Abstract
Transfer-messenger RNA (tmRNA) plays a dual role as a tRNA and an mRNA in trans-translation, during which the ribosome replaces mRNA with tmRNA encoding the tag-peptide. These processes have been suggested to involve several tmRNA-binding proteins, including SmpB and ribosomal protein S1. To investigate the molecular mechanism of trans-translation, we developed in vitro systems using purified ribosome, elongation factors, tmRNA and SmpB from Thermus thermophilus. A stalled ribosome in complex with polyphenylalanyl-tRNAPhe was prepared as a target of tmRNA. A peptidyl transfer reaction from polyphenylalanyl-tRNAPhe to alanyl-tmRNA was observed in an SmpB-dependent manner. The next peptidyl transfer to aminoacyl-tRNA occurred specifically to the putative resume codon for the tag-peptide, which was confirmed by introducing a mutation in the codon. Thus, the in vitro systems developed in this study are useful to investigate the early steps of trans-translation. Using these in vitro systems, we investigated the function of ribosomal protein S1, which has been believed to play a role in trans-translation. Although T. thermophilus S1 tightly bound to tmRNA, as in the case of Escherichia coli S1, it had little or no effect on the early steps of trans-translation.
Keywords: tmRNA, trans-translation, S1, Thermus thermophilus
INTRODUCTION
Transfer-messenger RNA (tmRNA, also known as SsrA RNA or 10Sa RNA) is a bifunctional RNA widely distributed among eubacteria. tmRNA has a tRNA-like domain (TLD) and a peptide-coding region (mRNA domain). Both terminal regions form a tRNA-like structure (Komine et al. 1994; Ushida et al. 1994), while the middle part typically forms four pseudoknots (PK) and some helices (Felden et al. 1996; Williams and Bartel 1996). The 3′ end of tmRNA, like tRNA, is aminoacylated by alanyl-tRNA synthetase (AlaRS) (Komine et al. 1994; Ushida et al. 1994). tmRNA was later shown to act also as mRNA, to facilitate a noncanonical translation, trans-translation (Keiler et al. 1996; Himeno et al. 1997). It produces a chimera polypeptide from two different RNAs, a problematic mRNA and tmRNA with a message for degradation. Thus, tmRNA plays a role in rescuing the stalled ribosome and tagging a signal for degradation to the incomplete protein from a terminator-less mRNA (Muto et al. 2000; Withey and Friedman 2003).
tmRNA forms a complex with several protein factors, such as SmpB, elongation factor Tu (EF-Tu), and S1, in vivo and in vitro (Karzai et al. 1999; Wower et al. 2000, 2004; Barends et al. 2001; Karzai and Sauer 2001; Okada et al. 2004). SmpB binds TLD (Hanawa-Suetsugu et al. 2002; Gutmann et al. 2003; Nameki et al. 2005) to perform functions in the processes of trans-translation, recruitment of tmRNA to the stalled ribosome (Karzai et al. 1999; Hanawa-Suetsugu et al. 2002), enhancement of alanylation of tmRNA (Barends et al. 2001; Hanawa-Suetsugu et al. 2002; Shimizu and Ueda 2002), and protection of tmRNA from degradation in the cell (Hanawa-Suetsugu et al. 2002). EF-Tu binds Ala-tmRNA to protect it from deacylation (Rudinger-Thirion et al. 1999; Barends et al. 2000, 2001; Hanawa-Suetsugu et al. 2001; Zvereva et al. 2001; Stepanov and Nyborg 2003; Valle et al. 2003).
S1 is a component of the small subunit of the ribosome (Subramanian 1983; Shiryaev et al. 2002). Due to the weak association with the ribosome, some of S1 seems to be apart from ribosomes in the cytoplasm. S1 specifically binds to the uridine-rich region upstream of the SD sequence (Boni et al. 1991; Zhang and Deutscher 1992) or the 5′-untranslated region of an mRNA without an SD sequence (Tzareva et al. 1994) and possesses a nucleic acid unwinding ability (Thomas and Szer 1982). It ensures the translation of mRNAs bearing no or weak SD sequences (Roberts and Rabinowitz 1989; Tzareva et al. 1994) of mRNAs that bear long SD sequences (Komarova et al. 2002) or of most natural mRNAs in vivo (Sorensen et al. 1998). The low-G+C group of Gram-positive bacteria lacks S1 but has a strong SD sequence in each mRNA instead (Isono and Isono 1976; Farwell et al. 1992). Thus, S1 is thought to help the association between mRNA and the ribosome besides the SD and anti-SD interaction, which is significant for efficient translation of mRNAs, especially those with a weak SD sequence (Boni et al. 1991; Zhang and Deutscher 1992; Tzareva et al. 1994; Tedin et al. 1997; Sengupta et al. 2001; Komarova et al. 2002).
Since S1 cross-links to the upstream region of the tag-encoding sequence of tmRNA that is significant for trans-translation, the interaction between S1 and tmRNA is presumed to be critical for initiation of tag-translation (Williams et al. 1999; Wower et al. 2000; Lee et al. 2001). Wower et al. (2000) have shown that Escherichia coli S1 enhances the binding of tmRNA to the ribosome in the absence of SmpB. E. coli S1 induces a conformational change in tmRNA, making the nucleotides around the resume codon accessible to a solvent (Bordeau and Felden 2002). A conformational change in tmRNA by S1 has also been suggested by a cryo-electron microscopic study on a Thermus thermophilus Ala-tmRNA•SmpB•EF-Tu•GDP•kirromycin•ribosome complex (Valle et al. 2003). Collectively, these findings raise the possibility that S1 binds tmRNA, guides it to the ribosome, anchors the upstream region of the resume codon on the ribosome, and destabilizes the surrounding structured region to precisely set the resume codon at the A site. On the other hand, some studies have argued against the significance of S1 for trans-translation. S1 is absent in the low-G+C group of Gram-positive bacteria, and tmRNA from Bacillus subtilis that lacks S1 can facilitate trans-translation in E. coli that has S1 (Ito et al. 2002). Overproduction of the N-terminal fragment inhibits general translation but not trans-translation in E. coli, also implying the insignificance of S1 for trans-translation (McGinness and Sauer 2004). S1 binds to the junction of the head, platform, and main body of the 30S subunit according to a cryo-electron microscopic study (Sengupta et al. 2001), which is reasonable for its function in binding and unwinding the upstream region of the initiation point of structured mRNA to initiate translation. However, upon tmRNA binding to the ribosome, the S1 binding region on tmRNA is apparently far from the position of S1 on the 30S subunit, leading to an idea that even if S1 has a role in trans-translation, that role may be independent of its role in canonical translation.
