, in turn, showed similarity to the ammonium transporters from bacteria, yeast (MEP), and plants (AMT).53 This provided the first testable hypothesis for the function for these proteins. The relationship of the Rh-glycoproteins to the AMT/MEP ammonium transporters from these other organisms has now been substantiated by functional transport data 9,54-57 and structural modeling 10,11,58. The Rh proteins reveal the power of comparative genomics and proteomics, in which sequence analysis and homology modeling can give important insight into mammalian protein function.
The non-erythroid Rh glycoproteins, RhBG and RhCG, are present in the kidney, liver, brain, and skin12,13 where ammonia production and elimination occur. In the kidney collecting segment and collecting duct, RhBG and RhCG are found on the basolateral and apical membranes, respectively, of the intercalated cells59 where they mediate transepithelial movement of ammonium from the interstitium to the lumen. 60,61 In the liver, RhBG is found on the basolateral membrane of perivenous hepatocytes, where it may function in ammonia uptake. RhCG is also present in bile duct epithelial cells, where it is positioned to contribute to ammonia secretion into the bile fluid.14
The mechanism of ammonia transport by Rh-glycoproteins is an active area of investigation. Expression of Rh-glycoproteins in heterologous systems indicated that mammalian transport is an electroneutral process that is driven by the NH4+ concentration and the transmembrane H+ gradient.9,55,56 Importantly, this mechanism effectively exchanges NH4+ for H+ in a process that results in transport of net NH3. Functional studies of the kidney, liver, and brain Rh homologs, along with the erythrocyte RhAG/Rh proteins, promises to lead to development of a unifying hypothesis of ammonia transport in mammals by the Rh family of proteins.
RhCE and RhD
The function of the more recently evolved erythrocyte blood group proteins, RhCE and RhD, has not yet been determined. When expressed in heterologous systems, they do not directly transport ammonia.62 Importantly, RhCE/D lack the highly conserved Histidine residues located in the membrane pore that are critical for transport.11,62 An attractive alternative hypothesis is that the RhCE/D may mediate movement of the uncharged molecule CO2, however, this remains to be determined. Evidence that RhCE/D may be evolving a new function in the RBC membrane comes from DIVERGE analysis (our observations) and evidence from others 63,64 that indicate the RhCE/D proteins are rapidly evolving, suggesting their function may be changing.
A structural role for the erythrocyte Rh proteins is suggested from the RBC shape defects, fragility, and the resulting anemia seen in patients with Rhnull disease.4,65 Rh/RhAG are part of a membrane protein complex that includes CD47, also known as integrin-associated protein (IAP), glycophorin B, and LW (ICAM-4).66,67 Band 3 may also be associated with the complex. 68 The Rhnull defect suggests the presence of a membrane-cytoskeletal attachment site in RBC mediated by Rh, RhAG, or a member of the Rh complex. Recent studies reveal that the Rh complex is linked to the membrane skeleton through CD47-protein 4.2 interactions 69, and through a novel Rh/RhAG-ankyrin cytoskeleton connection.7 RBC membrane protein-cytoskeleton and protein-protein interactions are an active area of investigation. Future studies are needed to determine the protein-protein associations and the dynamics of the assembly of the Rh-membrane complex to understand the Rhnull defect and to determine the functional role of the RhCE/D proteins.
Summary
The genetic basis of the Rh blood group proteins has been intensely investigated in the past decade, and the polymorphisms responsible for most of the antigens have now been determined. Presently, routine testing for RH is hampered because of the large number of Rh polymorphisms, as over one hundred RHD and forty-two RHCE gene variants have been reported 30, and additional variants are still being discovered. Currently, RH genetic testing is a powerful adjunct to serologic methods. It is being used to determine RHD zygosity, to type patients who are multiply transfused, to resolve D typing discrepancies, and to identify compatible donors for patients with antibodies to high incidence Rh antigens, primarily in SCD patients. The challenge is to develop automated platforms that sample numerous regions of the RH genes to detect the many hybrid genes, and to develop algorithms for unequivocal interpretation.
