Abstract
We present a simple approach to locate sites that undergo conformational changes upon crystallization by comparative structural mapping of the same RNA in three different environments. As a proof of principle, we probed the readily crystallized P4–P6ΔC209 domain from the Tetrahymena thermophila group I intron in a native solution, in a solution mimicking the crystallization drop, and in crystals. We chose the selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry, which monitors the flexibility and the conformation of each nucleotide. First, SHAPE successfully revealed the structural changes that occur during the crystallization process. Specifically, 64% of the nucleotides implicated in packing contacts and present in the portion of the molecule analyzed were identified. Second, reactivity differences for some of these nucleotides were already observed in the crystallization solution, suggesting that the crystallization buffer locked down a particular structure that was favorable to crystal formation. Third, the probing of a known structure extends our understanding of the structural basis for the SHAPE reaction by suggesting that reactivity is enhanced by a C2′-endo sugar pucker. Furthermore, by identifying local conformational changes of the RNA that take place during crystallization, SHAPE could be combined with the in vitro selection of stable mutants to rationalize the design of RNA candidates for crystallization.
Keywords: chemical probing, crystallization, structure mapping, X-ray crystallography
INTRODUCTION
Improvement in RNA-oriented biochemical technologies has facilitated structural analysis of large RNA molecules (herein, molecules beyond the length amenable to large-scale chemical synthesis, i.e., ∼35–50 nucleotides [nt]). After the determination of the tRNA three-dimensional structure (76 nt) in the mid-1970s (Kim et al. 1974; Robertus et al. 1974; Sundaralingam et al. 1975; Stout et al. 1976), another advance in RNA crystallography was made with the atomic resolution structure of the P4–P6 domain from a group I intron (160 nt) in 1996 (Cate et al. 1996). Then came a renaissance in the field of RNA crystallography that has now seen the structural determination of the archetypes of various RNA classes, such as, for example, ribosomal RNA (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000), group I intron ribozymes (Adams et al. 2004; Guo et al. 2004; Golden et al. 2005; Stahley and Strobel 2005), and riboswitches (Batey et al. 2004; Thore et al. 2006).
Notwithstanding these successes, obtaining crystals of large RNA molecules suitable for atomic-level diffraction still remains a significant hurdle. In fact, a single nucleotide change can dramatically affect diffraction quality and even crystallization (Anderson et al. 1996; Batey et al. 2001; Juneau et al. 2001; Vicens and Westhof 2001). As a consequence, the crystallization of a new RNA usually requires a trial-and-error approach that follows a double screening procedure: Up to 15–20 variants of the RNA sequence of interest are tested in sparse matrices containing 48–96 crystallization conditions (Doudna et al. 1993; Scott et al. 1995).
The pool of RNA variants being investigated usually comprises sequences that have been altered at the periphery (e.g., by adding base pairs and 5′/3′ overhangs [Scott et al. 1995; Batey et al. 2001] as well as by engineering stable tetraloops [Golden et al. 1997]), and/or in the core of the molecule of reference (e.g., by introducing thermostable mutations [Juneau et al. 2001; Guo et al. 2004]). Fortuitously, mutations at the periphery often affect crystal contacts, potentially leading to an improvement of resolution. The impact of mutations in the interior of the molecule is less well understood, but mutations giving improved crystals have been correlated with a stabilization of secondary (Juneau et al. 2001) or tertiary structure elements (Guo et al. 2006). In general, finding the particular nucleotide and the corresponding mutation that will improve the resolution is far from trivial. Hence, several rounds of trial-and-error optimization are usually needed to obtain high-quality crystals, with no guarantee of success. In order to speed up and possibly rationalize the design of the RNA constructs to be screened, crystallographers could benefit from knowing the parts of the RNA (in the periphery and at the core) that are potential sites for engineering.
In the present study, we performed footprinting of the 159-nt-long thermostable P4–P6ΔC209 mutant RNA from the Tetrahymena thermophila self-splicing intron (Juneau and Cech 1999; Juneau et al. 2001) in three different environments: a native solution, the crystallization solution, and in the crystal. Many studies have mapped the structure of RNA molecules under different solution conditions (Gesteland et al. 2005). However, to our knowledge, only one publication compares the chemical reactivity of a single modified nucleotide belonging to the same RNA in solution and in the crystal (Gherghe et al. 2005). We used the recently developed selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) method (Merino et al. 2005). The SHAPE chemistry takes advantage of the relationship between the local flexibility of each nucleotide and the reactivity of its 2′-hydroxyl group to N-methylisatoic anhydride (NMIA), with the consequence that residues constrained in Watson–Crick pairs are generally unreactive, while residues located outside canonical helices display a reactivity that depends on their local flexibility or accessibility to NMIA (Merino et al. 2005; Wilkinson et al. 2005).
SHAPE revealed nucleotides in the P4–P6ΔC209 RNA that undergo three types of local structural changes upon crystallization: (1) residues that acquire additional flexibility, (2) nucleotides that become constrained in reactive conformations by intramolecular interactions, and (3) nucleotides that are implicated in packing contacts. For an RNA whose structure has not yet been solved, such information could be useful in narrowing down the number of RNA constructs to screen before getting high-quality crystals. Furthermore, the comparison of the SHAPE data and the three-dimensional structure of the P4–P6ΔC209 RNA offered a direct correlation of the SHAPE reactivity of a particular nucleotide, its conformation, and its structural environment. This better understanding of the basis for SHAPE reactivity becomes important with the increasing popularity of the SHAPE methodology (Badorrek et al. 2006; Chen et al. 2006).
RESULTS
Adjusting the SHAPE protocol for in-crystal studies
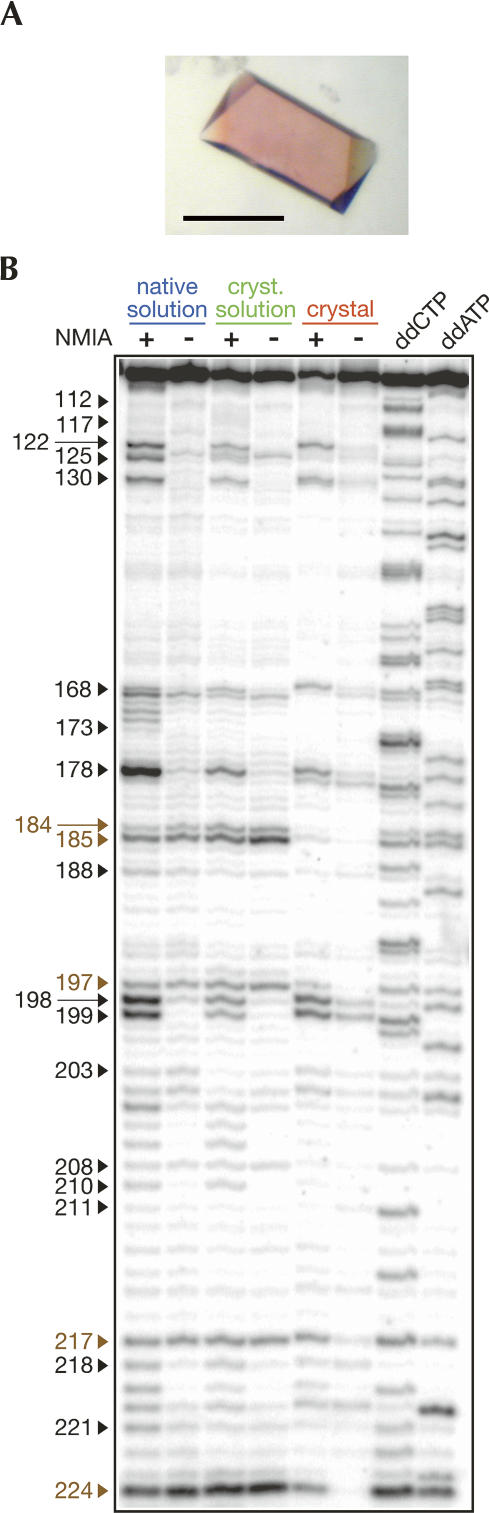
In order to distinguish among structural changes induced by the buffer change (from native to crystallization solution) and by the phase change (from crystallization solution to crystal), SHAPE mapping of the P4–P6ΔC209 domain was performed in three different contexts: (1) in a standard “native solution” in which the RNA is folded (6 mM MgCl2, 100 mM NaCl, 0.21 μM RNA) (Juneau et al. 2001), (2) in a solution containing the constituents of the crystallization drop (30 mM MgCl2, 43 mM NaCl, 0.17 mM cobalt hexamine, 30% 2-methyl-2,4-pentanediol [MPD], 0.21 μM RNA), and (3) in crystals. To perform the SHAPE reaction in the crystal, two or three crystals of 150–200 μm maximum length were sufficient to produce an interpretable signal (Fig. 1A). Crystals were soaked in a stabilizing solution containing the reactive agent NMIA or its solvent as a control (see Materials and Methods). Distinct positions showing a range of reactivities to NMIA were readily visible on the sequencing gel (e.g., residues 125, 168–173, and 178; the numbering system refers to the full-length intron; the P4–P6ΔC209 domain encompasses residues 102–260) (Fig. 1B).
