Abstract
RNA interference (RNAi) is an evolutionarily conserved process by which double-stranded small interfering RNA (siRNA) induces sequence-specific, post-transcriptional gene silencing. Unlike other mRNA targeting strategies, RNAi takes advantage of the physiological gene silencing machinery. The potential use of siRNA as therapeutic agents has attracted great attention as a novel approach for treating severe and chronic diseases. RNAi can be achieved by either delivery of chemically synthesized siRNAs or endogenous expression of small hairpin RNA, siRNA, and microRNA (miRNA). However, the relatively high dose of siRNA required for gene silencing limits its therapeutic applications. This review discusses several strategies to improve therapeutic efficacy as well as to abrogate off-target effects and immunostimulation caused by siRNAs. There is an in-depth discussion on various issues related to the (1) mechanisms of RNAi, (2) methods of siRNA production, (3) barriers to RNAi-based therapies, (4) biodistribution, (5) design of siRNA molecules, (6) chemical modification and bioconjugation, (7) complex formation with lipids and polymers, (8) encapsulation into lipid particles, and (9) target specificity for enhanced therapeutic effectiveness.
Keywords: RNA interference, small interfering RNA, bioconjugation, chemical modification, complex formation
INTRODUCTION
RNA interference (RNAi) is an evolutionarily conserved process by which double-stranded small interfering RNA (siRNA) induces sequence-specific, post-transcriptional gene silencing (Hannon 2002). The process of RNAi consists of an initiator step, in which long double-stranded RNA (dsRNA) is cleaved into siRNA fragments, and an effector step, in which these fragments are incorporated into a protein complex, dissociated, and used as a guiding sequence to recognize homologous mRNA that is subsequently cleaved. RNAi is considered a self-defense mechanism of eukaryotic cells to combat infection by RNA viruses and transposons. It is also assumed to tightly regulate protein levels in response to various environmental stimuli (Meister and Tuschl 2004).
The revolutionary finding of RNAi resulted from the work of Andrew Fire and coworkers at the Carnegie Institute in Washington, D.C., who demonstrated in 1998 that injection of dsRNA into Caenorhabditis elegans leads to efficient sequence-specific gene silencing (Fire et al. 1998). At that time, the state of the art in gene silencing was the use of antisense oligodeoxynucleotide (ODNs), which comprise single strands of short DNA or RNA complementary sequences that hybridize with the targeted mRNA (Mahato et al. 2005). However, the dsRNA seemed to induce silencing through a pathway distinct from classical antisense therapies due to the catalytic nature of RNAi, in which one siRNA can be used over and over to guide the cleavage of many mRNA molecules (Dykxhoorn et al. 2003). Bertrand et al. (2002) have compared the effects of antisense ODNs and siRNAs targeting green fluorescent protein (GFP) in vitro and in vivo. siRNA was quantitatively more efficient, and its effect lasted for a longer time in cell culture. In mice, siRNAs were able to silence gene expression, whereas no effect was observed in the presence of antisense ODNs.
The first evidence that siRNAs can mediate sequence-specific gene silencing in mammalian cells was provided in 2001 when the conversion of dsRNA into short RNA fragments was shown to be bypassed by the transfection of siRNA molecules into cells (Elbashir et al. 2001a). After that, various in vivo effects of siRNA and short hairpin RNA (shRNA) have been reported (Lewis et al. 2002; McCaffrey et al. 2002; Xia et al. 2002; Song et al. 2003b). For example, McCaffrey et al. (2002) showed that siRNA and shRNA reduce luciferase (Luc) expression in the liver in a sequence-specific manner. Song et al. (2003b) found that siRNA targeted to the Fas receptor protects mice from liver fibrosis.
Human clinical trials of RNAi-based drugs are currently under way by Acuity Pharmaceuticals and Sirna Therapeutics. Both companies are working on intravitreal administration of siRNA targeting vascular endothelial growth factor (VEGF), whose overexpression is the primary cause of age-related macular degeneration (AMD). The first results are encouraging in terms of tolerability of siRNA compounds. Other clinical trials involving siRNA for treating chronic myeloid leukemia and respiratory syncytial virus infection are also being carried out by the Hadassah Medical Organization and Alnylam Pharmaceutics, Inc., respectively.
Despite early excitement, siRNAs have shown to activate immune response in a sequence- and concentration-dependent manner, leading to nonspecific gene silencing (Jackson et al. 2003; Sioud 2005). In this respect, the introduction of chemical modifications and generation of designed siRNAs have become essential for achieving high gene silencing with a low degree of undesired effects. Chemical modifications of siRNAs also appear to stabilize these molecules in serum and show enhanced gene silencing (Braasch et al. 2003; Chiu and Rana 2003; Layzer et al. 2004). A breakthrough in this area has recently been achieved by linking cholesterol (Chol) to siRNAs (Soutschek et al. 2004), and by encapsulating them into stable nucleic acid lipid particles (Zimmermann et al. 2006). Undoubtedly, effective in vivo delivery of siRNAs will be a key factor in turning siRNA into a new class of therapeutics. In this review, we discuss several strategies to abrogate undesired effects and improve the efficiency of siRNA, including chemical modifications, siRNA sequence design, bioconjugation, complex formation, and encapsulation into lipid particles.
MECHANISM OF RNA INTERFERENCE
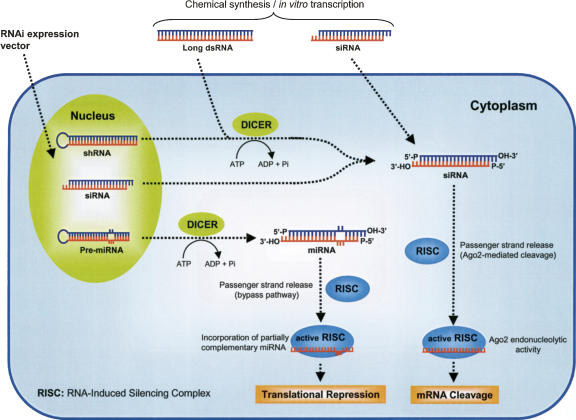
The initial studies that unraveled the RNAi machinery were obtained from experiments using Drosophila extracts. Figure 1 illustrates the mechanism of RNAi. It involves processing of long dsRNA of 500–1000 nucleotides (nt), which triggers into fragments of siRNA. Generally, siRNA is 21–23-nt dsRNA segments with 3′-overhangs of 2 nt on both strands, and unphosphorylated hydroxyl groups at the 2′ and 3′ sites. The siRNA structure is characteristic of an RNase III-like cleavage pattern and plays a crucial role in its recognition by other RNAi components. Dicer is the enzyme known to process dsRNA into siRNAs, and it contains dual catalytic domains and additional helicase and PAZ domains (Elbashir et al. 2001b; Zamore and Haley 2005). Processed siRNA is incorporated into a protein complex, termed RNA-induced silencing complex (RISC). Dicer is also involved in the early steps of RISC formation and may be required for siRNA entry into RISC (Lee et al. 2004). In mammalian cells, the protein Argonaute 2 (Ago2) is the catalytic component of RISC that cleaves target mRNAs (Liu et al. 2004b; Meister et al. 2004). In the RISC assembly process, siRNAs are initially loaded into Ago2 as duplexes, and then Ago2 cleaves the passenger strand, thereby liberating the guide strand from the siRNA duplex and producing active RISC capable of cleaving target mRNAs (Matranga et al. 2005; Leuschner et al. 2006). The guide strand serves as a template for the recognition of homologous mRNA, which upon binding to RISC is cleaved between bases 10 and 11 relative to the 5′ base of the guide siRNA strand by the catalytic activity of Ago2 (Elbashir et al. 2001c). The template siRNA is not affected by this reaction, so the RISC can undergo numerous cycles of mRNA cleavage that comprise the high efficiency of RNAi (Elbashir et al. 2001b; Meister and Tuschl 2004). The mechanism by which passenger strands are cleaved by Ago2 follows the same rules established for the siRNA-guided cleavage of a target mRNA (Leuschner et al. 2006).