Ideally, tests to determine D status would distinguish those with RBCs that lack, or have altered, D-epitopes and are at risk of sensitization to conventional D from those with reduce expression levels of D who are not at risk of producing anti-D. Unfortunately, serologic reagents cannot discriminate between these RBCs. These limitations suggest that genetic testing to determine D status may be commonplace in the future.
Understanding of RH gene variation can now be applied to more accurately assess alloimmunization risk for SCD patients. The diversity of RH genes in African-Americans often underlies development of allo- and auto-antibodies in this ethnic group, despite implementation of antigen- or phenotype-matching for transfusion. This suggests that identification of individuals homozygous for variant RH genes and at risk of alloimmunization, will improve transfusion management for these patients, and much more efficiently utilize minority donors.
The relationship of Rh and Rh-glycoproteins to the AMT/MEP ammonium transporters from bacteria, yeast, and plants has now been substantiated by functional transport data and structural modeling. The Rh proteins reveal the power of comparative genomics and proteomics, in which sequence analysis and homology modeling can give important insight into mammalian protein function.
The structural and physiologic role of Rh/RhAG in the RBCs remains to be more firmly established, but a role for the Rh complex in ammonia sequestration in RBCs, at the same time contributing to membrane shape and flexibility, can be imagined. The uncharged molecule NH3 is very toxic to brain cells and mitochondria,70 and the Rh/RhAG complex may play a role in keeping the total blood ammonia level low by trapping ammonium inside the RBC. In support, plasma total ammonia levels are low (0.1-0.2 mM), but total erythrocyte ammonia levels are three times greater. 71
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Levine P, Burnham L, Katzin WM, Vogel P. The role of isoimmunization in the pathogenesis of erythroblastosis fetalis. Am J Obstet Gynecol. 1941;42:925–937. [Google Scholar]
- 2.Mollison PL, Hughes-Jones NC, Lindsay M, Wessely J. Suppression of primary RH immunization by passively-administered antibody. Experiments in volunteers. Vox Sang. 1969;16:421–439. [PubMed] [Google Scholar]
- 3.Freda V, Gorman J, Pollack W. Rh factor: Prevention of isoimmunization and clinical trials in mothers. Science. 1966;151:828–830. doi: 10.1126/science.151.3712.828. [DOI] [PubMed] [Google Scholar]
- 4.Ballas S, Clark MR, Mohandas N, et al. Red cell membranes and cation deficiency in Rhnull syndrome. Blood. 1984;63:1046–1055. [PubMed] [Google Scholar]
- 5.Bruce LJ, Ghosh S, King MJ, et al. Absence of CD47 in protein 4.2-deficient hereditary spherocytosis in man: an interaction between the Rh complex and the band 3 complex. Blood. 2002;100:1878–1885. doi: 10.1182/blood-2002-03-0706. [DOI] [PubMed] [Google Scholar]
- 6.Dahl KN, Parthasarathy R, Westhoff CM, Layton DM, Discher DE. Protein 4.2 is critical to CD47-membrane skeleton attachment in human red cells. Blood. 2004;103:1131–1136. doi: 10.1182/blood-2003-04-1331. [DOI] [PubMed] [Google Scholar]
- 7.Nicolas V, Le Van Kim C, Gane P, et al. Rh-RhAG/ankyrin-R, a new interaction site between the membrane bilayer and the red cell skeleton, is impaired by Rh(null)-associated mutation. J Biol Chem. 2003;278:25526–25533. doi: 10.1074/jbc.M302816200. [DOI] [PubMed] [Google Scholar]