FIGURE 1.
From crystals to mapping. (A) Typical crystal of the P4–P6ΔC209 RNA used in this study. The scale bar corresponds to 100 μm. (B) Example of a SHAPE experiment performed under the three conditions tested as visualized by PAGE. Nucleotides identified as reactive under any condition tested are indicated in black. Nucleotides corresponding to pauses of the reverse transcriptase are indicated in brown. In this and subsequent figures, data originating from mapping in the native solution, the crystallization solution, and the crystal are color-coded using blue, green, and red, respectively.
The requirement to perform in-crystal probing precluded the insertion of the P4–P6ΔC209 RNA in the structure cassette that provides RNA linkers and a primer binding site, so that the complete sequence is accessible to SHAPE mapping (Merino et al. 2005). Instead, the last 19 nt at the 3′ end of the P4–P6ΔC209 RNA were used for primer binding. The absence of the structure cassette also prevented any normalization of the reactivities to the strong reactivity usually obtained at one particular cytosine located in a stem–loop outside of the RNA of interest (Merino et al. 2005; Wilkinson et al. 2005). We therefore designed a program—named “noRNAlize”—to normalize the integrated band intensities using a subset of the paired regions present in the structure (see Discussion and Materials and Methods).
The mapping observed on the gels was reproducible, as attested by an average correlation coefficient of 0.95 calculated for the normalized intensities of residues 100–240 from three independent experiments (the third experiment compared two independent sets of crystals with one set of native and crystallization solutions) (Fig. 2). Normalized intensities were subsequently merged, with the omission of sites where the band intensity was independent of NMIA, attributed to pausing of the reverse transcriptase (e.g., Fig. 1B, at residues 184, 185, 197, 217, 224). Usable information typically ranged from residue 110 (close to the top of the gel) to residue 230 (located ∼10 nt after the last nucleotide of the primer binding site), thereby covering ∼75% of the sequence of the P4–P6ΔC209 molecule (Fig. 3A).
FIGURE 2.
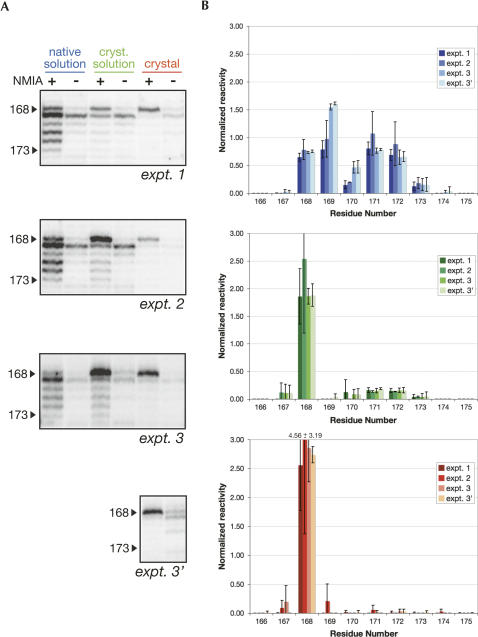
Reproducibility as attested by three independent experiments: example of nucleotides 168–173 that form the L5c loop. (A) Mapping results obtained for three distinct experiments as visualized by PAGE. The gels were rendered using SAFA (Das et al. 2005). Experiment 3 contained two different sets of crystals (the second set is referred to as “expt. 3′ ”). (B) Corresponding SHAPE reactivities after integration and normalization. Error bars were calculated from the merging of the different loads present on the same gel. Note that values for expt. 3 and expt. 3′ are close, as the normalization procedure was performed using the same lanes for the native and the crystallization solutions.
FIGURE 3.
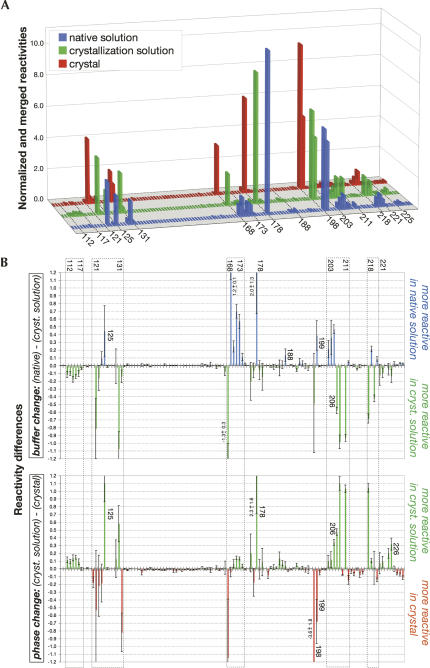
Normalized and merged reactivities and differences of reactivities. (A) Normalized and merged reactivities under the three contexts tested. Residues located in reactive regions are specified. (B) Differences of reactivities upon the transition from the native to the crystallization solution (buffer change, top panel) and upon the transition from the crystallization solution to the crystal (phase change, bottom panel). Error bars were calculated first from the merging of the different loads for each experiment and then from the merging of the three experiments. Dashed line boxes highlight regions of consecutive residues that underwent the main structural changes.