FIGURE 1.
Mechanisms of RNA interference (RNAi). RNAi is triggered by long double-stranded RNA (dsRNA), small interfering RNA (siRNA), plasmid or virus-based short hairpin RNA (shRNA), or microRNA (miRNA), and siRNA. Long dsRNA, shRNA, and pre-miRNA are processed by Dicer into 21–23-nt siRNA duplexes with symmetric 2-nt 3′ overhangs and 5′-phosphate groups. Processed siRNAs assemble with cellular proteins to form an RNA-induced silencing complex (RISC). During RISC assembly, one strand (passenger) is eliminated, while the other strand (guide) produces an active RISC, which eventually triggers the sequence-specific mRNA degradation (in case of total complementarity) or translational repression (in case of incorporation of partially complementary miRNA).
RNAi can also be induced by endogenous expression of small regulatory RNAs known as microRNAs (miRNAs) (Fig. 1). The generation of miRNAs occurs via sequential processing and maturation of long primary transcripts (pri-miRNA). Pre-microRNA (pre-miRNA) exits in the nucleus upon cleavage by Drosha and is recognized by the endonuclease Dicer, which processes the pre-miRNA into a 22-nt mature miRNA (Bartel 2004). The mature miRNA is subsequently incorporated into the silencing complex (RISC), where it mediates translational repression. During RISC assembly, the Ago2-mediated cleavage of the passenger strand is blocked by mismatches between the guide and passenger strands of miRNAs. In this case, a slower “backup” pathway dissociates and destroys the passenger strand, liberating active RISC (Matranga et al. 2005; Leuschner et al. 2006). In contrast to siRNA, miRNAs mediate RNAi through a translational inhibition mechanism involving imperfect complementarity to target mRNAs (Jackson et al. 2003; Bartel 2004).
One major difference between mammalian RNAi and RNAi in other eukaryotes is the lack of an amplification system for long-term persistence of RNAi in mammalian cells. For example, in Drosophila, ∼35 molecules of dsRNA can silence ∼1000 copies of a target mRNA per cell and can persist over the course of many generations (Nykanen et al. 2001). In contrast, in mammalian cells RNAi persists effectively only for an average of 66 h due to its dilution during cell divisions (Chiu and Rana 2002).
METHODS OF siRNA PRODUCTION
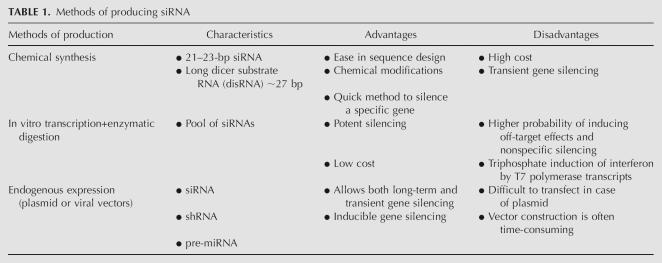
The methods of siRNA production can be classified into three different categories: (1) chemical synthesis, (2) in vitro transcription, and (3) endogenous expression. The choice of production method depends on the target gene, cell type, in vitro or in vivo use, and desired extent of gene silencing. Table 1 describes the advantages and disadvantages of each method of siRNA production.
TABLE 1.
Methods of producing siRNA
Chemical synthesis
Chemical synthesis is the most direct means of generating siRNAs and has several advantages, including precise control of the amount and purity of siRNA, ease in characterization and scale-up, and ease in chemical modifications for enhanced stability and target specificity (Elbashir et al. 2001a). The chemical synthesis of siRNA requires several steps including the generation of two homologous strands, annealing of the strands, addition of chemical entities to increase stability, and ensuring that 2-nt overhangs are present. The siRNA duplex requires a 3′-hydroxyl group and a 5′-phosphate group for functional activity (Dykxhoorn et al. 2003). Unlike DNA, RNA possesses an additional hydroxyl group at the 2′ position of each ribose building block, which destabilizes RNA under the basic conditions generally present in DNA synthesis reactions. Hence, the most difficult step in RNA synthesis is the simultaneous protection of the 5′- and 2′-hydroxyl groups during solid-phase chemistry.
One drawback of using chemically synthesized siRNA is that the most effective target sequence is unpredictable since gene silencing efficiency may vary depending on the segments of the transcripts that are targeted. For example, Holen et al. (2002) observed that only a few siRNAs resulted in a significant reduction of human tissue factor (HTF) expression after targeting its mRNA with several siRNAs synthesized against different sites of the same mRNA. The results suggested that accessible siRNA target sites may be rare in some human mRNAs. However, more effective gene silencing can be achieved by targeting different segments of the same transcript simultaneously. Significant enhancement in gene silencing was achieved by targeting the HIV-1 coreceptor CXCR4 and the apoptosis-inducing Fas ligand with two or more siRNAs against different sites of the same mRNA (Ji et al. 2003).
In vitro transcription and enzymatic digestion
Since chemically synthesized siRNAs are expensive, attempts are being made to produce siRNAs by in vitro transcription using T7 RNA polymerase. Synthetic DNA templates containing the T7 RNA polymerase promoter region followed by desired RNA sequence can be produced using an automated DNA synthesizer and then amplified using PCR. The T7 polymerase binds to the promoter sequence, initiates transcription, and then moves along the template strand toward its 5′ end, elongating the RNA transcript as it goes. Although a termination region on the DNA may be used to halt transcription, the runoff transcription, which stops when the 5′ end of the DNA template strand is reached, is also commonly used. Transcription of the PCR fragment by the T7 RNA polymerase produces both sense and antisense RNAs, which spontaneously anneal, forming full-length dsRNA (Milligan and Uhlenbeck 1989; Donze and Picard 2002).
The in vitro transcription approach is limited by specific sequence requirements related to T7 polymerase. The last guanosine of the T7 promoter is invariably the first ribonucleotide that is incorporated into the RNA by the T7 polymerase during transcription. Therefore, all siRNAs produced by this method start with a 5′-G residue and require a C-3′ residue at position 19 (i.e., 5′-G-N17-C-3′) to allow annealing with the complementary RNA, which also has to start with a 5′-G residue. Furthermore, since the efficacy of siRNAs targeted to different regions of a gene varies dramatically, the number of sequences that can be targeted using siRNAs generated by this method is limited (Donze and Picard 2002). Kim et al. (2004) demonstrated that siRNAs synthesized from the T7 polymerase system can trigger a potent induction of interferon α and β in a number of cell lines. The induction of these interferons was also seen by short single-stranded RNAs transcribed with T3, T7, and Sp6 RNA polymerases. These investigators further demonstrated that the presence of triphosphates on the 5′ end of T7 transcripts is required for interferon induction, since the treatment of T7 transcripts with phosphatase could abrogate this effect.