In the present study, we developed in vitro trans-translation systems composed of factors from T. thermophilus to study the role of S1 in trans-translation. Many biochemical and genetic studies on biological events in bacteria have been performed using E. coli, although structural studies have often been carried out using factors from thermophiles. T. thermophilus is a thermophile in which the structures of factors required for trans-translation, such as the ribosome (Yusupova et al. 1991; Carter et al. 2001) and SmpB (Someya et al. 2003), have been extensively studied. However, there have been only a few biochemical and no genetic studies (Yusupov et al. 1991; Yusupova et al. 1991; Uzawa et al. 2002; Stepanov and Nyborg 2003). In contrast, several kinds of cell-free trans-translation systems have been developed in E. coli (Himeno et al. 1997; Shimizu and Ueda 2002; Asano et al. 2005), although with few structural studies. Thus, T. thermophilus cell-free trans-translation systems might be expected to link structural studies to biochemical and genetic studies on trans-translation. The in vitro systems developed here can assess the early steps of trans-translation, which revealed that T. thermophilus S1 has little or no effect on these steps.
RESULTS AND DISCUSSION
In vitro poly (U)-dependent poly(Phe) synthesis
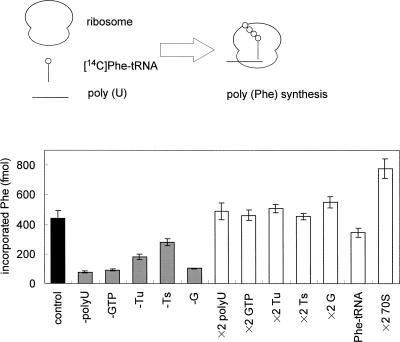
The ribosome translates poly (U) into poly (Phe) and probably stalls at the very 3′ end of the poly (U). This type of stalled ribosome has already been reported to be a target of tmRNA for trans-translation in E. coli (Himeno et al. 1997; Shimizu and Ueda 2002). A highly purified S1-free ribosome from T. thermophilus was incubated with poly (U), [14C]Phe-tRNAPhe, EF-Tu, EF-Ts, EF-G, and GTP. As expected, poly (U)-dependent [14C]phenylalanine incorporation was observed. Poly (U), GTP, and EF-G were all essential for the reaction (Fig. 1, -poly (U), -GTP, and -G). The efficiency decreased to 28% and 55% by exclusion of EF-Tu and EF-Ts, respectively, from the reaction mixture (Fig. 1, -Tu and -Ts). Considering that trans-translation can occur, but inefficiently, on the ribosomes stalled in the middle of mRNA (Ivanova et al. 2004; Asano et al. 2005), only a limited amount of ribosomes was added in the present in vitro poly (Phe) synthesis system (Fig. 1, ×2 70S) to reduce the ribosomes unexpectedly stalled in the middle of poly (U) relative to those expectedly stalled at the 3′ end of poly (U). The reaction mixture of poly (U)-dependent poly (Phe) synthesis was filtered with a 100-kDa cutoff membrane to remove unfavorable factors such as poly (U) and Phe-tRNAPhe and was concentrated to a half volume to enhance the sensitivity of the trans-translation assay system. It was used as the stalled ribosome for trans-translation.
FIGURE 1.
Poly (U)-directed poly (Phe) synthesis. Ribosome, poly (U), and [14C]Phe-tRNAPhe were incubated with elongation factors for 10 min at 65°C. (Gray bar) phenylalanine incorporation in the absence of any one of poly (U), GTP, EF-Tu, EF-Ts, and EF-G (-poly [U], -GTP, -Tu, -Ts and -G, respectively); (white bar) phenylalanine incorporation when any one of poly (U), GTP, EF-Tu, EF-Ts, EF-G, [14C]Phe-tRNAPhe, and ribosome was doubled (×2 poly [U], ×2 GTP, ×2 Tu, ×2 Ts, ×2 G, ×2 Phe-tRNAPhe, and ×2 ribosome, respectively). The average results of three independent experiments are shown with standard errors.
In vitro trans-translation systems
T. thermophilus tmRNA and SmpB were expressed in E. coli and purified. As expected, T. thermophilus tmRNA was aminoacylated with alanine by T. thermophilus AlaRS. Furthermore, T. thermophilus SmpB enhanced the aminoacylation of tmRNA by AlaRS, as in the case of E. coli SmpB (Barends et al. 2001; Hanawa-Suetsugu et al. 2002; Shimizu and Ueda 2002). Four micromolar SmpB enhanced the initial rate of aminoacylation of tmRNA by AlaRS from T. thermophilus 12-fold. The maximum plateau level was 3% in the absence of SmpB, while it was 50% in the presence of 4 μM SmpB. The binding of SmpB to tmRNA was confirmed by gel mobility shift assay (Nameki et al. 2005).
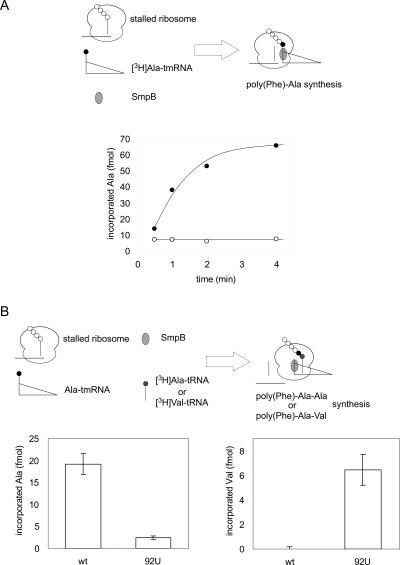
The stalled ribosome was incubated with Ala-tmRNA, SmpB, EF-Tu, and GTP to observe the reaction of peptidyl transfer from poly (Phe)-tRNAPhe to Ala-tmRNA, resulting in incorporation of Ala into poly (Phe)-Ala. This step is referred to as “trans-transfer” throughout this report. When [3H]Ala-tmRNA was incubated with the stalled ribosome, alanine incorporation was observed only in the presence of SmpB (Fig. 2A). This result indicates that the trans-transfer actually occurred as expected in consideration of the indispensability of SmpB in E. coli (Asano et al. 2005).