SHAPE achieves a precise monitoring of local RNA structure in the three environments tested
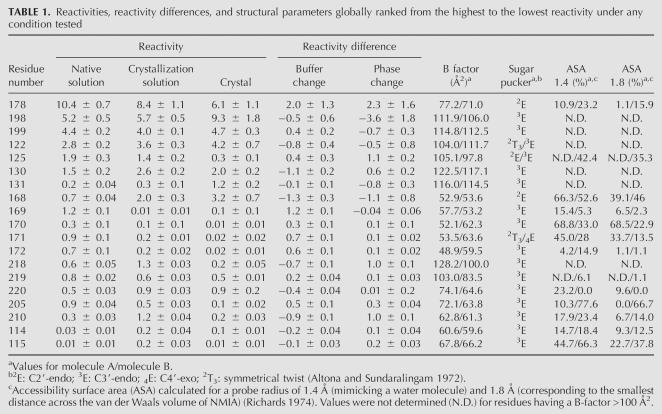
Most of the P4–P6ΔC209 RNA was unreactive to NMIA in the native solution and remained unreactive in the crystallization solution and in the crystal (Fig. 3A), as expected from previous studies in solution (Murphy and Cech 1993; Juneau and Cech 1999) and from its crystal structure (Juneau et al. 2001). This result also indicates that the lattice does not substantially alter the RNA three-dimensional structure. In at least one of the three environments tested, the strongest reactivities were observed at residues 178 (in the joining region between the P5a and P5c helices), 122, 125, 130, 131, 198, and 199 (all located in or close to the hinge region), and 168–173 (in the L5c loop) (Fig. 3A; Table 1). Most of these residues are found in single-stranded regions and bulges (Figs. 4, 5; Supplemental Fig. S1), consistent with the specificity of the NMIA reagent (Merino et al. 2005; Wilkinson et al. 2005). More moderate reactivities were detected for residues 203–211 and 112–117, which belong to the two strands of the coaxially stacked P4 and P5 helices, and for residues 218–230, which are part of the coaxially stacked P6a and P6b helices (Fig. 3A; Table 1).
TABLE 1.
Reactivities, reactivity differences, and structural parameters globally ranked from the highest to the lowest reactivity under any condition tested
FIGURE 4.
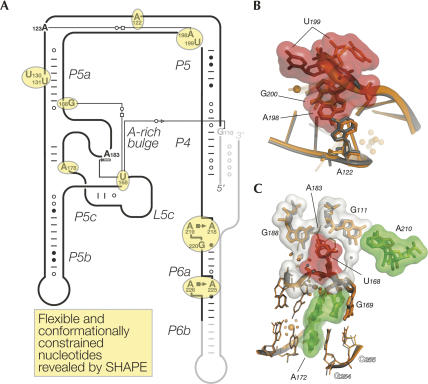
SHAPE reveals flexible and conformationally constrained nucleotides. (A) Schematic representation of the secondary structure of the P4–P6ΔC209 molecule indicating the flexible and conformationally constrained residues that are the most strongly reactive under any of the conditions tested (circled in yellow). Intramolecular interactions are shown according to the nomenclature for base pairing and base stacking (Leontis and Westhof 2001; Adams et al. 2004). The sections in light gray correspond to the regions not amenable to mapping in the current methodology. (B) Flexible residues at the hinge: U199 adopts two different conformations. Residues belonging to molecules A (in orange) and B (in gray) are superimposed. (C) Molecular details of the interactions involving the L5c loop. In this and subsequent figures, the following code is used: (1) residues that possess reactivity differences during the phase change are contoured by an accessible surface calculated for a 1.4 Å probe radius, which is colored according to Figure 3B (bottom panel); (2) solvent molecules (water molecules and magnesium ions) are shown as spheres colored according to the color code for molecules A and B; (3) labels in plain fonts refer to residues belonging to the reference molecules A and/or B; and (4) open labels refer to residues belonging to neighboring molecules.
FIGURE 5.
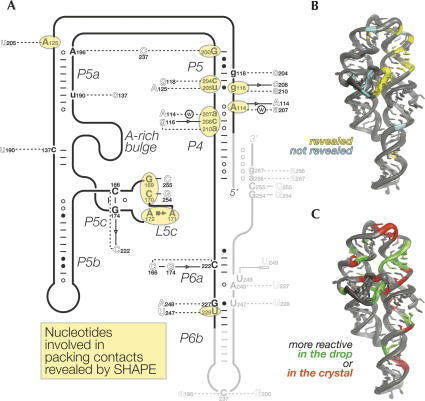
SHAPE reveals 64% of the packing contacts. (A) Schematic representation of the secondary structure of the P4–P6ΔC209 molecule indicating the location and the nature of the packing contacts. The nucleotides identified are circled in yellow. Lower- and uppercase letters refer to nucleotides involved in a packing interaction in 50% or 100% of the molecules, respectively. The nomenclature for base pairing and base stacking is used (Leontis and Westhof 2001; Adams et al. 2004). Dashed lines correspond to interactions involving H bonds that cannot be classified as base pairs. (B) Overall three-dimensional structure of the P4–P6ΔC209 molecule showing the residues involved in packing contacts that were revealed (in yellow) or not (in cyan) by the comparative SHAPE analysis. (C) Overall three-dimensional structure of the P4–P6ΔC209 molecule showing the residues that were more or less reactive upon the transition from the crystallization solution to the crystal (color coding as in Fig. 3B).
The eight most strongly reactive nucleotides (Table 1) match the two criteria previously proposed to cause a high SHAPE reactivity: high flexibility or tertiary interactions that constrain residues to adopt reactive conformations (Merino et al. 2005; Wilkinson et al. 2005). Six of these eight residues have high thermal agitation B factors (>100 Å2 for their 2′-hydroxyl groups in molecules A and/or B present in the asymmetric unit) (Supplemental Fig. S2), and some of these, like U199, even adopt two very different conformations in the two molecules (Fig. 4B). In contrast, the strong reactivities of A178 and U168 in the context of the crystal cannot be accounted for by flexibility. In fact, the B factors for the 2′-OH groups of A178 and U168 are close to or much lower than the average B factor (79.6 Å2 and 73.5 Å2 for all 2′-OH groups in molecules A and B, respectively), as A178 and U168 form extensive tertiary interactions in the crystal structure (Fig. 4C; Supplemental Fig. S3A). For example, U168 stacks against A183 of the A-rich bulge and forms H bonds in the shallow groove of the G111•A210 pair in P4, to G188 in P5a, and to water molecules coordinated to a magnesium ion (Fig. 4C). Additionally, both A178 and U168 are exposed to the solvent, as indicated by relatively high accessible surface areas (Table 1; Supplemental Fig. S3B). A C2′-endo sugar pucker could also account for their stronger reactivity (see Discussion). In conclusion, it appears that A178 and U168 are reactive in the crystal because of their uniquely constrained conformation. Both nucleotides form noncanonical interactions and are correctly scored by SHAPE as unpaired in the secondary structure.
Crystal contacts are revealed
Out of the eight most reactive residues (Table 1; Fig. 3), only A125 is involved in a crystal contact (just in molecule B) (Fig. 5). This is not surprising, because residues involved in crystal contacts are expected to become less reactive in the context of the crystal (as is the case for A125; see below). In fact, two additional results support that expectation. First, only 17 residues are more reactive in the crystal than in the crystallization solution versus 22 being more reactive in the crystallization solution than in the native solution (reactivity difference ≥ 0.05 or ≤−0.05; Fig. 3B, bottom panel). Noteworthy, none of these more reactive nucleotides is involved in any crystal contact (except for residues 200, 208, and 228, but their reactivity falls in the low ([−0.1]–0.1) range; Figs. 3B, 4, 5). Second, more residues have lower reactivities in the crystal than in the crystallization solution (with a few exceptions, e.g., for U168 and A178 described above): 67% of the reactive residues have a reactivity in the ([−0.1]–0.1) range in the crystal, compared to 58% in the crystallization solution (Fig. 3B, bottom panel) and to 33% in the native solution (Fig. 3B, top panel).