- 8.Marini AM, Matassi G, Raynal V, Andre B, Cartron JP, Cherif-Zahar B. The human Rhesus-associated RhAG protein and a kidney homologue promote ammonium transport in yeast. Nat Genet. 2000;26:341–344. doi: 10.1038/81656. [DOI] [PubMed] [Google Scholar]
- 9.Westhoff CM, Ferreri-Jacobia M, Mak D-OD, Foskett JK. Identification of the erythrocyte Rh blood group glycoprotein as a mammalian ammonium transporter. J Biol Chem. 2002;277:12499–12502. doi: 10.1074/jbc.C200060200. [DOI] [PubMed] [Google Scholar]
- 10.Khademi S, O'Connell J, 3rd, Remis J, Robles-Colmenares Y, Miercke LJ, Stroud RM. Mechanism of ammonia transport by Amt/MEP/Rh: structure of AmtB at 1.35 A. Science. 2004;305:1587–1594. doi: 10.1126/science.1101952. [DOI] [PubMed] [Google Scholar]
- 11.Conroy MJ, Bullough PA, Merrick M, Avent ND. Modelling the human rhesus proteins: implications for structure and function. Br J Haematol. 2005;131:543–551. doi: 10.1111/j.1365-2141.2005.05786.x. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Chen Y, Mo R, et al. Characterization of human RhCG and mouse Rhcg as novel nonerythroid Rh glycoprotein homologues predominantly expressed in kidney and testis. J Biol Chem. 2000;275:25641–25651. doi: 10.1074/jbc.M003353200. [DOI] [PubMed] [Google Scholar]
- 13.Liu Z, Peng J, Mo R, Hui CC, Huang CH. Rh type B glycoprotein is a new member of the Rh superfamily and a putative ammonia transporter in mammals. J Biol Chem. 2001;276:1424–1433. doi: 10.1074/jbc.M007528200. [DOI] [PubMed] [Google Scholar]
- 14.Weiner ID, Verlander JW. Renal and hepatic expression of the ammonium transporter proteins, Rh B Glycoprotein and Rh C Glycoprotein. Acta Physiol Scand. 2003;179:331–338. doi: 10.1046/j.0001-6772.2003.01210.x. [DOI] [PubMed] [Google Scholar]
- 15.Weiner ID, Miller RT, Verlander JW. Localization of the ammonium transporters, Rh B glycoprotein and Rh C glycoprotein, in the mouse liver. Gastroenterology. 2003;124:1432–1440. doi: 10.1016/s0016-5085(03)00277-4. [DOI] [PubMed] [Google Scholar]
- 16.Cherif-Zahar B, Mattei MG, Le Van Kim C, Bailly P, Cartron J-P, Colin Y. Localization of the human Rh blood group gene structure to chromosome 1p34.3-1p36.1 region by in situ hybridization. Hum Genet. 1991;86:398–400. doi: 10.1007/BF00201843. [DOI] [PubMed] [Google Scholar]
- 17.Arce MA, Thompson ES, Wagner S, Coyne KE, Ferdman BA, Lublin DM. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood. 1993;82:651–655. [PubMed] [Google Scholar]
- 18.Simsek S, de Jong CAM, Cuijpers HTM, et al. Sequence analysis of cDNA derived from reticulocyte mRNAs coding for Rh polypeptides and demonstration of E/e and C/c polymorphism. Vox Sang. 1994;67:203–209. doi: 10.1111/j.1423-0410.1994.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 19.Cherif-Zahar B, Bloy C, Le Van Kim C, et al. Molecular cloning and protein structure of a human blood group Rh polypeptide. Proc Natl Acad Sci USA. 1990;87:6243–6247. doi: 10.1073/pnas.87.16.6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wagner FF, Flegel WA. RHD gene deletion occurred in the Rhesus box. Blood. 2000;95:3662–3668. [PubMed] [Google Scholar]
- 21.Race RR, Sanger R. Blood groups in man. ed Sixth Blackwell; Oxford, England: 1975. [Google Scholar]
- 22.Colin Y, Cherif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 23.Singleton BK, Green CA, Avent ND, et al. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in Africans with the Rh D-negative blood group phenotype. Blood. 2000;95:12–18. [PubMed] [Google Scholar]