Sohail et al. (2003) reported an alternative approach for producing a desired siRNA sequence using T7 polymerase in vitro transcription followed by transcript digestion by deoxyribozymes, which are known as “DNAzymes” or catalytic DNA. The cleavage of RNA by DNAzymes occurs between two specific nucleotides, and the requirement for this dinucleotide sequence is different for different DNAzyme groups, making them flexible tools for digesting a variety of sequences (Breaker and Joyce 1994; Feldman and Sen 2001). The siRNAs produced by this method caused dose-dependent inhibition of type 1 insulin-like growth factor receptor in human breast cancer cells comparable to that induced by chemically synthesized siRNAs of the same sequence (Sohail et al. 2003).
Both recombinant human Dicer (re-hDicer) and Escherichia coli RNAse III are used to cleave long dsRNA produced by in vitro transcription. Treatment with E. coli RNase III or re-hDicer randomly cleaves the RNA into a population of siRNA molecules that are effective mediators of gene silencing in a manner similar to that observed when using synthetic siRNA. Usually, the cleavage products of E. coli enzyme RNase III range from 12 to 15 base pairs (bp) in length with termini identical to those produced by Dicer (Amarasinghe et al. 2001). Although these short dsRNAs are not long enough to trigger an RNAi in mammalian cells, the average product length generated by RNase III digestion can be increased by altering digestion conditions. Yang et al. (2002) have shown that 20–25-bp siRNAs produced by RNase III digestion efficiently inhibited various endogenous genes in different mammalian cell lines without nonspecific effects. In addition, RNase III can digest dsRNA faster than re-hDicer and can be easily overexpressed and purified (Amarasinghe et al. 2001; Bernstein et al. 2001). The siRNAs produced by re-hDicer are 21–23 bp in length and contain 2-nt 3′ overhangs with 5′-phosphate and 3′-hydroxyl termini, which are essential for RNAi activity. Dicer-generated siRNAs have been shown to be effective in silencing exogenous and endogenous gene expression in mammalian cells (Kawasaki et al. 2003; Myers et al. 2003).
The enzymatic synthesis of siRNA may provide more effective gene silencing than chemically synthesized siRNA, as the enzymatically generated siRNA can correspond to sequences overlapping the entire gene. However, complex siRNA populations may have a higher probability of targeting other genes, and therefore promoting nonspecific effects, than do discrete siRNAs. Additionally, enzymatic synthesis of siRNA requires separation of siRNA from uncleaved RNA duplexes and residual nucleic acids (Yang et al. 2002; Myers et al. 2003).
Endogenous expression
The application of synthetic siRNAs is restricted by both low-to-moderate transfection efficiency and the short-term persistence of transient gene expression. A single transfection of siRNA may not provide a sufficient window of functional depletion for proteins with long half-lives. Another potential problem inherent in chemically synthesized siRNA is variability in transfection efficiency, especially in difficult-to-transfect cells. To circumvent these limitations, expression vectors currently in use employ siRNA or shRNA expression cassettes that resemble pre-miRNAs and undergo processing by Dicer. Like synthetic siRNAs, they are designed to pair perfectly with the target mRNA to induce RNAi. These shRNAs are designed for either transient or long-term gene silencing and can be produced from plasmid or viral expression vectors (Amarzguioui et al. 2005).
Plasmid vectors
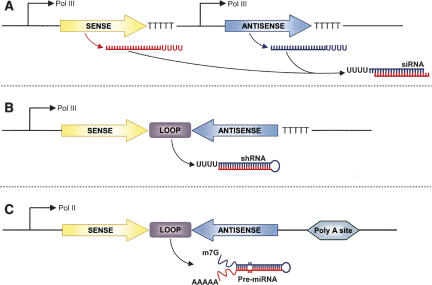
shRNA, siRNA, and miRNA can be produced from plasmid vectors containing promoters that are dependent on either RNA polymerase (Pol) II or Pol III. Among them, Pol III promoters are used most frequently because it is possible to express small RNAs that carry the structural feature of siRNA. Figure 2 is a schematic representation of different strategies used to create expression cassettes using RNA polymerase promoters for generation of siRNA, shRNA, and miRNA. In the first strategy, the sense and antisense strands are expressed as two independent transcripts that hybridize within the cells to form functional siRNA duplexes (Fig. 2A). In the second strategy, the sense and antisense strands are expressed as a single transcript separated by a short loop of 4–10 nt of sequence. The transcript forms a hairpin structure that can be processed by Dicer into functional siRNA (Fig. 2B). In the third strategy, miRNAs that are complementary to the target gene are expressed using the Pol II promoter (Fig. 2C; Dykxhoorn et al. 2003; Sioud 2004).
FIGURE 2.
Schematic representation of expression cassettes using RNA polymerase promoters for generation of small-interfering RNAs (siRNA). (A) Tandem-type promoters express sense and antisense strands individually. After transcription, both strands hybridize forming a duplex siRNA. (B) Short hairpin RNAs (shRNA) are expressed as a single transcript separated by a short loop of 4–10 nt of sequence. The transcripts form a hairpin structure that can be processed by Dicer into functional siRNAs. (C) Expression of imperfect duplex hairpin structures that is based on pre-microRNA (pre-miRNA) structures. pre-miRNAs are processed by Dicer into a mature miRNA, which can direct gene silencing.
Two different Pol III promoters predominantly being used for shRNA expression are the U6 promoter and the H1 promoter (Brummelkamp et al. 2002; Paddison et al. 2002). In addition to these two promoters, other Pol III promoters such as 5S, 7SK, and tRNA promoters have also been used for expressing siRNAs endogenously (Paul et al. 2002; Czauderna et al. 2003b; Kawasaki and Taira 2003). The activities of these promoters vary from cell type to cell type. To optimize shRNA expression, it is beneficial to create expression vectors with at least two different promoters and transfect them into the cells being targeted for gene knockdown. Construction of shRNA expression vectors poses two serious technical challenges. First, it is difficult to sequence constructs that contain a hairpin region, probably because of the tight palindromic structure. Second, ∼20%–40% of constructs get mutated within the hairpin region (Brummelkamp et al. 2002; Paddison et al. 2002; Paul et al. 2002).
Viral vectors
The introduction of siRNA expression plasmids into cells often requires electroporation, microinjection, or complex formation with synthetic carriers (lipids, polymers, or peptides). While most rapidly dividing cell lines are easily transfected using shRNA expression plasmids, these plasmid vectors are not easily transfected into primary cells, stem cells, and nondividing cells. In the absence of cell division, the siRNA expression plasmids cannot be introduced into the nucleus, where the DNA is transcribed. To overcome this limitation, different viral vectors encoding shRNA including retroviral, adenoviral, and adeno-associated viral (AAV) are being developed. Typically, these vectors use a Pol III promoter, such as U6, H1, or transfer RNA promoters (Dykxhoorn et al. 2003).
Retroviral vectors have been reported to mediate an efficient and stable siRNA expression (Rubinson et al. 2003; Liu et al. 2004a). Unlike Moloney murine leukemia virus (MoMLV), lentiviral vectors efficiently integrate into the genome of nondividing cells, such as pancreatic islets, hematopoietic stem cells, or terminally differentiated cells. A lentiviral vector encoding shRNA has been shown to effectively silence GFP, BCL-2, and Interleukin (IL) 12 receptor (CD25) genes (Tiscornia et al. 2003; Schomber et al. 2004; Wong et al. 2005). Lentiviral vectors encoding shRNA have also been shown to inhibit HIV-1 infection in hematopoietic stem cells and human CD4+ T-cells (Li et al. 2003; Nishitsuji et al. 2006). However, lentiviral vectors are associated with infrequent insertional mutagenesis. Consequently, the use of adenoviral vectors is being explored since these vectors do not integrate into the host genome and efficiently transduce both dividing and nondividing cells (Wu et al. 2003). Adenoviruses contain a linear double-stranded DNA genome that remains episomal after infection. Recombinant adenoviral vectors containing expression cassettes of interest are readily generated and can be purified to very high titers (up to 1013 infection units/mL) (Huang and Kochanek 2005).