A total of 14 different nucleotides involved in packing are revealed by the present approach (reactivity ≥0.05 or ≤−0.05), thereby encompassing 64% of the nucleotides involved in lattice formation that reside in the part of the molecule analyzed here (residues 110–230) (Figs. 3B, 5). The precision level attained by SHAPE allowed the successful mapping of several kinds of lattice contacts, except for those involving residues embedded in Watson–Crick or G•U wobble pairs in each of the three environments tested and those that engage a single hydrogen bond in 50% of the molecules only (Fig. 5A).
As an example of crystal contacts revealed by SHAPE, one of the strongest differences already apparent by visual inspection of the gels (Figs. 1, 2) maps to residues 168–173. Positions 169–173 were moderately to highly reactive in solution, but became less reactive in the crystallization solution and even less in the crystal (Table 1; Fig. 3). G169 and C170 of molecule A form Watson–Crick pairs, respectively, with C255 and G254 of molecule B in the asymmetric unit, and reciprocally for molecule B (Juneau et al. 2001). These two intermolecular pairs stack on an A platform involving A171 and A172 followed by a noncanonical A•U pair involving A173 (Fig. 4C). SHAPE was therefore successful at uncovering the nucleotides directly engaged in Watson–Crick pairs, but also the adjacent adenines that participate in the local structural change resulting from packing interactions.
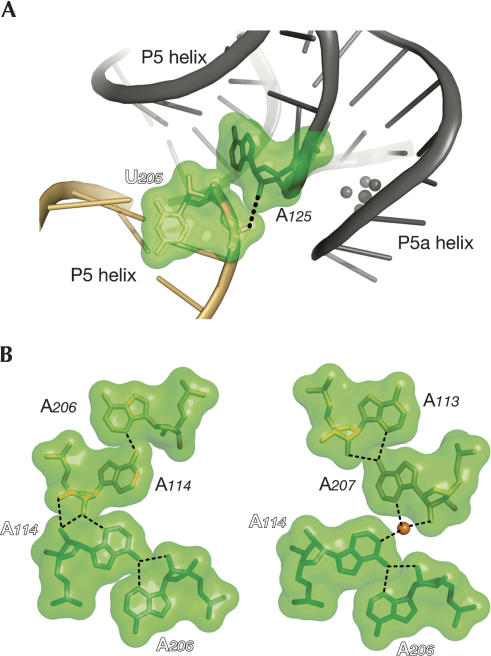
Additional examples of packing contacts revealed by SHAPE comprise hydrogen bonds involving the ribose 2′-hydroxyl group in different structural contexts (Fig. 6; Supplemental Fig. S4). For instance, the reactivity of A125 became progressively suppressed in the transition from native solution to crystallization solution to crystal (Table 1; Fig. 3), suggesting that A125 became protected from modification by NMIA. This result seemed contradictory to the high flexibility and accessibility of the 2′-OH of A125 suggested by the crystal structure (Table 1). However, in molecule B, the 2′-OH of A125 is involved in a direct hydrogen bond to one of the phosphate oxygen atoms of U205 from a symmetry-related molecule (Fig. 6A). This suggests that the involvement of the 2′-OH in a packing contact, even in only 50% of the molecules, was sufficient to decrease its average SHAPE reactivity in the crystal.
FIGURE 6.
Examples of crystal contacts involving the 2′-OH group. (A) Crystal contact involving the 2′-OH group of A125 in molecule B and a phosphate oxygen of U205 from a crystallographically related molecule (in light orange). (B) Atomic-level details of the interactions engaging the sugar edge of the adenines forming the tandem A•A pairs.
In two different structural contexts, SHAPE monitored crystal contacts involving shallow groove recognition of the P4/P5 and P6a/P6b regions. About half of the corresponding residues were most reactive in the crystallization solution, relative to both the native solution and the crystal (Fig. 3B). Residues 203–211, which are part of one strand of the P4/P5 region, were globally more reactive during the changes than residues 112–117 on the second strand. Reactivity differences of residues 225–230, which belong to one strand of the P6a/P6b region, tended to be more moderate (residues 244–249 of the other strand were part of the primer-binding site). The crystal structure shows that the P4/P5 and P6a/P6b helical domains are engaged in distinct shallow groove–shallow groove packing interactions. The shallow grooves of both domains are widened by either tandem A113•A207/A114•A206 pairs, the unpaired A115 and the G116•U205 pair, or by two A platforms (at positions 218–219 in molecule B only and at positions 225–226 in both molecules) followed by G•U pairs. These structural characteristics allowed the P4/P5 and P6a/P6b domains to serve as docking platforms for helical regions from symmetry-related molecules (Supplemental Fig. S4). As expected from interactions in the shallow groove, most of the 2′-OH groups are implicated in direct H bonds (e.g., A114 participating in ribose zippers), as well as water-mediated H bonds (e.g., between A207 and A114 of a symmetry-related molecule) (Fig. 6B). Overall, the stabilization resulting from dense crystal packing in these two helical regions was effectively assessed by the SHAPE chemistry.
DISCUSSION
A general method for the identification of local changes of RNA structure
In the present work we contrast the chemical reactivity of individual nucleotides from the same RNA molecule in two different solutions and in the crystal, with the purpose of identifying promising sites for engineering. Studies performed ∼20 yr ago compared the accessibility of a cytosine in the anticodon of the yeast tRNAAsp to dimethyl sulfate (DMS), under native (10 mM MgCl2) and semidenaturing (no MgCl2) conditions that both contained 1.6 M ammonium sulfate, like the crystallization solution (Moras et al. 1986). Additionally, comparative DMS mapping in solution and in the crystals had been performed for the P4–P6 domain, revealing that residues belonging to the L5c loop were somehow involved in crystal contacts (B. Golden, pers. comm.). Yet these and other similar studies performed on larger constructs were limited by the strong specificity of DMS for certain chemical functional groups and by nicks in the RNA that prevented efficient primer extension (B. Golden and E. Podell, pers. comm.). More recently, crystals of a self-complementary 16 base pair (bp) duplex containing one 2′-amine substitution per strand were soaked in a solution containing an activated ester to produce a 2′-amide at that position (Gherghe et al. 2005). The three-dimensional structures of the unmodified and the modified RNA were compared to understand the molecular basis of this particular reactivity.
The sensitivity of the SHAPE method enabled us to characterize the general structural stabilization that results from crystal formation at the nucleotide level. Particularly, comparative mapping revealed residues at the core and the periphery that undergo structural changes upon the formation of intra- and intermolecular interactions. The crystal structure of the P4–P6ΔC209 RNA (Juneau et al. 2001) provided a basis for the understanding of these reactivity differences. For example, U168 was interpreted to be stabilized in a strongly reactive conformation by extensive intramolecular interactions at the core (Fig. 4C), whereas A125 became sequestered away from modification by being engaged in a crystal contact at the periphery (Fig. 6A). Importantly, SHAPE goes beyond the flexible versus conformationally constrained distinction and reveals how residues within one category can be very differently influenced by the buffer and the phase changes (Fig. 3B). For instance, the two flexible bulged nucleotides U130 and U199 are strongly reactive, as expected, but the reactivity of U130 increases upon the transition from the native to the crystallization solution, and decreases upon the phase change, while the reverse is observed for U199 (Table 1; Fig. 3B). Additionally, the SHAPE probing identified residues adjacent to the most strongly affected positions, such as residue 131 adjacent to the flexible bulged U130, as well as residues 112, 113, 117, and 206 that are buried by the close packing of the P4/P5 helical domains in the lattice (Supplemental Fig. S4). Residues involved in particular motifs were also uncovered (e.g., residues 218–219 and 225–226 that form two A platforms) (Fig. 4A).