- 24.Chiu RW, Murphy MF, Fidler C, Zee BC, Wainscoat JS, Lo YM. Determination of RhD zygosity: comparison of a double amplification refractory mutation system approach and a multiplex real-time quantitative PCR approach. Clin Chem. 2001;47:667–672. [PubMed] [Google Scholar]
- 25.Lo YM, Hjelm NM, Fidler C, et al. Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma. N Engl J Med. 1998;339:1734–1738. doi: 10.1056/NEJM199812103392402. [DOI] [PubMed] [Google Scholar]
- 26.Rijnders RJ, Van Der Luijt RB, Peters ED, et al. Earliest gestational age for fetal sexing in cell-free maternal plasma. Prenat Diagn. 2003;23:1042–1044. doi: 10.1002/pd.750. [DOI] [PubMed] [Google Scholar]
- 27.Bianchi DW, Avent ND, Costa JM, van der Schoot CE. Noninvasive prenatal diagnosis of fetal Rhesus D: ready for Prime(r) Time. Obstet Gynecol. 2005;106:841–844. doi: 10.1097/01.AOG.0000179477.59385.93. [DOI] [PubMed] [Google Scholar]
- 28.Daniels G. Human Blood Groups. ed Second Blackwell Science; Cambridge, Mass: 2002. [Google Scholar]
- 29.Wagner FF, Gassner C, Muller TH, Schonitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood. 1999;93:385–393. [PubMed] [Google Scholar]
- 30.Blumenfeld OO, Patnaik SK. Allelic genes of blood group antigens: a source of human mutations and cSNPs documented in the Blood Group Antigen Gene Mutation Database. Hum Mutat. 2004;23:8–16. doi: 10.1002/humu.10296. [DOI] [PubMed] [Google Scholar]
- 31.Wagner FF, Flegel WA. doi: 10.1159/000366176. The Rhesus Site at http://www.uniulm.de/∼fwagner/RH/RB/weakD.htm. [DOI] [PMC free article] [PubMed]
- 32.Shao CP, Maas JH, Su YQ, Kohler M, Legler TJ. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang. 2002;83:156–161. doi: 10.1046/j.1423-0410.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 33.Sun CF, Chou CS, Lai NC, Wang WT. RHD gene polymorphisms among RhD-negative Chinese in Taiwan. Vox Sang. 1998;75:52–57. [PubMed] [Google Scholar]
- 34.Beck ML, Harding J. Incidence of D category VI among Du donors in the USA. Transfusion. 1991;31(S):25. [Google Scholar]
- 35.Lacey PA, Caskey CR, Werner DJ, Moulds JJ. Fatal hemolytic disease of a newborn due to anti-D in an Rh-positive Du variant mother. Transfusion. 1983;23:91–94. doi: 10.1046/j.1537-2995.1983.23283172867.x. [DOI] [PubMed] [Google Scholar]
- 36.Krumpel B. Are weak D RBCs really immunogenic. Transfusion. 2006;46:1061–1066. doi: 10.1111/j.1537-2995.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 37.Garratty G. Do we need to be more concerned about weak D antigens? Transfusion. 2005;45:1547–1551. doi: 10.1111/j.1537-2995.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 38.Flegel WA. Homing in on D antigen immunogenicity. Transfusion. 2005;45:466–468. doi: 10.1111/j.0041-1132.2005.05001.x. [DOI] [PubMed] [Google Scholar]
- 39.Wagner T, Kormoczi GF, Buchta C, et al. Anti-D immunization by DEL red blood cells. Transfusion. 2005;45:520–526. doi: 10.1111/j.0041-1132.2005.04256.x. [DOI] [PubMed] [Google Scholar]
- 40.Yasuda H, Ohto H, Sakuma S, Ishikawa Y. Secondary anti-D immunization by Del red blood cells. Transfusion. 2005;45:1581–1584. doi: 10.1111/j.1537-2995.2005.00579.x. [DOI] [PubMed] [Google Scholar]
- 41.Westhoff CM. Review: the Rh blood group D antigen… dominant, diverse, and difficult. Immunohematol. 2005;21:155–163. [PubMed] [Google Scholar]