FIGURE 2.
In vitro trans-translation systems. (A) Trans-transfer reaction. Stalled ribosomes with poly (Phe)-tRNAPhe were incubated with [3H]Ala-tmRNA in the presence (closed circles) or absence (open circles) of 0.5 μM SmpB, and the incorporation of [3H]Ala into poly (Phe)-[3H]Ala was measured. (B) Resume codon decoding. Similarly, stalled ribosomes were incubated with unlabeled Ala-tmRNAwt or Ala-tmRNA92U in combination with [3H]Ala-tRNAAla (left panels) or [3H]Val-tRNAVal (right panels). Error bars show the standard error of the average of three independent experiments.
After the trans-transfer reaction and presumably translocation of tmRNA from the A site to the P site, the first codon of the mRNA domain should be set at the A site. The resume codon has been predicted to be G91C92C93, an alanine codon, for T. thermophilus tmRNA from a sequence comparison (Gueneau de Novoa and Williams 2004). In order to monitor the decoding of the resume codon, the stalled ribosome was incubated with unlabeled Ala-tmRNA and [3H]Ala-tRNAAla together. This reaction was expected to result in the production of poly (Phe)-Ala-[3H]Ala. To ascertain the validity of this system, a mutant tmRNA (tmRNA92U), in which the putative resume codon was altered from GCC to GUC encoding a valine, was constructed, and it was used for the resume codon decoding assay in combination with [3H]Ala-tRNAAla or [3H]Val-tRNAVal.
First, [3H]Ala-tRNAAla together with Ala-tmRNAwt or Ala-tmRNA92U was added to the stalled ribosomes (Fig. 2B, left panel). [3H]Ala was incorporated into poly (Phe)-Ala-[3H]Ala when wild-type tmRNA was used, whereas no incorporation of [3H]Ala was observed when tmRNA92U was used. Next, [3H]Val-tRNAVal was added instead of [3H]Ala-tRNAAla (Fig. 2B, right panel). In this case, [3H]Val was incorporated into poly (Phe)-Ala-[3H]Val only when tmRNA92U was used. Taken together, the results show that the resume codon decoding occurred only when the predicted resume codon matched the anticodon of the aminoacyl-tRNA used. These results confirmed that our in vitro system is valid for monitoring the initial steps of trans-translation, including the two steps (1) binding of Ala-tmRNA to the ribosome to undergo a peptide transfer reaction, and (2) setting of the resume codon to the A site to be translated. These results also indicate that the resume codon of T. thermophilus tmRNA is G91C92C93 (an alanine codon), and the amino acid sequence of the tag-peptide of T. thermophilus is therefore (A)ANTNYALAA, as predicted from a phylogenetic study (Gueneau de Novoa and Williams 2004).
Effect of S1 on translation and trans-translation
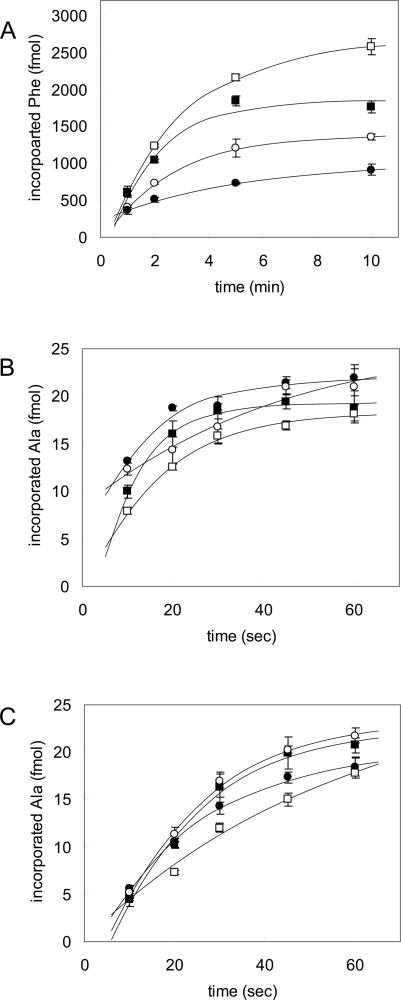
Almost all populations of the ribosomes used in the present study were free of S1, which was checked by SDS polyacrylamide gel electrophoresis. No evident band corresponding to S1 was detected in the ribosome fraction (data not shown). It has been reported that poly (U)-dependent poly (Phe) synthesis occurred in E. coli even in the absence of S1 and that the addition of S1 to the reaction promoted the incorporation of phenylalanine (Van Dieijen et al. 1975; Yokota et al. 1977). Poly (Phe) synthesis without S1 was also observed in the present T. thermophilus system (Figs. 1, 3A). Exogenous addition of S1 to the reaction remarkably promoted both the initial rate and the plateau level of polyphenylalanine synthesis (Fig. 3A), and 1.2 μM S1 enhanced the rate by fourfold. This result indicates that S1 is functional in the in vitro translation system.
FIGURE 3.
Effect of S1 on translation and trans-translation in vitro. (A) Effect of S1 on poly (U)-dependent poly (Phe) synthesis was examined by performing the reaction with 0 (closed circles), 0.3 (open circles), 0.6 (closed triangles), and 1.2 (open triangles) μM S1. The reaction mixture including S1 was prepared on ice without GTP, and the reaction was started by the addition of GTP. (B) The effect of S1 on trans-transfer was examined in the system shown in Fig. 1A. [3H]Ala-tmRNA and SmpB with 0 (closed circles), 1.5 (open circles), 3.0 (closed triangles), or 6.0 (open triangles) μM S1 were added to the stalled ribosome and incubated (final concentration of S1 in the reaction mixture: 0, 0.3, 0.6, or 1.2 μM). The values of experiments without SmpB were subtracted as backgrounds. (C) The effect of S1 on the resume codon decoding was examined. Unlabeled Ala-tmRNA, [3H]Ala-tRNAAla, and 0 (closed circles), 1.5 (open circles), 3.0 (closed triangles), or 6.0 (open triangles) μM S1 were added to the stalled ribosome. The average results of at least three independent experiments are shown with standard errors.