Overall, the SHAPE reactivity differences obtained for the P4–P6ΔC209 molecule suggest qualitative guidelines to predict the local structural changes induced by crystallization in the case of an unknown structure. First, nucleotides implicated in crystal contacts should show a decrease in reactivity upon the transition from the native solution to the crystal. In principle, an occasional nucleotide involved in packing interactions could be constrained to adopt a more reactive conformation, even though this was not observed in the case of the P4–P6ΔC209 RNA. Second, reactivity differences of residues located in regions forming crystal contacts should be more moderate than that of nucleotides that become more flexible or conformationally constrained. For intermolecular interactions between helices, such reactivity differences should come by patches, as exemplified by the interactions in the shallow grooves of helices P4/P5 and P6a/P6b that span 5–7 adjacent residues (Fig. 3B). Third, formation of intermolecular Watson–Crick pairs involving residues in loops (e.g., involving G169 and C170) or interactions involving the 2′-hydroxyl group of a bulged residue (e.g., involving A125) should generally give a stronger signal than that of helical interactions. It seems a priori difficult to distinguish between strong reactivity differences (>0.5) provoked by the formation of a crystal contact (e.g., for A125) or of a particular motif (e.g., for A218 participating in an A platform). However, only six such residues are observed during the buffer and the phase changes, half of which are implicated in crystal contacts (Fig. 3B). Since mutating nucleotides not directly involved in crystal contacts can also contribute to improving crystallographic resolution (Guo et al. 2004), we are currently investigating whether mutating such residues revealed by SHAPE improves crystal quality.
An unexpected finding of this study is that some of these structural changes were already observed in solution at low RNA concentration (0.21 μM) upon changing the buffer to crystallization conditions. For example, similar trends in the reactivity change of residues 168–173 (which are involved in the pairing between the two molecules in the asymmetric unit) were observed during the buffer change and the phase change (Fig. 3B). This result might signify that a preorganization of the L5c loop in the crystallization buffer precedes the docking of residues 254–255 from the second molecule, or that dimer formation could occur even at the low RNA concentration tested. Inversely, some residues were seen to behave differently upon the transitions to the crystallization solution and to the crystalline phase. For instance, the reactivities of residues 206, 207, and 210 were enhanced in the crystallization solution compared to the native solution, whereas they were reduced in the context of the crystal, as expected, since they are embedded in interactions that would be predicted to preclude any reactivity (Figs. 3B, 5, 6B). Such behavior could indicate the formation of particular local structures that ultimately get either disrupted or masked by the crystal packing, similarly to the transient interactions that were shown by SHAPE to form during thermal unfolding of tRNAAsp (Wilkinson et al. 2005).
Because the P4–P6ΔC209 molecule was shown to crystallize in a variety of buffers (Juneau et al. 2001), it could be worth mapping the RNA in these solutions to observe whether these conditions lock down a similar or a different structure. Furthermore, it would be interesting to probe additional RNA molecules, as well as the RNA components of RNP assemblies, such as those crystallizing in the presence of the U1A protein (Ferré-D'Amaré et al. 1998; Rupert and Ferré-D'Amaré 2001; Adams et al. 2004). Overall, SHAPE could potentially become a useful method to screen the behavior of RNA molecules in different crystallization buffers, without requiring any prior knowledge of their three-dimensional structures.
Correlation with previous in vitro selection experiments
In previous work aimed at improving diffraction quality, in vitro selection was employed to identify single mutations that increase thermostability (Juneau and Cech 1999; Guo and Cech 2002). The selection was performed outside the context of the crystal, without any need for information about which residues were involved in intra- and intermolecular interactions. Significantly, several mutations that were reported to stabilize the folding of the P4–P6 molecule (e.g., U130C, as well as the deletion of A207 and A210) (Juneau and Cech 1999) coincide with positions of altered SHAPE reactivity in the P4–P6ΔC209 RNA. Hence, the comparative SHAPE mapping and the in vitro selection for thermostable mutants can be seen as being complementary: The first enables the localization of the RNA sites that undergo local conformational changes during the crystallization process, while the second selects for mutants that stabilize folding. It seems reasonable to hypothesize that a combination of these two approaches would facilitate the design of RNA constructs with superior chances to yield crystals diffracting to high resolution.
Insights into the structural requirements for SHAPE reactivity: Role of the sugar pucker
By comparing the reactivity of individual nucleotides in the crystal with the crystal structure, the present study offers the opportunity to correlate SHAPE reactivity and a particular structural context. As inferred from previous mapping studies of nucleotide flexibility (Chamberlin and Weeks 2000), the temperature B factors are a good indicator of whether the reactivity of a residue is due to its flexibility (e.g., for A198 and U199) or to a particularly constrained conformation (e.g., for U168 and A178). The example of the P4–P6ΔC209 molecule shows that a high B factor is necessary but not sufficient to account for a strong SHAPE reactivity (Table 1; Supplemental Fig. S2). The absence of any SHAPE reactivity in regions that possess a high B factor (e.g., residues 119–121, 132–134, and 190–195; Supplemental Fig. S2) can be explained by the inaccessibility of these residues to NMIA (they are embedded in Watson–Crick pairs), and possibly by an overestimation of the B factors during refinement. For residues that possess a low B factor (relative to the average), the three-dimensional structure therefore provides a structural basis for improving our understanding of the requirements for reactivity to NMIA.
Distances measured in the three-dimensional structure between the 2′-OH group of residue n and the 5′ phosphate oxygen atoms of residue n+1 suggested that in general a distance >4.0 Å is necessary to allow a significant reactivity to NMIA. This agrees with the postulate that SHAPE reactivity is influenced by the proximity of the 3′-phosphodiester anion (Merino et al. 2005). However, this distance requirement is not sufficient to account for reactivity, as both unreactive (e.g., for paired residues in the P5b helix) and reactive 2′-OH groups (e.g., for A178 and U168) were found to be within 4.4–4.9 Å of a phosphate oxygen atom.
We propose that the C2′-endo sugar pucker adopted by A178 and U168, instead of the C3′-endo conformation typical of RNA, accounts for their strong reactivity (Table 1). Noteworthy, G17 and G18, located in the D loop of the yeast tRNAAsp, were previously shown to be strongly reactive to NMIA in solution (Chamberlin and Weeks 2000; Merino et al. 2005), as well as to be involved in multiple tertiary interactions. Furthermore, they adopt a C2′-endo sugar pucker in the crystal structure (Westhof et al. 1985; Comarmond et al. 1986). Additionally, some of the nucleotide analogs used in structure–reactivity assays of NMIA could adopt C2′-endo conformations (Merino et al. 2005). A C2′-endo conformation has been previously shown to favor an intraresidue H bond between the 2′-OH and the 3′-oxygen atom (Auffinger and Westhof 1997). Formation of such an interaction could enhance the reactivity of the 2′-OH by increasing its nucleophilicity.