A variety of properties make AAV vectors an interesting tool for organ-directed shRNA expression, including the lack of pathogenicity, and ease of vector production. The AAV-2 type is highly prevalent in the human population and frequently neutralized by antibodies. Therefore, it is important to evaluate different AAV serotypes for organ-directed shRNA expression. AAV-8 vectors expressing shRNA are reported to transduce almost 100% of hepatocytes after intravenous injection into mice (Grimm and Kay 2006). However, overexpression of shRNA caused length- and dose-dependent liver injury and ultimate death. Morbidity was associated with the down-regulation of liver-derived miRNA, indicating possible competition between miRNA and shRNA for nuclear Krayopherin exportin 5 (Grimm et al. 2006). Therefore, it is important to optimize siRNA dose, length, and sequence to avoid oversaturation of endogenous small RNA pathways.
Inducible expression vectors
Plasmid- and viral-vector-based constitutive expressions of shRNAs by RNA Pol III U6 and H1 promoters often fail to adjust the levels of gene silencing necessary for cell survival and growth. Besides, gene silencing for longer periods may result in nonphysiological responses. This problem can be circumvented by generating inducible regulation of RNAi (Czauderna et al. 2003b; Wiznerowicz and Trono 2003; Gupta et al. 2004). Ohkawa and Taira (2000) described the successful regulation of gene silencing by the integration of the bacterial tetracycline operon sequence (tetO) into the U6 promoter. Matsukura et al. (2003) have applied this tetracycline-inducible U6 promoter for in vivo transcription of shRNA, which enables stable transfection followed by conditional expression of shRNA. The main drawback of the tetracycline-inducible system is a relatively high background expression in the uninduced state in certain cell lines (No et al. 1996; Van Craenenbroeck et al. 2001). In contrast, the ecdysone-inducible system is tightly regulated, with no expression in the uninduced state and a rapid inductive response (Gupta et al. 2004).
Owing to the time-consuming process of cloning siRNA into plasmid constructs and the need for verification of the cloned sequence, an easier approach of screening sequences involving the production of siRNA expression cassettes (SECs) was developed (Castanotto et al. 2002). SECs are PCR-derived siRNA expression templates introduced directly into the cells. SECs consist of a Pol III promoter, a sequence encoding an shRNA, and an RNA polymerase III termination site. The final result is a PCR product that contains a Pol III promoter, a DNA sequence that, once transcribed, forms shRNA, and a terminator sequence. After transcription, the shRNA is intracellularly cleaved into siRNA by the RNAi machinery. By incorporating restriction sites at their ends, SECs effectively elicit gene silencing and can be cloned into a plasmid to create an siRNA expression vector. The expediency and low cost of this procedure lends itself to mass screening of siRNA libraries as well as identification of siRNA target sites (Chang 2004; Wooddell et al. 2005).
BARRIERS TO RNAi-BASED THERAPIES
RNAi intersects a number of pathways and siRNA transfection often activates immune response leading to unintended and off-target effects. Therefore, the potential induction of inflammatory cytokines and interferon (IFN) responses by siRNAs represent a major obstacle for their use as inhibitors of gene expression. Therapeutic applications of siRNAs require a better understanding of the mechanisms that trigger such unwanted effects.
Immune response activation
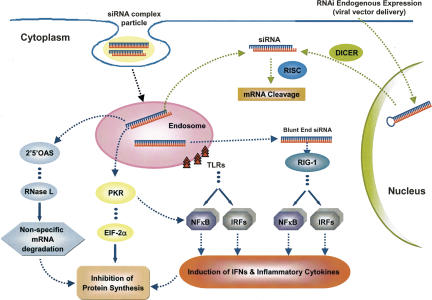
Figure 3 is a schematic representation of the immune response activation by siRNAs. Non-immune and immune cells get activated in the presence of long dsRNA, leading to the activation of cytoplasmic receptors such as the dsRNA-dependent protein kinase R (PKR) and the retinoic acid-inducible gene-I (RIG-I) (Saunders and Barber 2003; Yoneyama et al. 2004). PKR is activated by autophosphorylation following binding to dsRNA. Once activated, it phosphorylates the eukaryotic translation initiation factor (EIF-2)-α, leading to the global suppression of protein biosynthesis and subsequent programmed cell death. PKR can also activate nuclear factor κB (NF-κB) with consequent induction of type-I IFN production. A family of 2′-5′-oligoadenylate synthetases (2′5′-OAS) is also activated by dsRNA. This leads to the activation of RNase L, which eventually triggers the nonspecific degradation of mRNA (Marques and Williams 2005).
FIGURE 3.
Activation of immune response by siRNAs. siRNA/shRNA can be delivered by two different ways: (1) Delivery of viral vector followed by endogenous expression of shRNA in the nucleus, which is transported to the cytoplasm and processed by Dicer into siRNAs. The siRNA causes sequence-specific mRNA degradation and does not induce an interferon (IFN) response. (2) Synthetic delivery systems forming an siRNA complex particle, for example, liposome. In this way, the siRNA is taken up by the cell via endocytosis, causing sequence-specific mRNA cleavage through RNAi and activation of the immune response. The immune response can be activated by different pathways: (a) recognition of siRNAs by Toll-like receptors (TLRs); (b) activation of retinoic acid-inducible gene I (RIG-I) by blunt-end siRNAs; (c) activation of dsRNA-dependent protein kinase R (PKR); (d) activation of 2′-5′-oligoadenylate synthetases (2′5′-OAS). Generally, activation of these pathways leads to the induction of several cellular factors, including nuclear factor κB (NF-κB), interferon regulatory factors (IRFs), eukaryotic translation initiation factor 2α (EIF-2α), and RNase L. Induction of interferons (IFNs), inflammatory cytokines, nonspecific mRNA degradation, and general inhibition of protein synthesis are some undesired effects of the immunostimulation caused by siRNAs.
RIG-I is an intracellular dsRNA sensor capable of triggering IFN production. Diverse cell types derived from RIG-I knockout mice have impaired responses to viral dsRNA, establishing the essential role of RIG-I in the mammalian antiviral response (Yoneyama et al. 2004; Marques and Williams 2005). The helicase RIG-I is necessary for recognizing blunt-ended siRNAs, leading to the activation of dsRNA signaling. RIG-I unwinds siRNAs containing blunt ends more efficiently than siRNAs containing 3′ overhangs. The efficiency in duplex unwinding is then translated into signaling downstream to interferon regulatory factor 3 (IRF-3) and NF-κB activation (Marques et al. 2006).
Mammalian immune cells express a family of toll-like receptors (TLRs), which recognize pathogen-associated molecular patterns including unmethylated cytosine-guanine motifs (commonly known as CpG motifs) and viral dsRNA. TLR3, the receptor for dsRNA, was a logical candidate for recognizing siRNA in the context of immunostimulation. Indeed, TLR3 overexpressed in cultured human embryonic kidney (HEK) 293 cells was capable of recognizing siRNAs (Marques and Williams 2005). However, the activation of immune cells by siRNAs is sequence dependent, and either sense or antisense strands individually can induce inflammatory cytokine production as efficiently as duplex siRNAs (Judge et al. 2005; Sioud 2005, 2006). Thus, as TLR3 neither recognizes single-stranded RNA (ssRNA) nor requires sequence specificity, it is unlikely involved in activation of the immune system by siRNAs (Marques and Williams 2005; Sioud 2005).