E. coli S1 has been reported to bind tightly to tmRNA (Wower et al. 2000; Hanawa-Suetsugu et al. 2001). In this study, we examined the binding property of S1 from T. thermophilus to tmRNA by gel mobility shift assay (data not shown). Shifted bands were observed with increasing concentration of S1. The Kd value was estimated to be 5–10 nM from the free tmRNA bands. Thus, S1 from T. thermophilus binds tmRNA as tightly as that from E. coli, implying a significance of S1 for trans-translation. We reproducibly detected plural shifted bands with increase in the concentration of S1 as in the case of E. coli S1, which may be reflected by binding of multiple molecules of S1 to a single tmRNA (Bordeau and Felden 2002).
The effect of S1 on the trans-transfer step was examined by adding exogenous S1 to the Ala-tmRNA•SmpB complex prior to the addition of the complex to the stalled ribosomes free of S1. Since the concentration of S1 used was far higher than the apparent Kd value estimated from the gel mobility shift assay, Ala-tmRNA was expected to predominantly form a complex with S1. The reaction was started by adding a mixture containing [3H]Ala-tmRNA, SmpB, EF-Tu, GTP, and S1 to S1-free stalled ribosome. As already shown in Figure 2A, alanine incorporation was observed even in the absence of S1 (Fig. 3B, closed circles). Addition of S1 to the reaction showed no enhancement of trans-transfer, although a slight inhibition was observed at high concentration (Fig. 3B). The effect of the lower concentration of S1 was also examined, but no effect was observed (data not shown).
The effect of S1 on the setting of the resume codon at the A site to be decoded after trans-transfer was examined. It should be noted that because Ala-tmRNA and Ala-tRNAAla were added together to the stalled ribosome, the observed effect of S1 includes the effects not only on the resume codon decoding but also on trans-transfer. A mixture containing [3H]Ala-tRNAAla and a complex of unlabeled Ala-tmRNA, SmpB, and various concentrations of S1 was added to the stalled ribosome. Incorporation of [3H]Ala into a peptide was observed in the absence of S1 (Figs. 2B, 3B). S1 slightly inhibited rather than enhanced this reaction, which can be attributed to the effect of S1 on the trans-transfer step. No effect of S1 at its lower concentration was observed. These results strongly suggest that S1 is not involved in the step of the resume codon decoding.
The above results from the cell-free trans-translation systems demonstrate that T. thermophilus S1, even if it causes a structural change around the resume codon, does not contribute to the early steps of trans-translation involving tmRNA recognition by the ribosome, peptidyl-transfer from peptidyl-tRNA to Ala-tmRNA, setting of the resume codon to the A site, and peptidyl-transfer from peptidyl-tmRNA to Ala-tRNAAla. It is possible that S1 functions in later steps of trans-translation, such as in the exit of TLD from the E site. Alternatively, it may function only in the protection of tmRNA from degradation in the cell.
In the present study, we demonstrated that T. thermophilus S1 was not essential for the early steps of trans-translation, although it tightly bound to tmRNA. This suggests that S1 has no role in the functional interaction between tmRNA and the stalled ribosome or in a structural link between the tRNA domain and the mRNA domain for functional accommodation of tmRNA in the stalled ribosome. SmpB, another tmRNA-binding protein, may be deeply involved in this mechanism. Our T. thermophilus cell-free system should contribute to further clarification of the molecular mechanism of trans-translation in concert with recent and future progress of structural studies.
MATERIALS AND METHODS
Cloning and purification of tmRNA
The ssrA gene was amplified from genomic DNA from T. thermophilus HB8 by polymerase chain reaction with two primers, 5′-TTATCTAGAcacattGGGGGTGAAACGGTCTCGACGGGGGTC-3′ and 5′-GGTGCATGCcgatggagaatttTGGTGGAGGTGGGGGGAGTC-3′, in which the sense or anti-sense sequence of either end of the gene for tmRNA from T. thermophilus (GenBank, Y15063) is underlined. For proper processing of the primary transcript into a mature tmRNA in E. coli, the flanking sequences of the E. coli ssrA gene (GenBank, D12501) shown as lowercase letters, were added. A restriction enzyme cleavage sequence, shown in italics, was also added to each primer. The amplified DNA fragment was ligated into a plasmid vector, pGEM-EX2 (Promega), after digestion with XbaI and SphI. The resulting plasmid was termed pTtmRNAwt. The sequence of the recombinant tmRNA was revealed to be version 2 of the T. thermophilus tmRNA sequence in the tmRNA Web site that has G310 instead of A310 (Gueneau de Novoa and Williams 2004). The plasmid for preparation of a mutant tmRNA92U was constructed by PCR using the primers 5′-GGCTGCGCGCCGAGGTGCGGGTGGCCTCGTAAAAACCCGCAACGGCATAACTGTCAACACCAACTACGC-3′ and 5′-GGTGCATGCCGATGGAGAATTTTGGTGGAGGTGGGGGGAGTC-3′. The amplified DNA and pTtmRNAwt were ligated after digestion with HssHII and XbaI, resulting in pTtmRNA92U. The plasmids were transformed into E. coli strain BL21 (DE3) (ssrA::kan) constructed by P1 transduction from W3110 ΔssrA (Komine et al. 1994). T. thermophilus tmRNAwt and tmRNA92U were overexpressed and purified as described previously (Hanawa-Suetsugu et al. 2001).
Preparation of EF-Tu, EF-Ts, EF-G, S1, AlaRS, SmpB, and ribosome from T. thermophilus
Expression vectors of EF-Tu, EF-Ts, and EF-G were kind gifts from Dr. Sprinzl. Recombinant proteins were overexpressed in the E. coli strain BL21 (DE3) and purified as previously reported (Blank et al. 1995). The genes for SmpB, AlaRS, His-tagged EF-Tu (EF-Tu-His), and ribosomal protein S1 were cloned from the T. thermophilus HB8 genome by the RIKEN Structural Genomics Initiative (Yokoyama et al. 2000).