- 42.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion. 2003;43:1248–1254. doi: 10.1046/j.1537-2995.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 43.Chen Q, Hustinx H, Flegel WA. The RHCE allele ceSL: the second example for D antigen expression without D-specific amino acids. Transfusion. 2006;46:766–772. doi: 10.1111/j.1537-2995.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- 44.Beckers EA, Porcelijn L, Ligthart P, et al. The RoHar antigenic complex is associated with a limited number of D epitopes and alloanti-D production: a study of three unrelated persons and their families. Transfusion. 1996;36:104–108. doi: 10.1046/j.1537-2995.1996.36296181919.x. [DOI] [PubMed] [Google Scholar]
- 45.Daniels GL, Faas BH, Green CA, et al. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion. 1998;38:951–958. doi: 10.1046/j.1537-2995.1998.381098440860.x. [DOI] [PubMed] [Google Scholar]
- 46.Adams RJ, Brambilla DJ, Granger S, et al. Stroke and conversion to high risk in children screened with transcranial Doppler ultrasound during the STOP study. Blood. 2004;103:3689–3694. doi: 10.1182/blood-2003-08-2733. [DOI] [PubMed] [Google Scholar]
- 47.Lee MT, Piomelli S, Granger S, et al. Stroke Prevention Trial in Sickle Cell Anemia (STOP): Extended Follow-up and Final Results. Blood. 2006 doi: 10.1182/blood-2005-10-009506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vichinsky E, Fischer R, Fung E. A randomized, controlled phase II trial in sickle cell disease patients with chronic iron overload demonstrates that once-daily oral iron chelator deferasirox (Exjade, ICL670) is well tolerated and reduces iron burden [abstract] Blood. 2005;106:95a. [Google Scholar]
- 49.Ness PM. To match or not to match: the question for chronically transfused patients with sickle cell anemia. Transfusion. 1994;34:558–560. doi: 10.1046/j.1537-2995.1994.34794330007.x. [DOI] [PubMed] [Google Scholar]
- 50.Wayne AS, Kevy SV, Nathan DG. Transfusion management of sickle cell disease. Blood. 1993;81:1109–1123. [PubMed] [Google Scholar]
- 51.Cherif-Zahar B, Raynal V, Gane P, et al. Candidate gene acting as a suppressor of the RH locus in most cases of Rh-deficiency. Nature Genet. 1996;12:168–173. doi: 10.1038/ng0296-168. [DOI] [PubMed] [Google Scholar]
- 52.Huang C-H. Molecular insights into the Rh protein family and associated antigens. Curr Opin Hematol. 1997;4:94–103. doi: 10.1097/00062752-199704020-00004. [DOI] [PubMed] [Google Scholar]
- 53.Marini A-M, Urrestarazu A, Beauwens R, Andre B. The Rh (Rhesus) blood group polypeptides are related to NH4+ transporters. TIBS. 1997;22:460–461. doi: 10.1016/s0968-0004(97)01132-8. [DOI] [PubMed] [Google Scholar]
- 54.Westhoff CM, Siegel DL, Burd CG, Foskett JK. Mechanism of genetic complementation of ammonium transport in yeast by human erythrocyte Rh-associated glycoprotein. J Biol Chem. 2004;279:17443–17448. doi: 10.1074/jbc.M311853200. [DOI] [PubMed] [Google Scholar]
- 55.Mak DO, Dang B, Weiner ID, Foskett JK, Westhoff CM. Characterization of ammonia transport by the kidney Rh glycoproteins RhBG and RhCG. Am J Physiol Renal Physiol. 2006;290:F297–305. doi: 10.1152/ajprenal.00147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ludewig U. Electroneutral ammonium transport by basolateral rhesus B glycoprotein. J Physiol. 