TLR7 and TLR8 were initially shown to mediate the recognition of RNA viruses and small synthetic antiviral compounds referred to as imidazoquinolines. TLR7, TLR8, and TLR9 are expressed in endosomes and require endosomal maturation for efficient signaling (Marques and Williams 2005). siRNA recognition by TLR7, TLR8, and TLR9 results in activation of NF-κB and IRFs, which induce inflammatory cytokines and IFNs, respectively (Fig. 3; Marques and Williams 2005). Interestingly, not all ssRNA molecules are capable of activating TLR7 and TLR8, but, rather, U- and G-rich ssRNAs seem to be preferentially recognized (Sioud 2006). It has now become clear that TLR7 and TLR8 mediate the recognition of siRNAs in a sequence-dependent manner (Heil et al. 2004; Hornung et al. 2005; Judge et al. 2005).
Recognition of siRNAs by TLRs takes place in the endosome, before the siRNAs enter the cytoplasm. Therefore, it is expected that if siRNAs could enter the cytoplasm avoiding the endosome, they should bypass the activation of immune systems but still mediate gene silencing. Robbins et al. (2006) tested the immunostimulatory effects of lipid-delivered siRNAs versus Pol III promoter-expressed shRNAs in primary CD34+ progenitor-derived hematopoietic cells. They observed that in this system, cationic lipid/siRNA complexes are potent inducers of IFN-α and type-I IFN gene expression, whereas the same sequences when expressed endogenously are nonimmunostimulatory.
The method of siRNA delivery also has a great influence on the immunostimulatory activity of siRNAs (Fig. 3). Injection of siRNAs after complex formation with cationic liposomes into mice induces the release of inflammatory cytokines, including IL-6, tumor necrosis factor-α (TNF-α), and IFN-α (Judge et al. 2005; Sioud 2005). In contrast, injection of naked, unmodified siRNAs or siRNAs conjugated to cholesterol has no significant effect on immunostimulation (Heidel et al. 2004; Marques and Williams 2005). There are two possible explanations for the absence of immunostimulation by naked siRNAs. First, unmodified siRNAs have a short half-life in serum and may be degraded before being recognized by specific receptors. Second, cationic lipid/siRNA complexes are more readily recognized by immune cells than are naked siRNAs. In fact, Sioud (2005) reported that naked siRNAs containing a phosphorothioate (PS) backbone were not immunostimulatory. However, these results should be interpreted carefully considering that naked CpG ODNs are known to be immunostimulatory independent of backbone modifications (Marques and Williams 2005).
Off-target effects and nonspecific gene silencing
For its therapeutic applications, siRNA must not cause any unintended effect other than sequence-specific gene silencing. However, recent studies indicate that there are off-target effects associated with the use of siRNA (Jackson et al. 2003). Off-target effects consist of any gene silencing effect caused by siRNAs in nontarget mRNAs through the RNAi mechanism. Generally, it occurs due to lack of complementarity between siRNAs and target mRNAs. An explanation for the fact that siRNAs can induce silencing of nontarget genes can be found in the RNAi machinery. Although the actual substrate specificity of individual siRNAs appears to be very high, RNAi machinery can tolerate single mutations located in the center of the siRNA molecule without losing the ability to induce gene silencing. It makes some siRNAs able to silencing other than those genes related to their homologous mRNA. However, complete inactivation of the RNAi mechanism can occur if four or more mutations are introduced in the guide strand, making the active RISC unable to recognize the target mRNA (Jackson et al. 2003; Persengiev et al. 2004; Sioud 2004).
siRNAs and miRNAs were found to be functionally interchangeable. siRNAs can act as miRNAs depending on the degree of complementarity with the target mRNA. If synthetic siRNAs bear a sufficiently low degree of complementary bases, target mRNA translation occurs instead of mRNA degradation, whereas miRNAs will lead to mRNA degradation if perfect complementarity with target mRNA exists. This process is one of the reasons for the off-target effects of siRNAs (Jackson et al. 2003; Bartel 2004). Another reason for the occurrence of off-target effects is that not only the antisense but also the sense strand of an siRNA can direct gene silencing of nontarget genes, and it has been documented to occur when as few as 15 bp of complementarity exists between the siRNA and mRNA (Jackson et al. 2003). Birmingham et al. (2006) also demonstrated that the presence of one or more perfect matches between the hexamer or heptamer seed (positions 2–7 or 2–8 of the antisense strand) of an siRNA and the 3′-untranslated region (UTR), but not the 5′-UTR or open reading frame, is associated with off-targeting. A high proportion of “off-target” transcripts silenced by siRNAs have been shown to have 3′-UTR sequence complementarity to the seed region of the siRNA, since the base mismatches within the siRNA seed region reduced the set of original off-target transcripts. Since there is no algorithm that can eliminate significant numbers of 7–8-base matches of siRNAs to the transcriptome, it will be difficult to achieve perfect specificity (Jackson et al. 2006). These findings suggest a strong mechanistic parallel between siRNA off-targeting and miRNA-mediated gene regulation.
Nonspecific gene silencing is those effects caused by siRNAs through any pathway other than RNAi. Semizarov et al. (2003) investigated the overall cellular effects of siRNAs on transcription levels. In a concentration-dependent manner, siRNAs nonspecifically interfered in the expression of a significant number of genes, many of which are known to be involved in apoptosis and stress response. Persengiev et al. (2004) reported that treatment with siRNA may result in diverse cellular activities, such as cell signaling, cytoskeletal organization, gene expression, metabolism, and cell adhesion. Jackson et al. (2003) attributed the nonspecific effects to cross-hybridization of transcripts containing regions of partial homology with the siRNA sequence. However, the nonspecific effects observed by Persengiev et al. (2004) cannot be explained by off-target regulation because the siRNAs tested lacked significant sequence similarity to any human gene. Moreover, for all of the genes tested, expression was affected by two siRNAs with sequences that are completely unrelated. Bridge et al. (2003) also reported that shRNAs can affect the expression of many genes, including several IFN targets. The activation of sequence-independent inhibition of gene expression by siRNA or shRNA-expression vectors seems to be, at least in part, due to the induction of type-I IFN, PKR, and signaling through TLR3 (Kariko et al. 2004; Marques and Williams 2005).
BIODISTRIBUTION OF siRNA
To develop effective delivery systems of chemically synthesized siRNA, it is important to gain in-depth understanding of its biodistribution and pharmacokinetic properties at whole-body, organ, and cellular levels.
siRNA duplexes are much more stable than ssRNAs. However, since they are similar to single-stranded antisense oligonucleotides, they still tend to degrade on incubation with serum, contributing to their short half-lives in vivo. 125I-labeled siRNAs are widely distributed in the body after intravenous injection into mice, with preferential accumulation in the liver and kidney (Braasch et al. 2004). Radiolabeled siRNA is also distributed to the heart, spleen, and lung. However, very little siRNA is detected in the brain (Braasch et al. 2004; Soutschek et al. 2004; Santel et al. 2006). Intravenous administration of naked siRNA in rats showed a short half-life of 6 min and a clearance of 17.6 mL/min. The poor pharmacokinetic properties of siRNAs are related to their low in vivo stability, since they can be degraded by endogenous RNases. Another reason is their fast elimination by kidney filtration because of their small molecular mass (∼7 kDa) (Soutschek et al. 2004). Consequently, improving the pharmacokinetic properties of siRNA duplexes is an important goal for developing siRNA-mediated gene silencing.