Ribosomal protein S1 cloned into pET11b (Novagen) was expressed in E. coli strain BL21 (DE3) codonplus RIL (Stratagene). Recombinant S1 was precipitated by the addition of 50%-saturated ammonium sulfate to the supernatant, which was obtained after heat treatment of the cell lysate for 20 min at 70°C. The precipitate was dissolved in 20 mM HEPES-NaOH buffer (pH 7.0) containing 150 mM NaCl and 0.9 M ammonium sulfate, and then subjected to hydrophobic column chromatography (HiPrep Buthyl: GE Healthcare). Further purification was performed by ion exchange chromatography (MONO Q and MONO S: GE Healthcare), and finally, by size-exclusion chromatography equilibrated with 20 mM HEPES-NaOH buffer (pH 7.0) containing 150 mM NaCl and 4 mM 2-mercaptoethanol. The protein yield was 1 mg per 1 g of frozen cells.
EF-Tu-His cloned into pET15b was overexpressed in E. coli strain BL21 (DE3). Cells were disrupted with a French Press in buffer A (20 mM Tris-HCl [pH 7.6], 10 mM MgCl2, 4.6 mM 2-mercaptoethanol, and 20 μM GDP) containing 100 mM NH4Cl. The supernatant of the cell extract that had been heat treated at 60°C was subjected to anion-exchange chromatography (HiTrap Q HP). EF-Tu-His fractions enriched by a 50–500 mM linear gradient elution of KCl were loaded onto Ni-NTA agarose (QIAGEN). Pooled fractions were dialyzed against buffer A containing 50 mM KCl. The protein yield was 15 mg per 1 g frozen cells.
AlaRS cloned into pET15b was overexpressed in E. coli strain BL21 (DE3). Cells were disrupted with a French Press in 50 mM Tris-HCl (pH 8.0) buffer containing 100 mM NH4Cl, 0.5 mM EDTA, 4.6 mM 2-mercaptoethanol, and 5% glycerol. The supernatant of the cell extract that had been subjected to heat treatment at 65°C was loaded on Ni-NTA agarose. After the resin had been washed with 20 mM Tris-HCl (pH 7.6) buffer containing 10 mM imidazole and 1 M NH4Cl, absorbed proteins were eluted with 200 mM imidazole, pooled, and dialyzed against buffer B (HEPES-NaOH [pH 7.0] and 100 mM KCl). Further purification was performed by anion-exchange chromatography (Resource Q) and size-exclusion chromatography (Superdex-200). The protein yield was 0.5 mg per 1 g frozen cells.
Ribosomes from T. thermophilus were prepared from frozen cells harvested in the middle-log phase as previously reported (Gogiya et al. 1986) with slight modification. 70S ribosome was purified with successive purification steps: rinse with CsCl-cushion buffer, 6%–38% sucrose density gradient centrifugation, and hydrophobic chromatography (HiPrep Butyl). The final buffer consisted of 20 mM HEPES-KOH (pH 7.5), 20 mM MgSO4, 100 mM NH4Cl, 4 mM 2-mercaptoethanol, and 0.5 mM spermine. 70S ribosome yield was 30 A260 units per 1 g cells.
Preparation of aminoacyl-RNA
AlaRS, valyl-tRNA synthetase (ValRS), and phenylalanyl-tRNA synthetase (PheRS) were prepared from E. coli cells using DEAE-Toyopearl 650M (Tosoh) (Tamura et al. 1991, 1994). AlaRS was further purified by hydroxyapatite column chromatography, Gigapite (Seikagaku Corporation).
For preparation of Ala-tmRNA, tmRNA was aminoacylated by incubating 1 μM tmRNA and AlaRS from E. coli with 8 μM L-[3-3H]alanine (15 TBq/mmol, PerkinElmer) or 30 μM unlabeled alanine in aminoacylation buffer for 10 min at 37°C. Twenty-eight percent of total tmRNA was aminoacylated with [3H]alanine. Ala-tmRNA was further purified by an immobilized EF-Tu column (Ribeiro et al. 1995) with slight modifications as follows: The immobilized EF-Tu column was prepared by incubating 2 mL of Ni-NTA (Qiagen) and 8 mg of His-tagged EF-Tu from T. thermophilus in a buffer (50 mM Tris-HCl [pH 7.5], 50 mM NH4Cl, 50 mM KCl, 10 mM MgCl2, 1 mM GTP, 5 mM phospho[enol]pyruvic acid, 15 units of pyruvate kinase [Sigma], and 10 mM 2-mercaptoethanol) for 10 min at 37°C. The Ni-NTA/EF-Tu/GTP was mixed with 1.6 nmol of aminoacylated RNA and incubated for 2 min on ice. After pouring the mixture into an empty column (Poly-Prep Chromatography Columns, Bio-Rad Laboratories), deacylated RNA was eluted with 4 mL of buffer B (50 mM Tris-HCl [pH 7.5], 50 mM NH4Cl, 50 mM KCl, 10 mM MgCl2, 1 mM GTP, and 5 mM 2-mercaptoethanol) and then with 4 mL of buffer C (50 mM HEPES-Na [pH 7.5], 50 mM NH4Cl, 150 mM NaCl, 10 mM MgCl2, 50 μM GTP, and 5 mM 2-mercaptoethanol). Then, aminoacylated RNA was eluted with 4 mL of buffer D (50 mM Tris-HCl [pH 7.5], 1 M NaCl, 10 mM MgCl2, 1 mM GDP, and 5 mM 2-mercaptoethanol) and precipitated with ethanol. We confirmed that nonaminoacylated tmRNA did not bind to the immobilized EF-Tu column.
For Phe-tRNAPhe preparation, tRNAPhe from E. coli (Sigma) was aminoacylated by PheRS with 30 μM L-[14C(U)]phenylalanine (18.4 GBq/mmol, PerkinElmer) or 10 μM L-[3-3H]phenylalanine (1.96 TBq/mmol, Amersham Biosciences) in aminoacylation buffer. Ninety-three percent and 86% of total tRNAPhe were aminoacylated with [14C]phenylalanine and [3H]phenylalanine, respectively. Phe-tRNAPhe was extracted with acidic phenol and precipitated with ethanol followed by dry-up and resolution with distilled water.