2004;559:751–759. doi: 10.1113/jphysiol.2004.067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ripoche P, Bertrand O, Gane P, Birkenmeier C, Colin Y, Cartron JP. Human Rhesus-associated glycoprotein mediates facilitated transport of NH(3) into red blood cells. Proc Natl Acad Sci U S A. 2004;101:17222–17227. doi: 10.1073/pnas.0403704101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng L, Kostrewa D, Berneche S, Winkler FK, Li XD. The mechanism of ammonia transport based on the crystal structure of AmtB of Escherichia coli. Proc Natl Acad Sci U S A. 2004;101:17090–17095. doi: 10.1073/pnas.0406475101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verlander JW, Miller RT, Frank AE, Royaux IE, Kim YH, Weiner ID. Localization of the ammonium transporter proteins RhBG and RhCG in mouse kidney. Am J Physiol. 2003;284:F323–337. doi: 10.1152/ajprenal.00050.2002. [DOI] [PubMed] [Google Scholar]
- 60.Handlogten ME, Hong SP, Westhoff CM, Weiner ID. Basolateral ammonium transport by the mouse inner medullary collecting duct, mIMCD-3, cell. Am J Physiol Renal Physiol. 2004 doi: 10.1152/ajprenal.00363.2003. [DOI] [PubMed] [Google Scholar]
- 61.Handlogten ME, Hong SP, Westhoff CM, Weiner ID. Apical ammonia transport by the mouse inner medullary collecting duct cell (mIMCD-3) Am J Physiol Renal Physiol. 2005;289:F347–358. doi: 10.1152/ajprenal.00253.2004. [DOI] [PubMed] [Google Scholar]
- 62.Westhoff CM, Wylie DE. Transport characteristics of mammalian Rh and Rh glycoproteins expressed in heterologous systems. Transfus Clin Biol. 2006;13:132–138. doi: 10.1016/j.tracli.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 63.Matassi G, Cherif-Zahar B, Pesole G, Raynal V, Cartron JP. The members of the RH gene family (RH50 and RH30) followed different evolutionary pathways. J Mol Evol. 1999;48:151–159. doi: 10.1007/pl00006453. [DOI] [PubMed] [Google Scholar]
- 64.Kitano T, Sumiyama K, Shiroishi T, Saitou N. Conserved evolution of the Rh50 gene compared to its homologous Rh blood group gene. Biochem Biophy Res Comm. 1998;249:78–85. doi: 10.1006/bbrc.1998.9074. [DOI] [PubMed] [Google Scholar]
- 65.Sturgeon P. Hematological observations on the anemia associated with blood type Rhnull. Blood. 1970;36:310–320. [PubMed] [Google Scholar]
- 66.Huang CH, Liu PZ, Chen JG. Molecular biology and genetics of the Rh blood group system. Semin Hematol. 2000;37:150–165. doi: 10.1016/s0037-1963(00)90040-4. [DOI] [PubMed] [Google Scholar]
- 67.Avent ND, Reid ME. The Rh blood group system: a review. Blood. 2000;95:375–387. [PubMed] [Google Scholar]
- 68.Bruce LJ, Beckmann R, Ribeiro ML, et al. A band 3-based macrocomplex of integral and peripheral proteins in the RBC membrane. Blood. 2003;101:4180–4188. doi: 10.1182/blood-2002-09-2824. [DOI] [PubMed] [Google Scholar]
- 69.Dahl KN, Westhoff CM, Discher DE. Fractional attachment of CD47 (IAP) to the erythrocyte cytoskeleton and visual colocalization with Rh protein complexes. Blood. 2003;101:1194–1199. doi: 10.1182/blood-2002-04-1187. [DOI] [PubMed] [Google Scholar]
- 70.Kosenko E, Kaminsky YG, Felipo V, Minana MD, Grisolia S. Chronic hyperammonemia prevents changes in brain energy and ammonia metabolites induced by acute ammonium intoxication. Biochim Biophys Acta. 1993;1180:321–326. doi: 10.1016/0925-4439(93)90057-8. [DOI] [PubMed] [Google Scholar]
- 71.Huizenga JR, Tangerman A, Gips CH. Determination of ammonia in biological fluids. Ann Clin Biochem. 1994;31:529–543. doi: 10.1177/000456329403100602. [DOI] [PubMed] [Google Scholar]
- 72.Reid ME, Lomas-Francis C. The Blood Group Antigen Facts Book. ed Second Academic Press; San Diego, CA: 2004. [Google Scholar]