STRATEGIES FOR IMPROVING RNAi-BASED THERAPIES
To develop strategies for improving RNAi, we need to address several problems, including the enzymatic stability, cellular uptake, pharmacokinetic properties, and potential for off-target effects and immunoactivation of siRNAs. The sequence design of siRNA molecules is extremely important to improve their efficacy as well as to reduce the potential for off-target effects and activation of the immune response (Reynolds et al. 2004). Backbone modifications and bioconjugation with lipids and peptides are known to improve the stability and cellular uptake of siRNAs (Chiu and Rana 2003; Lorenz et al. 2004). Several reagents have also been employed for systemic administration of siRNAs as an alternative to the covalent modifications of the siRNA molecule (Simeoni et al. 2003; Morrissey et al. 2005b; Urban-Klein et al. 2005; Santel et al. 2006). Cationic liposomes are routinely used for transfection of nucleic acids into mammalian cells. Positively charged carriers, such as protamine-antibody fusion proteins, polyethyleneimine (PEI), and cyclodextrin-containing polycation, have been tested for complex formation with siRNAs. To achieve cell-specific delivery of siRNAs, several targeting ligands, including cell membrane receptors and antibodies, have been explored (Song et al. 2005).
Design of siRNAs
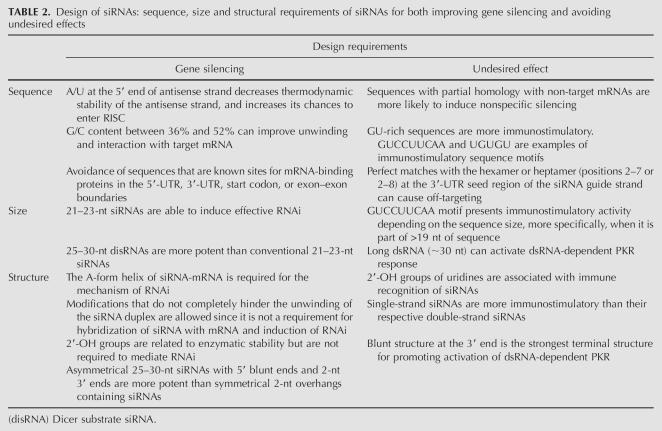
A thorough understanding of the sequence, size, and structural requirements of siRNAs is essential to effectively mediate RNAi. Table 2 summarizes the most important siRNA design rules to improve gene silencing and avoid undesired effects.
TABLE 2.
Design of siRNAs: sequence, size and structural requirements of siRNAs for both improving gene silencing and avoiding undesired effects
siRNA sequence
Some research groups have elaborated several guidelines for designing siRNAs that can potentially silence gene expression (Elbashir et al. 2002; Paddison et al. 2002; Reynolds et al. 2004). Several sequence motifs are consistent with effective siRNAs, including AAN19TT, NAN19NN, NARN17YNN, and NANN17YNN (where N is any nucleotide, R is a purine, and Y is a pyrimidine). In principle, any region of the mRNA can be targeted. However, it should be pointed out that complementarity is often found in the 3′-UTR of the off-target sequences. Furthermore, sequence complementarity between the 5′ end of the guide strand and the mRNA is known to be the key to off-target silencing. Additionally, intronic sequences must be avoided since RNAi is a cytoplasmic process (Dykxhoorn et al. 2003).
Ui-Tei et al. (2004) proposed that highly effective RNAi can be achieved if siRNA satisfying the following four sequence conditions at the same time is used: (1) AU richness in the 5′-terminal, 7-bp-long region of the antisense strand; (2) G/C at the 5′ end of the sense strand; and (3) the absence of any long GC stretch of >9 bp in length. Even though Elbashir et al. (2002) reported that the target region should be at least 50–100 nt downstream from the start codon, Dykxhoorn et al. (2003) suggest that there is a predisposition for effective siRNA-directed silencing toward the 3′ portion of the gene. Moreover, sequences with even representation of all nucleotides on the antisense strand are favored, and those regions with stretches of a single nucleotide, especially G, should be avoided as G-rich oligonucleotides have a tendency to form quartets. Reynolds et al. (2004) analyzed 180 siRNAs targeting every other position of two 197-base regions of firefly luciferase and human cyclophilin B mRNA in a total of 90 siRNAs per gene. A two-base shift in the target position was sufficient to significantly alter siRNA activity, suggesting that its functionality is determined by the siRNA-specific properties. Most potent siRNA has a G/C content ranging from 36% to 52%. siRNA duplexes with a GC content >52% may have difficulty in dissociating into passenger and guide strands, while siRNAs with a GC content <30% may interact less well with the mRNA recognition site (Dykxhoorn and Lieberman 2006).
Low internal stability of the siRNA at the 5′ antisense end is a prerequisite for effective silencing and probably important for duplex dissociation after assembly with RISC (Schwarz et al. 2003). The asymmetric RISC formation model proposed by Schwarz et al. (2003) predicts that duplex siRNA dissociation preferably occurs at an “easier” duplex end, possessing A:U, G:U, or unpaired bases at its 5′-end position and being thermodynamically less stable. Consequently, the end of the siRNA with the lower thermodynamic stability will favor selection of the antisense strand into RISC as the guide strand. The importance of thermodynamically unstable or flexible base pairs at or near the guide strand for siRNA duplex dissociation in HEK 293 cells has also been pointed out by Khvorova et al. (2003).
The immunostimulatory activity of an siRNA duplex is, at least in part, a function of its nucleotide sequence. The GU-rich sequence seems to be a common feature among the immunostimulatory motifs: UGUGU and GUCCUUCAA are present in the immunostimulatory ssRNAs. A single U-to-C base substitution in the 5′-UGUGU-3′ siRNA sequence substantially reduces the siRNA-mediated induction of IFN-α by human peripheral blood mononuclear cells. In contrast, the opposite substitution forming a 5′-UGU-3′ motif renders the siRNA immunostimulatory activity. The sequence recognition mechanism is stringent enough that minimal base substitutions can have profound effects on the immunostimulatory capacity of the siRNAs. However, it is important to note that several immunostimulatory siRNAs do not contain either of these motifs, whereas others are not stimulatory despite being U- and G-rich (Heil et al. 2004; Judge et al. 2005).
Targeting the human TLR-9 gene with a pool of siRNAs in plasmacytoid dendritic cells (PDC) (also called IFN producing cells), Hornung et al. (2005) found that nine bases at the 3′ end (5′-GUCCUUCAA-3′) of the sense strand of siRNA are responsible for its immunostimulatory activity. The immunostimulation and gene silencing were shown to be two independent functional activities. Regardless of the GU content, it seems that TLR7 and/or TLR8 can also recognize dsRNA with similar efficacy (Heil et al. 2004). Indeed, Hornung et al. (2005) argued that the mechanism involved in single-strand siRNA recognition by TLRs is also responsible for recognizing the immunostimulatory motifs within the siRNA duplexes. Nonetheless, to date, there are only two siRNA motifs known to effectively activate innate immunity in the context of double-strand siRNA to a level comparable to that obtained with free single-strand RNA. siRNA duplexes seem to be less effective than siRNA in activating innate immunity. The intracellular receptors for single-strand siRNA, in particular TLR8 and TLR7, do not effectively recognize most immunostimulatory RNA motifs in the context of siRNA duplexes (Sioud 2006).