For Ala-tRNAAla and Val-tRNAVal, crude tRNAs were partially purified from T. thermophilus cells by phenol extraction and treatment with RNase-free DNase I (Takara) followed by fractionation by electrophoresis on a 15% polyacrylamide gel containing 7 M urea. The resulting partially purified tRNAs were aminoacylated by AlaRS and ValRS from E. coli with L-[3-3H]alanine (3.15 TBq/mmol, PerkinElmer) and L-[3,4(n)-3H]valine (1.52 TBq/mmol, Amersham Biosciences), respectively, in aminoacylation buffer described above. After phenol extraction and ethanol precipitation, aminoacylated tRNAs were further purified using the immobilized EF-Tu column as described above.
In vitro translation and trans-translation
Poly (Phe) synthesis was performed by incubating 0.5 pmol of S1-free ribosome, 1.2 μg of poly (U) (Sigma), 5 pmol of [3H]Phe-tRNAPhe, 5 pmol of EF-Tu, and 2 pmol of EF-G in 40 μL of TMNDS buffer containing 20 mM Tris-HCl (pH 7.5), 4 mM MgCl2, 50 mM NH4Cl, 1 mM dithiothreitol, 0.2 mM spermine, and 0.5 mM GTP at 65°C. Aliquots were withdrawn, diluted in 4 mL of 5% trichloro acetic acid, and incubated for 10 min at 90°C. The precipitated peptide was recovered by filtration with a mixed cellulose membrane (Advantec). The filter was dried, and the radioactivity on the filter was counted by a liquid scintillation counter (Aloka).
The stalled ribosome was prepared from the reaction mixture of poly (U)-dependent poly (Phe) synthesis as described above. After reaction for 10 min, the mixture was filtered through a Microcon YM-100 (Millipore) and washed with 100 μL of TMNDS buffer four times. The ribosome fraction that remained on the filter was recovered, and the volume was adjusted to half of the poly (Phe) synthesis reaction volume.
The trans-transfer reaction was initiated by adding a 10 μL aliquot of a mixture containing 0.2 μM [3H]Ala-tmRNA, 2 μM SmpB, and 0.5 μM EF-Tu with or without S1 in TMNDS buffer to 40 μL of stalled ribosomes. The reaction mixture was incubated at 65°C and aliquots were withdrawn. The product was detected by the same procedure as that used for the detection of poly (Phe) described above.
The resume codon decoding was performed by mixing and incubating the stalled ribosome (40 μL) and a mixture (10 μL) containing 0.5 μM unlabeled Ala-tmRNA, 0.2 μM [3H]Ala-tRNAAla or [3H]Val-tRNAVal, 2 μM SmpB, 1 μM EF-Tu, and 0.2 μM EF-G in TMNDS buffer. The product was detected by trichloroacetic acid precipitation and filtration as described above.
ACKNOWLEDGMENTS
We thank the staff of the Gene Research Center of Hirosaki University for the use of the facility. This work was supported by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to A.M. and H.H. (No. 14035201); a grant-in-aid for scientific research from the Japan Society for the Promotion of Science to A.M. and H.H. (No. 17380061); Grant for Priority Research Designated by the President of Hirosaki University to A.M. and H.H.; Research Fellowships of Japan Society for the Promotion of Science for Young Scientists to K.T.; and by the RIKEN Structural Genomics/Proteomics Initiative of the National Project on Protein Structural and Functional Analyses, Ministry of Education, Culture, Sports, Science and Technology of Japan to S.Y.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.363207.
REFERENCES
- Asano, K., Kurita, D., Takada, K., Konno, T., Muto, A., Himeno, H. Competition between trans-translation and termination or elongation of translation. Nucleic Acids Res. 2005;33:5544–5552. doi: 10.1093/nar/gki871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barends, S., Wower, J., Kraal, B. Kinetic parameters for tmRNA binding to alanyl-tRNA synthetase and elongation factor Tu from Escherichia coli . Biochemistry. 2000;39:2652–2658. doi: 10.1021/bi992439d. [DOI] [PubMed] [Google Scholar]
- Barends, S., Karzai, A.W., Sauer, R.T., Wower, J., Kraal, B. Simultaneous and functional binding of SmpB and EF-Tu-GTP to the alanyl acceptor arm of tmRNA. J. Mol. Biol. 2001;314:9–21. doi: 10.1006/jmbi.2001.5114. [DOI] [PubMed] [Google Scholar]
- Blank, J., Grillenbeck, N.W., Kreutzer, R., Sprinzl, M. Overexpression and purification of Thermus thermophilus elongation factors G, Tu, and Ts from Escherichia coli . Protein Expr. Purif. 1995;6:637–645. doi: 10.1006/prep.1995.1084. [DOI] [PubMed] [Google Scholar]
- Boni, I.V., Isaeva, D.M., Musychenko, M.L., Tzareva, N.V. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeau, V., Felden, B. Ribosomal protein S1 induces a conformational change of tmRNA; more than one protein S1 per molecule of tmRNA. Biochimie. 2002;84:723–729. doi: 10.1016/s0300-9084(02)01442-6. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons W.M., Jr., Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., Ramakrishnan, V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Farwell, M.A., Roberts, M.W., Rabinowitz, J.C. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis . Mol. Microbiol. 1992;6:3375–3383. doi: 10.1111/j.1365-2958.1992.tb02205.x. [DOI] [PubMed] [Google Scholar]
- Felden, B., Himeno, H., Muto, A., Atkins, J.F., Gesteland, R.F. Structural organization of Escherichia coli tmRNA. Biochimie. 1996;78:979–983. doi: 10.1016/s0300-9084(97)86720-x. [DOI] [PubMed] [Google Scholar]
- Gogiya, Z.V., Yusupov, M.M., Spirina, T.N. Structure of Thermus thermophilus ribosomes. Mol. Biologia (Russia) 1986;20:519–526. [Google Scholar]