The other ordered residues in the structure of P4–P6ΔC209 for which a C2′-endo was modeled during the refinement (Juneau et al. 2001) have a much lower reactivity (e.g., 0.2 ± 0.03 for A206 in molecule A, 0.1 ± 0.03 for C138, and 0.1 ± 0.04 for U182 in both molecules) or no reactivity (e.g., U179) in the crystal. The inaccessibility of these residues to the solvent and/or the proximity of electronegative or negatively charged groups could account for the decrease in reactivity (e.g., the 2′-OH of U179 is within 2.3 Å and 3.6 Å of the O1P of G180 in molecules A and B, respectively). The study of additional RNA molecules in the context of the crystal would be required to understand precisely the influence on the reactivity to NMIA of a C2′-endo sugar pucker and of the subsequent intraresidue H bonds that engage the 2′-OH. A deeper understanding of the structural basis for the chemical reactivity to NMIA would further improve the use of chemical modification data to constrain secondary structure predictions (Badorrek and Weeks 2006; Mathews and Turner 2006).
Finally, in order to facilitate the design of RNA constructs for crystallization, it might be useful to be able to differentiate a strong SHAPE reactivity due to a high flexibility from that due to a C2′-endo sugar pucker. By offering a biochemical signature of C2′-endo sugar puckers, nucleotide analog interference mapping using 2′-fluoro and 2′-deoxy derivatives could provide that distinction (Ortoleva-Donnelly et al. 1998).
A new application of the SHAPE method
Overall, the present study further supports the “user-friendliness” of the SHAPE chemistry: (1) The original SHAPE protocol could be modified to incorporate different ionic concentrations and the addition of precipitants such as MPD, (2) NMIA diffused through the solvent channels of RNA crystals without any noticeable interference with the efficiency of the reaction, and (3) similar mapping patterns were obtained for crystals grown at pH 6.0 but subsequently soaked at pH 8.0, as for crystals directly grown at pH 8.0 (data not shown). Additionally, the time to complete one experiment (from the set up of the SHAPE reaction to obtaining the normalized reactivities) was about 2 d.
Finally, we developed a universal normalization procedure that does not require the presence of the structure cassette at the 3′ end of the RNA molecule being probed (Merino et al. 2005). In our case, the addition of the structure cassette would have most likely prevented crystallization. However, we thereby lost the possibility to normalize the reactivities to that of a particular cytosine located in the cassette (Merino et al. 2005; Wilkinson et al. 2005; Badorrek et al. 2006). Normalizing reactivities to one of the most strongly reactive positions within the P4–P6ΔC209 molecule was considered, yet in the absence of additional independent experimental data, it did not seem reasonable to assume that the reactivity of any of these nucleotides would remain unchanged in the three contexts tested.
Normalization was eventually performed using 10% of the lowest intensity sites. The corresponding 10–12 nt are embedded in secondary structure elements, which should remain unaffected by the various environments tested (see Materials and Methods). Any signal equal to or lower than that of these particular residues would be made equal to zero by this procedure. However, the 10–12 sites were selected to be among the lowest signals; therefore, the only information lost was near the noise level and did not have any significant signal/noise ratio. Using that particular threshold, we found reactivity differences <−0.05 or >0.05 to correspond to meaningful signals. Modification of either the number of sites chosen for normalization, the threshold in reactivity difference, or both enabled the identification of more nucleotides undergoing local structural changes, but introduced a larger number of false positive results. A deeper statistical analysis of these parameters would be required to optimize the detection of the nucleotides impacted by crystallization. However, this will be possible only once different RNA molecules have been analyzed using the same methodology.
This normalization procedure allowed the inclusion of the control lanes ([−] NMIA), which are normalized separately from the lanes containing the signal ([+] NMIA) in other protocols (Wilkinson et al. 2006). Consequently, the reactions in the six lanes that correspond to one experiment (Figs. 1, 2A) were normalized in noRNAlize as one set of data, assuming the mean NMIA reactivity was conserved under the three environments tested (i.e., the ratio of strong/weak reactivities was conserved). This assumption was borne out by the bootstrap procedure used during normalization: Different normalizations performed using the same data set but different selections of lowest intensity sites yielded highly reproducible results with low error (see error estimates in Table 1). A further indication of the absence of a major bias in the normalization was that some SHAPE reactivities increased while others decreased during the buffer change and the phase change (Fig. 3A, cf., residues 178 and 198;). Additionally, sampling of the lowest intensities throughout the gel averaged out irregularities due to background measurement. In conclusion, the comprehensive and automated normalization procedure offered by noRNAlize can be seen as being universally applicable to any RNA of unknown three-dimensional structure, as it requires only minimal understanding about the structure being probed (i.e., whether the low-intensity sites selected correspond to residues in Watson–Crick pairs).
CONCLUSION
We applied the SHAPE method to probe the P4–P6ΔC209 RNA in solution and in crystals. In the process, we developed a comprehensive and automated normalization tool to facilitate computational processing of SHAPE reactivities. SHAPE mapping revealed nucleotides undergoing local structural changes upon crystallization, including the majority of the nucleotides involved in crystal packing contacts. Thus, we suggest that comparative SHAPE mapping could serve as a guide for site-directed mutagenesis, a critical step on the hazardous path to getting high-resolution RNA crystals. Furthermore, comparisons with structural parameters retrieved from the three-dimensional structure (thermal disorder, sugar pucker, solvent accessibility, and local environment around the 2′-hydroxyl group) widened our understanding of the basis of SHAPE reactivity. The analysis of additional RNA molecules should confirm whether a C2′-endo sugar pucker correlates with an enhancement of modification by NMIA.
MATERIALS AND METHODS
In vitro transcription, purification of transcripts, and crystallization
The pUC19-derived plasmid containing the P4–P6ΔC209 domain from Tetrahymena thermophila (Juneau and Cech 1999) was linearized using the restriction enzyme EarI (New England Biolabs, R0528S). One hundred micrograms of linearized plasmid were transcribed for 16 h at room temperature (∼22°C) in a 5 mL reaction mixture containing 4 mM of each rNTP, 30 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 1 μg inorganic pyrophosphatase (Sigma, I-1891), 800 U RNasin Plus RNase inhibitor (Promega, N2611), 300 μL T7 polymerase (prepared in house), and 40 mM tris-HCl (pH 8.0).
The RNA was purified by denaturing polyacrylamide gel electrophoresis (PAGE): 4% acrylamide/bisacrylamide (19:1), 8 M urea, 1× TBE (100 mM tris-base, 83 mM boric acid, 1 mM ethylenediaminetetraacetic acid [EDTA]). Gel slices were crushed and soaked in 1× TE (10 mM tris-HCl at pH 7.5, 1 mM EDTA) for 2 h at 4°C with one buffer exchange. The RNA eluate was filtered and concentrated on an Amicon-Ultra centrifugal filter unit (MWCO 10,000; Millipore, UFC901024), precipitated in ethanol, and resuspended in 5 mM Na cacodylate (pH 6.0). The RNA was refolded at a final concentration of 4 mg/mL in 5 mM MgCl2 and 5 mM Na cacodylate (pH 6.0). The sample was incubated at 65°C for 10 min in a heating block, then slow cooled to room temperature.
Crystals were grown at room temperature using the hanging drop vapor diffusion method, with three drops/cover slip, corresponding to three ratios (1 μL:2 μL, 1 μL:1 μL, 2 μL:1 μL) of the solution containing the RNA and the reservoir solution (15%–20% MPD, 30 mM MgCl2, 0.3 mM spermine, 0.2 mM cobalt hexamine, 60 mM HEPES at pH 8.0). The drops were equilibrated against 1 mL of 18–22.5% MPD in the well (Juneau et al. 2001).