A number of academic and commercially affiliated Web-based softwares have been developed to assist researchers in the identification of efficient siRNA sequences (Naito et al. 2004; Yuan et al. 2004). Since the rules that govern efficient siRNA-directed silencing are still unknown, researchers seeking to silence gene expression should synthesize several siRNAs to a gene and validate empirically the efficiency of each one. To ensure that the chosen siRNA sequence targets a single gene, a search of the selected sequence should be carried out against sequence databases. This can be done using the Smith–Waterman algorithm or the Basic Local Alignment Search Tool (BLAST) located at the National Center of Biotechnology Information Web site (http://www.ncbi.nlm.gov). Sequences in these databases that share partial homology with siRNAs might be targeted for silencing by the siRNAs. Potential off-target effects of the siRNAs might be minimized by choosing an siRNA with maximal sequence divergence from the list of genes with partial sequence identity to the intended mRNA target.
siRNA size
Synthetic RNA duplexes of 25–30 nt in length (more specifically, 27 nt), which are Dicer substrates, have been shown to be up to 100-fold more potent than corresponding conventional 21-nt siRNAs (Kim et al. 2005a). The use of 27-nt dsRNAs, also called Dicer substrate dsRNA (disRNA), allows targeting of some sites within a given sequence that are refractory to suppression with traditional 21-nt siRNAs. The use of disRNAs to trigger RNAi can result in enhanced and prolonged gene silencing at lower concentrations than those required for conventional 21-nt siRNAs applications (Kim et al. 2005a). It is important to note that in vitro Dicer cleavage of 27-nt disRNAs before transfection does not significantly enhance gene silencing efficiency. Additionally, it has been shown that chemically synthesized shRNAs that are Dicer substrates are more potent inducers of RNAi than conventional siRNAs. Moreover, a two-base 3′ overhang directs Dicer cleavage (Siolas et al. 2005). The enhanced potency of the longer duplexes is attributed to the fact that they are substrates of the Dicer endonuclease, directly linking the production of siRNAs for incorporation into the RISC (Kim et al. 2005a).
The size of siRNA also plays an important role in the activation of immune response in a sequence-independent manner. Although it was initially postulated that a minimum of 30 nt was necessary to activate dsRNA signaling through PKR activation, shorter siRNAs, as few as 21–23 nt, are capable of binding and activating PKR in vitro (Marques et al. 2006). In addition, Hornung et al. (2005) observed that 12-nt ssRNAs containing the immunostimulatory motif (GUCCUUCAA) were poor inducers of IFN-α in PDCs. However, the investigators observed that the motif identified needs to be part of a longer ODN sequence, at least 19 bases, to induce IFN-α activation. The approach reported by Kim et al. (2005a), which consists of Dicer substrate 27-nt dsRNA with increased RNAi potency relative to conventional 21-nt siRNAs, may facilitate the use of lower concentrations of duplex RNA. Therefore, since the undesired effects can occur in an siRNA dose-dependent manner, the use of Dicer substrate 27-nt dsRNA may be an alternative to overcome this problem without losing efficacy.
siRNA structure
According to Chiu and Rana (2002), the A-form helix of the guide strand-mRNA duplex is required for the mechanism of RNAi. Chemical modifications disrupting the functional groups of the major groove of the A-form helix formed by the guide strand and its mRNA specifically at the cleavage site inhibit RNAi. The major groove is also required for promoting RNA–RISC interactions that subsequently lead to mRNA cleavage (Chiu and Rana 2002, 2003).
The 2′-OH of the ribonucleotide of RNAs is required for the nucleophilic attack during the hydrolysis of the RNA backbone, the reaction catalyzed by degradative RNases (Chiu and Rana 2003). However, 2′-OH is not required for RNAi-mediated degradation, and, even more specifically, it is not required for nucleotide base-pairing with nucleotides lining the mRNA cleavage site (Chiu and Rana 2003). Additionally, Sioud (2006) has shown that the 2′-OH of uridines is required for immune recognition and signaling of siRNAs, being responsible, at least in part, for activation of the immune response.
New design approaches that improve the performance of long dsRNAs as Dicer substrates have been reported (Kim et al. 2005a). These new approaches include 25–30-nt asymmetric dsRNAs with a 5′ blunt end and a 2-nt 3′ overhang on the other end. The improved efficacy of disRNAs is postulated to result from their recognition and cleavage by Dicer, followed by their subsequent incorporation into the RISC complex. This interpretation is supported by observations that Drosophila Dicer is not only instrumental in handing over siRNA to nascent RISC, but it is itself a component of the latter (Lee et al. 2004). Providing the RNAi machinery with a Dicer substrate presumably results in more efficient incorporation of the active 21-nt siRNA strand into RISC. These modifications also direct the way Dicer processes the dsRNA substrate, resulting in a more homogeneous pool of siRNA products.
The extremities of the siRNA duplex are critical determinants of its capacity to trigger immune response activation. A blunt structure at the 3′end is the strongest terminal structure for promoting activation of dsRNA signaling through the PKR pathway, followed by a 5′overhang (Marques et al. 2006). In contrast, 3′overhangs, normally present in endogenous Dicer products such as miRNAs, allow RNAi to proceed without activation of dsRNA-dependent PKR (Marques et al. 2006). The presence of the 3′ overhangs also precludes activation of dsRNA signaling by siRNAs and reveals an important basis for discrimination between self and nonself dsRNAs. Interestingly, this type of discrimination appears to take place after RIG-I binds to dsRNAs (Marques et al. 2006).
Chemical modifications
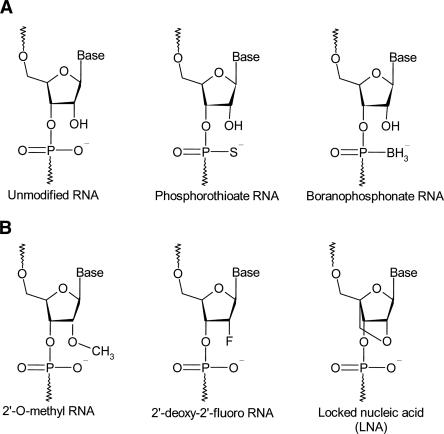
Chemical modifications to sugars, backbones, or bases of oligonucleotides can be used to control their pharmacokinetic profiles and reduce nonspecific effects without affecting their biological activity. The valuable know-how of backbone modifications of antisense ODNs can be readily adapted to develop new siRNA technologies (Marshall and Kaiser 2004). Although siRNA duplexes used for silencing are inherently more stable than ssRNAs, there are reasons to chemically modify one or both strands. Apart from improving stability and reducing off-target effects, chemical modifications can aid in broadly targeting siRNAs into cells and tissues, and certain conjugates can enhance uptake in specific cell types. Multiple chemical modifications can be introduced at various positions with the siRNA duplexes. Figure 4 shows the most common chemical modifications introduced in siRNAs.
FIGURE 4.
Most common chemical modifications introduced in siRNAs. (A) Phosphodiester modifications: unmodified siRNA (phosphodiester RNA); phosphorothioate RNA; boranophosphonate. (B) 2′-Sugar modifications: 2′-O-Methyl RNA; 2′-deoxy-2′-fluoro RNA; locked nucleic acid (LNA).
Phosphodiester modifications
One of the simplest modifications is the introduction of phosphorothioate (PS) linkages, which can be prepared by replacing one of the two nonbridging oxygen atoms in the internucleotide linkage of RNA by a sulfur atom. PS linkages are known to reduce cleavage by nucleases and increase the half-life of ODNs in vivo. However, oligonucleotides with extensive PS linkages are also known to increase binding to serum proteins and can be toxic in vivo (Manoharan 2004; Mahato et al. 2005).