- Gueneau de Novoa, P., Williams, K.P. The tmRNA website: Reductive evolution of tmRNA in plastids and other endosymbionts. Nucleic Acids Res. 2004;32:D104–D108. doi: 10.1093/nar/gkh102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann, S., Haebel, P.W., Metzinger, L., Sutter, M., Felden, B., Ban, N. Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature. 2003;424:699–703. doi: 10.1038/nature01831. [DOI] [PubMed] [Google Scholar]
- Hanawa-Suetsugu, K., Bordeau, V., Himeno, H., Muto, A., Felden, B. Importance of the conserved nucleotides around the tRNA-like structure of Escherichia coli transfer-messenger RNA for protein tagging. Nucleic Acids Res. 2001;29:4663–4673. doi: 10.1093/nar/29.22.4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawa-Suetsugu, K., Takagi, M., Inokuchi, H., Himeno, H., Muto, A. SmpB functions in various steps of trans-translation. Nucleic Acids Res. 2002;30:1620–1629. doi: 10.1093/nar/30.7.1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himeno, H., Sato, M., Tadaki, T., Fukushima, M., Ushida, C., Muto, A. In vitro trans-translation mediated by alanine-charged 10Sa RNA. J. Mol. Biol. 1997;268:803–808. doi: 10.1006/jmbi.1997.1011. [DOI] [PubMed] [Google Scholar]
- Isono, K., Isono, S. Lack of ribosomal protein S1 in Bacillus stearothermophilus . Proc. Natl. Acad. Sci. 1976;73:767–770. doi: 10.1073/pnas.73.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, K., Tadaki, T., Lee, S., Takada, K., Muto, A., Himeno, H. Trans-translation mediated by Bacillus subtilis tmRNA. FEBS Lett. 2002;516:245–252. doi: 10.1016/s0014-5793(02)02561-9. [DOI] [PubMed] [Google Scholar]
- Ivanova, N., Pavlov, M.Y., Felden, B., Ehrenberg, M. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 2004;338:33–41. doi: 10.1016/j.jmb.2004.02.043. [DOI] [PubMed] [Google Scholar]
- Karzai, A.W., Sauer, R.T. Protein factors associated with the SsrA•SmpB tagging and ribosome rescue complex. Proc. Natl. Acad. Sci. 2001;98:3040–3044. doi: 10.1073/pnas.051628298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karzai, A.W., Susskind, M.M., Sauer, R.T. SmpB, a unique RNA-binding protein essential for the peptide-tagging activity of SsrA (tmRNA) EMBO J. 1999;18:3793–3799. doi: 10.1093/emboj/18.13.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiler, K.C., Waller, P.R., Sauer, R.T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science. 1996;271:990–993. doi: 10.1126/science.271.5251.990. [DOI] [PubMed] [Google Scholar]
- Komarova, A.V., Tchufistova, L.S., Supina, E.V., Boni, I.V. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine, Y., Kitabatake, M., Yokogawa, T., Nishikawa, K., Inokuchi, H. A tRNA-like structure is present in 10Sa RNA, a small stable RNA from Escherichia coli . Proc. Natl. Acad. Sci. 1994;91:9223–9227. doi: 10.1073/pnas.91.20.9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Ishii, M., Tadaki, T., Muto, A., Himeno, H. Determinants on tmRNA for initiating efficient and precise trans-translation: Some mutations upstream of the tag-encoding sequence of Escherichia coli tmRNA shift the initiation point of trans-translation in vitro. RNA. 2001;7:999–1012. doi: 10.1017/s1355838201010342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinness, K.E., Sauer, R.T. Ribosomal protein S1 binds mRNA and tmRNA similarly but plays distinct roles in translation of these molecules. Proc. Natl. Acad. Sci. 2004;101:13454–13459. doi: 10.1073/pnas.0405521101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muto, A., Fujihara, A., Ito, K., Matsuno, J., Ushida, C., Himeno, H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells. 2000;5:627–635. doi: 10.1046/j.1365-2443.2000.00356.x. [DOI] [PubMed] [Google Scholar]
- Nameki, N., Someya, T., Okano, S., Suemasa, R., Kimoto, M., Hanawa-Suetsugu, K., Terada, T., Shirouzu, M., Hirao, I., Takaku, H., et al. Interaction analysis between tmRNA and SmpB from Thermus thermophilus . J. Biochem. 2005;138:729–739. doi: 10.1093/jb/mvi180. [DOI] [PubMed] [Google Scholar]
- Okada, T., Wower, I.K., Wower, J., Zwieb, C.W., Kimura, M. Contribution of the second OB fold of ribosomal protein S1 from Escherichia coli to the recognition of tmRNA. Biosci. Biotechnol. Biochem. 2004;68:2319–2325. doi: 10.1271/bbb.68.2319. [DOI] [PubMed] [Google Scholar]
- Ribeiro, S., Nock, S., Sprinzl, M. Purification of aminoacyl-tRNA by affinity chromatography on immobilized Thermus thermophilus EF-Tu.GTP. Anal. Biochem. 1995;228:330–335. doi: 10.1006/abio.1995.1359. [DOI] [PubMed] [Google Scholar]
- Roberts, M.W., Rabinowitz, J.C. The effect of Escherichia coli ribosomal protein S1 on the translational specificity of bacterial ribosomes. J. Biol. Chem. 1989;264:2228–2235. [PubMed] [Google Scholar]
- Rudinger-Thirion, J., Giege, R., Felden, B. Aminoacylated tmRNA from Escherichia coli interacts with prokaryotic elongation factor Tu. RNA. 1999;5:989–992. doi: 10.1017/s135583829999101x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta, J., Agrawal, R.K., Frank, J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. 2001;98:11991–11996. doi: 10.1073/pnas.211266898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, Y., Ueda, T. The role of SmpB protein in trans-translation. FEBS Lett. 2002;514:74–77. doi: 10.1016/s0014-5793(02)02333-5. [DOI] [PubMed] [Google Scholar]
- Shiryaev, V.M., Selivanova, O.M., Hartsch, T., Nazimov, I.V., Spirin, A.S. Ribosomal protein S1 from Thermus thermophilus: Its detection, identification, and overproduction. FEBS Lett. 2002;525:88. doi: 10.1016/s0014-5793(02)03092-2. [DOI] [PubMed] [Google Scholar]