Primer labeling
A primer complementary to the 3′ end of the P4–P6ΔC209 molecule was purchased from Sigma-Proligo (sequence: 5′-TGAACTGCATCCATATCAAC). Radioactive labeling of 50 nmol primer was performed for 30 min at 37°C in a 10 μL reaction mixture containing 50 nmol γ-32ATP (6000 Ci/mmol; Perkin-Elmer, NEG035C005MC) and 10 U T4 polynucleotide kinase (New England Biolabs, M0201S) in the commercial reaction buffer. The labeled primer was purified by denaturing PAGE (20% acrylamide/bisacrylamide (19:1), 8 M urea, 1× TBE) and eluted as described above. The labeled primer was precipitated in ethanol and resuspended in 1 mM HEPES (pH 8.0).
SHAPE mapping in solution and in crystal
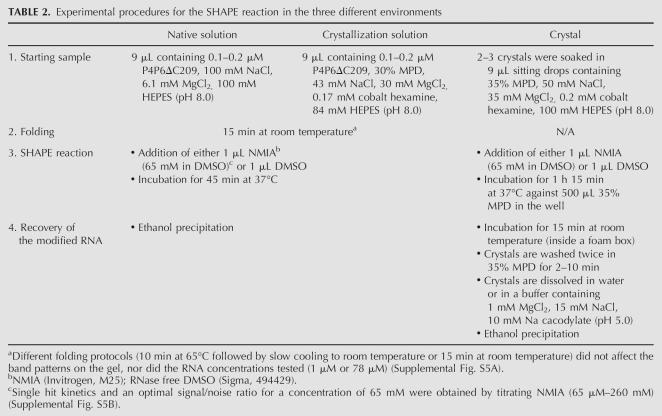
The protocol for the SHAPE reaction was adapted from Merino et al. (2005) and Wilkinson et al. (2006) to take into account the larger size of the RNA tested, to incorporate the crystallization components in the crystallization solution, and to allow in-crystal probing (Table 2). The subsequent primer extension steps of the RNA in the three environments were done concomitantly, essentially as described (Merino et al. 2005; Wilkinson et al. 2006), except for lower incubation temperatures (48°C–50°C) and for a longer extension step (8 min).
TABLE 2.
Experimental procedures for the SHAPE reaction in the three different environments
Sequencing PAGE
Dideoxy sequencing markers were produced as described (Merino et al. 2005; Wilkinson et al. 2006). Extended products were heated for 5 min at 95°C prior to being loaded on sequencing gels (15% acrylamide/bisacrylamide [19:1], 7 M urea, 1× TBE). Samples were typically loaded two or three times at ∼2–2.5- h intervals. Gels were scanned using a phosphorimager (Amersham Biosciences/GE).
Data integration and normalization
Intensities of the bands corresponding to modification by NMIA and of the control lanes were integrated using semiautomated footprinting analysis (SAFA) for the three environments (Das et al. 2005). We developed the program noRNAlize (available at https://simtk.org/home/nornalize) under Matlab (Mathworks) to normalize these raw intensities and to allow averaging of multiple experiments and error analysis. noRNAlize takes as input the SAFA output and normalizes the data in an automated fashion, by assuming that the SHAPE reactivities of the weakest 10% of the total number of bands per gel are constant in the three environments tested (this threshold is annotated in the source code and can be modified). The corresponding residues that were selected were different for each experiment (e.g., for Experiment 1: 146, 156, 158, 191, 194, 211, 212, 225, 229, 230, 232).
To assess the error introduced by this assumption, noRNAlize performs a bootstrap and error propagation analysis on the data, returning an average reactivity value with error (Efron 1979; Efron and Tibshirani 1993). The bootstrap procedure leaves out randomly selected weak intensities and repeats the normalization procedure. Finally, values are averaged to obtain mean and standard deviation values, and SHAPE reactivities are normalized to the mean raw intensity, assuming that the mean NMIA reactivity is approximately the same under all solution and crystal conditions tested.
Pearson product-moment correlation coefficients were calculated in order to evaluate the agreement between five independent experiments and the reproducibility of the normalization procedure. A Pearson correlation coefficient close to 1.0 indicated high overall agreement between the different data sets, allowing for the SHAPE reactivities of three experiments (one experiment containing two different sets of crystals) to be merged in Excel (Microsoft).
Structural analysis
SHAPE reactivities and reactivity differences were mapped on the three-dimensional structure of the P4–P6ΔC209 molecule (Juneau et al. 2001, PDB ID: 1HR2). Crystallographically related neighbors were compiled using CNS (Brünger et al. 1998). Crystal contacts were analyzed using CCP4i (Potterton et al. 2003) and SwissPDB Viewer (Guex and Peitsch 1997). Figures were rendered using PyMOL (pymol.sourceforge.net).
SUPPLEMENTAL DATA
Supplemental Figures S1–S5, as well as input and output files from SAFA and NORNALIZE used in this study, are freely available for download at https://simtk.org/home/nornalize.
ACKNOWLEDGMENTS
We thank Kevin Weeks, Kevin Wilkinson, and Chris Badorrek for sharing their SHAPE protocol and for regular discussions, Barbara Golden and Elaine Podell for sharing unpublished data, Marvin Caruthers and Rob Batey for helpful discussions, David Zappulla, Chrysa Latrick, Eric Westhof, Barbara Golden, Kevin Wilkinson, Colby Stoddard, and Rob Batey for insightful comments on the manuscript, and Kevin Weeks for reviewing the article. This work was partially funded by the NIH through NIGMS P01-GM66275 and an NIH Roadmap grant U54 GM072970 for the National Centers for Biomedical Computation (Simbios). A.L. is funded by a Damon Runyan Cancer Research Foundation post-doctoral fellowship. Q.V., A.R.G., and T.R.C. are supported by the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.400207.
REFERENCES
- Adams, P.L., Stahley, M.R., Gill, M.L., Kosek, A.B., Wang, J., Strobel, S.A. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altona, C., Sundaralingam, M. Conformational-analysis of sugar ring in nucleosides and nucleotides—New description using the concept of pseudorotation. J. Am. Chem. Soc. 1972;94:8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Anderson, A.C., Earp, B.E., Frederick, C.A. Sequence variations as a strategy for crystallizing RNA motifs. J. Mol. Biol. 1996;259:696–703. doi: 10.1006/jmbi.1996.0351. [DOI] [PubMed] [Google Scholar]
- Auffinger, P., Westhof, E. Rules governing the orientation of the 2′-hydroxyl group in RNA. J. Mol. Biol. 1997;274:54–63. doi: 10.1006/jmbi.1997.1370. [DOI] [PubMed] [Google Scholar]
- Badorrek, C.S., Weeks, K.M. Architecture of a γ retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- Badorrek, C.S., Gherghe, C.M., Weeks, K.M. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc. Natl. Acad. Sci. 2006;103:13640–13645. doi: 10.1073/pnas.0606156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Sagar, M.B., Doudna, J.A. Structural and energetic analysis of RNA recognition by a universally conserved protein from the signal recognition particle. J. Mol. Biol. 2001;307:229–246. doi: 10.1006/jmbi.2000.4454. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Gilbert, S.D., Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature. 2004;432:411–415. doi: 10.1038/nature03037. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cate, J.H., Gooding, A.R., Podell, E., Zhou, K., Golden, B.L., Kundrot, C.E., Cech, T.R., Doudna, J.A. Crystal structure of a group I ribozyme domain: Principles of RNA packing. Science. 1996;273:1678–1684. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- Chamberlin, S.I., Weeks, K.M. Mapping local nucleotide flexibility by selective acylation of 2′-amine substituted RNA. J. Am. Chem. Soc. 2000;122:216–224. [Google Scholar]
- Chen, Y., Fender, J., Legassie, J.D., Jarstfer, M.B., Bryan, T.M., Varani, G. Structure of stem–loop IV of Tetrahymena telomerase RNA. EMBO J. 2006;25:3156–3166. doi: 10.1038/sj.emboj.7601195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comarmond, M.B., Giege, R., Thierry, J.C., Moras, D., Fischer, J. 3-dimensional structure of yeast transfer RNAAsp. 1. Structure determination. Acta Crystallogr. B. 1986;42:272–280. [Google Scholar]
- Das, R., Laederach, A., Pearlman, S.M., Herschlag, D., Altman, R.B. SAFA: Semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna, J.A., Grosshans, C., Gooding, A., Kundrot, C.E. Crystallization of ribozymes and small RNA motifs by a sparse matrix approach. Proc. Natl. Acad. Sci. 1993;90:7829–7833. doi: 10.1073/pnas.90.16.7829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron, B. Rietz Lecture—Bootstrap methods—Another look at the jackknife. Ann. Stat. 1979;7:1977. 1–26. [Google Scholar]
- Efron, B., Tibshirani, R.J. An introduction to the bootstrap. Chapman & Hall/CRC; Boca Raton, FL: 1993. [Google Scholar]
- Ferré-D'Amaré, A.R., Zhou, K.H., Doudna, J.A. Crystal structure of a hepatitis δ virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Gesteland, R.F., Cech, T.R., Atkins, J.F. The RNA world. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2005. [Google Scholar]
- Gherghe, C.M., Krahn, J.M., Weeks, K.M. Crystal structures, reactivity and inferred acylation transition states for 2′-amine substituted RNA. J. Am. Chem. Soc. 2005;127:13622–13628. doi: 10.1021/ja053647y. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Podell, E.R., Gooding, A.R., Cech, T.R. Crystals by design: A strategy for crystallization of a ribozyme derived from the Tetrahymena group I intron. J. Mol. Biol. 1997;270:711–723. doi: 10.1006/jmbi.1997.1155. [DOI] [PubMed] [Google Scholar]
- Golden, B.L., Kim, H., Chase, E. Crystal structure of a phage Twort group I ribozyme-product complex. Nat. Struct. Mol. Biol. 2005;12:82–89. doi: 10.1038/nsmb868. [DOI] [PubMed] [Google Scholar]
- Guex, N., Peitsch, M.C. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guo, F., Cech, T.R. Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nat. Struct. Biol. 2002;9:855–861. doi: 10.1038/nsb850. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gooding, A.R., Cech, T.R. Structure of the Tetrahymena ribozyme: Base triple sandwich and metal ion at the active site. Mol. Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Guo, F., Gooding, A.R., Cech, T.R. Comparison of crystal structure interactions and thermodynamics for stabilizing mutations in the Tetrahymena ribozyme. RNA. 2006;12:387–395. doi: 10.1261/rna.2198206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K., Cech, T.R. In vitro selection of RNAs with increased tertiary structure stability. RNA. 1999;5:1119–1129. doi: 10.1017/s135583829999074x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juneau, K., Podell, E., Harrington, D.J., Cech, T.R. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Suddath, F.L., Quigley, G.J., McPherson, A., Sussman, J.L., Wang, A.H., Seeman, N.C., Rich, A. Three-dimensional tertiary structure of yeast phenylalanine transfer RNA. Science. 1974;185:435–440. doi: 10.1126/science.185.4149.435. [DOI] [PubMed] [Google Scholar]
- Leontis, N.B., Westhof, E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews, D.H., Turner, D.H. Prediction of RNA secondary structure by free energy minimization. Curr. Opin. Struct. Biol. 2006;16:270–278. doi: 10.1016/j.sbi.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Merino, E.J., Wilkinson, K.A., Coughlan, J.L., Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Moras, D., Dock, A.C., Dumas, P., Westhof, E., Romby, P., Ebel, J.P., Giegé, R. Anticodon–anticodon interaction induces conformational changes in tRNA: Yeast tRNAAsp, a model for tRNA–mRNA recognition. Proc. Natl. Acad. Sci. 1986;83:932–936. doi: 10.1073/pnas.83.4.932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy, F.L., Cech, T.R. An independently folding domain of RNA tertiary structure within the Tetrahymena ribozyme. Biochemistry. 1993;32:5291–5300. doi: 10.1021/bi00071a003. [DOI] [PubMed] [Google Scholar]
- Ortoleva-Donnelly, L., Szewczak, A.A., Gutell, R.R., Strobel, S.A. The chemical basis of adenosine conservation throughout the Tetrahymena ribozyme. RNA. 1998;4:498–519. doi: 10.1017/s1355838298980086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potterton, E., Briggs, P., Turkenburg, M., Dodson, E. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- Richards, F.M. The interpretation of protein structures: Total volume, group volume distributions and packing density. J. Mol. Biol. 1974;82:1–14. doi: 10.1016/0022-2836(74)90570-1. [DOI] [PubMed] [Google Scholar]
- Robertus, J.D., Ladner, J.E., Finch, J.T., Rhodes, D., Brown, R.S., Clark, B.F., Klug, A. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature. 1974;250:546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Rupert, P.B., Ferré-D'Amaré, A.R. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Scott, W.G., Finch, J.T., Grenfell, R., Fogg, J., Smith, T., Gait, M.J., Klug, A. Rapid crystallization of chemically synthesized hammerhead RNAs using a double screening procedure. J. Mol. Biol. 1995;250:327–332. doi: 10.1006/jmbi.1995.0380. [DOI] [PubMed] [Google Scholar]
- Stahley, M.R., Strobel, S.A. Structural evidence for a two-metal-ion mechanism of group I intron splicing. Science. 2005;309:1587–1590. doi: 10.1126/science.1114994. [DOI] [PubMed] [Google Scholar]
- Stout, C.D., Mizuno, H., Rubin, J., Brennan, T., Rao, S.T., Sundaralingam, M. Atomic coordinates and molecular-conformation of yeast phenylalanyl transfer-RNA—Independent investigation. Nucleic Acids Res. 1976;3:1111–1123. doi: 10.1093/nar/3.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaralingam, M., Stout, C.D., Mizuno, H., Rubin, J., Rao, S.T., Mallikarjunan, M., Brennan, T. Structure of yeast phenylalanine transfer-RNA. Acta Crystallogr. A. 1975;31:S39. [Google Scholar]
- Thore, S., Leibundgut, M., Ban, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Vicens, Q., Westhof, E. Crystal structure of paromomycin docked into the eubacterial ribosomal decoding A site. Structure. 2001;9:647–658. doi: 10.1016/s0969-2126(01)00629-3. [DOI] [PubMed] [Google Scholar]
- Westhof, E., Dumas, P., Moras, D. Crystallographic refinement of yeast aspartic-acid transfer-RNA. J. Mol. Biol. 1985;184:119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.A., Merino, E.J., Weeks, K.M. RNA SHAPE chemistry reveals nonhierarchical interactions dominate equilibrium structural transitions in tRNAAsp transcripts. J. Am. Chem. Soc. 2005;127:4659–4667. doi: 10.1021/ja0436749. [DOI] [PubMed] [Google Scholar]
- Wilkinson, K.A., Merino, E.J., Weeks, K.M. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): Quantitative RNA structure analysis at single nucleotide resolution. Nature Protocols. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons, W.M., Jr., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]