- Someya, T., Nameki, N., Hosoi, H., Suzuki, S., Hatanaka, H., Fujii, M., Terada, T., Shirouzu, M., Inoue, Y., Shibata, T., et al. Solution structure of a tmRNA-binding protein, SmpB, from Thermus thermophilus . FEBS Lett. 2003;535:94–100. doi: 10.1016/s0014-5793(02)03880-2. [DOI] [PubMed] [Google Scholar]
- Sorensen, M.A., Fricke, J., Pedersen, S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- Stepanov, V.G., Nyborg, J. tmRNA from Thermus thermophilus. Interaction with alanyl-tRNA synthetase and elongation factor Tu. Eur. J. Biochem. 2003;270:463–475. doi: 10.1046/j.1432-1033.2003.03401.x. [DOI] [PubMed] [Google Scholar]
- Subramanian, A.R. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Asahara, H., Himeno, H., Hasegawa, T., Shimizu, M. Identity elements of Escherichia coli tRNAAla . J. Mol. Recognit. 1991;4:129–132. doi: 10.1002/jmr.300040404. [DOI] [PubMed] [Google Scholar]
- Tamura, K., Nameki, N., Hasegawa, T., Shimizu, M., Himeno, H. Role of the CCA terminal sequence of tRNAVal in aminoacylation with valyl-tRNA synthetase. J. Biol. Chem. 1994;269:22173–22177. [PubMed] [Google Scholar]
- Tedin, K., Resch, A., Blasi, U. Requirements for ribosomal protein S1 for translation initiation of mRNAs with and without a 5′ leader sequence. Mol. Microbiol. 1997;25:189–199. doi: 10.1046/j.1365-2958.1997.4421810.x. [DOI] [PubMed] [Google Scholar]
- Thomas, J.O., Szer, W. RNA-helix-destabilizing proteins. Prog. Nucleic Acid Res. Mol. Biol. 1982;27:157–187. doi: 10.1016/s0079-6603(08)60600-5. [DOI] [PubMed] [Google Scholar]
- Tzareva, N.V., Makhno, V.I., Boni, I.V. Ribosome-messenger recognition in the absence of the Shine–Dalgarno interactions. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- Ushida, C., Himeno, H., Watanabe, T., Muto, A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis . Nucleic Acids Res. 1994;22:3392–3396. doi: 10.1093/nar/22.16.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uzawa, T., Yamagishi, A., Oshima, T. Polypeptide synthesis directed by DNA as a messenger in cell-free polypeptide synthesis by extreme thermophiles, Thermus thermophilus HB27 and Sulfolobus tokodaii strain 7. J. Biochem. 2002;131:849–853. doi: 10.1093/oxfordjournals.jbchem.a003174. [DOI] [PubMed] [Google Scholar]
- Valle, M., Gillet, R., Kaur, S., Henne, A., Ramakrishnan, V., Frank, J. Visualizing tmRNA entry into a stalled ribosome. Science. 2003;300:127–130. doi: 10.1126/science.1081798. [DOI] [PubMed] [Google Scholar]
- Van Dieijen, G., Van Der Laken, C.J., Van Knippenberg, P.H., Van Duin, J. Function of Escherichia coli ribosomal protein S1 in translation of natural and synthetic messenger RNA. J. Mol. Biol. 1975;93:351–366. doi: 10.1016/0022-2836(75)90282-x. [DOI] [PubMed] [Google Scholar]
- Williams, K.P., Bartel, D.P. Phylogenetic analysis of tmRNA secondary structure. RNA. 1996;2:1306–1310. [PMC free article] [PubMed] [Google Scholar]
- Williams, K.P., Martindale, K.A., Bartel, D.P. Resuming translation on tmRNA: A unique mode of determining a reading frame. EMBO J. 1999;18:5423–5433. doi: 10.1093/emboj/18.19.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Withey, J.H., Friedman, D.I. A salvage pathway for protein structures: tmRNA and trans-translation. Annu. Rev. Microbiol. 2003;57:101–123. doi: 10.1146/annurev.micro.57.030502.090945. [DOI] [PubMed] [Google Scholar]
- Wower, I.K., Zwieb, C.W., Guven, S.A., Wower, J. Binding and cross-linking of tmRNA to ribosomal protein S1, on and off the Escherichia coli ribosome. EMBO J. 2000;19:6612–6621. doi: 10.1093/emboj/19.23.6612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower, I.K., Zwieb, C., Wower, J. Contributions of pseudoknots and protein SmpB to the structure and function of tmRNA in trans-translation. J. Biol. Chem. 2004;279:54202–54209. doi: 10.1074/jbc.M410488200. [DOI] [PubMed] [Google Scholar]
- Yokota, T., Arai, K.I., Kaziro, Y. Involvement of 30S ribosomal protein S1 in poly(U)-directed polyphenylalanine synthesis. J. Biochem. 1977;82:1485–1489. doi: 10.1093/oxfordjournals.jbchem.a131838. [DOI] [PubMed] [Google Scholar]
- Yokoyama, S., Hirota, H., Kigawa, T., Yabuki, T., Shirouzu, M., Terada, T., Ito, Y., Matsuo, Y., Kuroda, Y., Nishimura, Y., et al. Structural genomics projects in Japan. Nat. Struct. Biol. 2000;7(Suppl.):943–945. doi: 10.1038/80712. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Garber, M.B., Vasiliev, V.D., Spirin, A.S. Thermus thermophilus ribosomes for crystallographic studies. Biochimie. 1991;73:887–897. doi: 10.1016/0300-9084(91)90130-s. [DOI] [PubMed] [Google Scholar]
- Yusupova, G., Yusupov, M., Spirin, A., Ebel, J.P., Moras, D., Ehresmann, C., Ehresmann, B. Formation and crystallization of Thermus thermophilus 70S ribosome/tRNA complexes. FEBS Lett. 1991;290:69–72. doi: 10.1016/0014-5793(91)81228-z. [DOI] [PubMed] [Google Scholar]
- Zhang, J., Deutscher, M.P. A uridine-rich sequence required for translation of prokaryotic mRNA. Proc. Natl. Acad. Sci. 1992;89:2605–2609. doi: 10.1073/pnas.89.7.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zvereva, M.I., Ivanov, P.V., Teraoka, Y., Topilina, N.I., Dontsova, O.A., Bogdanov, A.A., Kalkum, M., Nierhaus, K.H., Shpanchenko, O.V. Complex of transfer-messenger RNA and elongation factor Tu. Unexpected modes of interaction. J. Biol. Chem. 2001;276:47702–47708. doi: 10.1074/jbc.M106786200. [DOI] [PubMed] [Google Scholar]