Braasch et al. (2003) reported that PS siRNAs containing 12–21 PS substitutions per strand were stable during extended incubation in fetal calf serum media, but their stability was not higher than unmodified siRNA duplexes. The investigators also monitored the inhibition of human caveolin-1 (hCav) expression by PS siRNAs. The modified siRNAs were able to silence the hCav gene, but less inhibition was observed by fully modified siRNAs. The nuclear uptake of PS siRNAs was greater than that of unmodified siRNAs, which can explain their reduced activity. Although backbone modifications did not reduce the silencing activity of siRNAs targeting the HTF gene in human keratinocytes, most extensively PS siRNAs proved to be cytotoxic, resulting in ∼70% cell death compared with mock-transfected cells (Amarzguioui et al. 2003). The introduction of PS linkages to siRNA yields mixed results for siRNA distribution to various organs. In another study, Braasch et al. (2004) observed an increase in the distribution of siRNAs to spleen, heart, and lung, while the distribution to kidney and liver decreased. However, the effects were modest, suggesting that the introduction of PS linkages may not play a major role in determining the distribution of siRNA.
An alternative backbone modification that confers increased biological stability to nucleic acids is the boranophosphonate linkage. In boranophosphonate ODNs, the nonbridging phosphodiester oxygen is replaced with an isoelectronic borane (–BH3) moiety. The charge distribution of boranophosphonate ODNs also differs from that of normal phosphate and phosphorothioate ODNs. Thus, it changes the polarity and increases the hydrophobicity of the molecule. While boranophosphonate modification has been less widely studied, it has shown more advantages than PSs. Targeting GFP in HeLa cells, Hall et al. (2004) observed that the activity of boranophosphonate siRNAs exceeds that of PS siRNAs, and it was often greater than that of unmodified siRNAs, irrespective of the base or strand modified. However, boranophosphonate modifications placed at the center of the antisense strand reduced RNAi activity. Boranophosphonate siRNAs modified at minimal levels also showed improved stability over unmodified siRNAs against nuclease degradation. Additionally, increasing the number of modifications yielded more stable siRNAs.
2′-Sugar modifications
Modifications of RNA at the 2′-position of the ribose ring have been shown to increase siRNA stability against endonucleases and reduce immune response activation. These modifications include 2′-O-methyl (2′-OMe), 2′-deoxy-2′-fluoro modifications, and locked nucleic acid.
Fluoro and methyl linkages
The siRNA motif consisting of 2′-OMe and 2′-fluoro nucleotides has enhanced plasma stability and increased in vivo potency. The 2′-OMe sugar modification retains the canonical right-handed A-form helical geometry, which is required for siRNA activity. This modification has also been shown to increase the nuclease resistance of ODNs and siRNA duplexes (Chiu and Rana 2003). The effect of 2′-OMe modification has been found to be dependent on both position and extent of incorporation. siRNAs with fully 2′-OMe-substituted sense strands were functional when the duplexes were of 20-bp blunt construction, but not for canonical duplexes of 19-bp constructs with 3′ overhangs (Kraynack and Baker 2006). In contrast, siRNAs with alternating 2′-OMe and unmodified nucleotides (Czauderna et al. 2003a), or alternating 2′-OMe or 2′-O-fluoro nucleotides (Allerson et al. 2005), had activity similar to that of the unmodified duplexes. This suggests that minimal chemical modifications are compatible with siRNA function. Jackson et al. (2006) reported that 2′-OMe modifications to specific positions within the siRNA seed region reduce both the number of off-target transcripts and the magnitude of their regulation, without significantly affecting silencing of the intended targets. Current evidence suggests that 2′-OMe sugar modifications decrease the free energy of hybridization, which would tend to compensate for, rather than impair, weaker base-pairing (Inoue et al. 1987; Lesnik et al. 1993). Additionally, the effects on off-target silencing of the 2′-OMe modification show a sharp position dependence, in contrast to the broader position dependence of base substitutions (Jackson et al. 2006). It is important to note that 2′-OMe modifications impairing the cleavage of the passenger strand also impair target RNA cleavage. However, 2′-O-methyl ribose groups positioned 1 or 2 nt downstream from the cleavage site did not impair target RNA cleavage (Matranga et al. 2005; Leuschner et al. 2006).
Chiu and Rana (2003) targeted the EGFP gene in HeLa cells using different siRNAs modified at the 2′-OH site. The effects of 2′-OH modifications on RNAi were studied by replacing uridine and cytidine in the antisense strand of siRNA by 2′-fluoro-uridine (2′-FU) and 2′-fluoro-cytidine (2′-FC), respectively. These modifications increased the siRNA stability upon exposure to HeLa cell extracts, without losing gene silencing activity. On the other hand, modifying the 2′-OH to a bulky methyl group to create 2′-OMe nucleotides in the sense or antisense strand greatly diminished EGFP gene silencing, whereas double-stranded 2′-OMe-modified siRNAs completely abolished RNAi. One reasonable explanation is that the methyl group, as a bulky group, may severely limit the interactions among siRNA, target mRNA, and the RNAi machinery that are required to successfully mediate gene silencing.
Layzer et al. (2004) observed that modification of siRNA with 2′-fluoro pyrimidines does not have an adverse effect on gene silencing and target specificity, as seen in its ability to distinguish between the GL2 and GL3 luciferase target mRNAs. In addition, 2′-F-modified siRNA showed a dramatic increase in the stability over 2′-OH siRNAs in human plasma. More than 50% of the 2′-OH siRNA was degraded within a minute of exposure to plasma, and practically all molecules were degraded after 4 h. In contrast, >50% of the 2′-F siRNA molecules remained full length after 24 h of exposure to plasma. However, after transfecting 2′-F GL2 siRNA into HeLa R19-Luc cells, which stably express the GL2 luciferase gene, little difference was observed in both the duration and magnitude of suppression between 2′-F and 2′-OH siRNAs. These results are consistent with those of Song et al. (2003a), who demonstrated that 2′-OH siRNA activity is limited by dilution in cell culture, but not by intracellular stability. Additionally, both 2′-OH and 2′-F siRNAs reduced luciferase expression in vivo by >85% when injected in rodents, but 2′-F modification of the siRNA did not grant any advantage to either the magnitude or the interval of response. Although 2′-F-modified siRNAs have much greater stability in plasma than 2′-OH siRNAs, these data suggest that once siRNAs are localized within the cell, modifications that enhance nuclease stability do not influence activity or persistence.
Locked nucleic acid
Locked nucleic acid (LNA), also referred to as inaccessible RNA, is a family of conformationally locked nucleotide analogs that displays unprecedented hybridization affinity toward complementary DNA and RNA. Commonly used LNA contains a methylene bridge connecting the 2′ oxygen with the 4′ carbon of the ribose ring. This bridge locks the ribose ring in the 3′-endo conformation characteristic of RNA (Bondensgaard et al. 2000; Braasch and Corey 2001). LNA has been shown to be compatible with siRNA intracellular machinery, preserving molecule integrity while offering several improvements that are relevant to the development of siRNA technology (Braasch et al. 2003; Elmen et al. 2005).
LNA may be used to increase the functional half-life of siRNA in vivo by two different mechanisms: (1) enhancing the resistance of the constituent RNA strands against degradation by ssRNases, and (2) stabilizing the siRNA duplex structure that is crucial for silencing activity. Elmen et al. (2005) reported that the stability of siRNA can be enhanced by conjugating LNA at the 3′ ends of the sense strand of siRNA. Introduction of LNA modifications at the 3′ overhangs in either one or both strands of siRNA against firefly luciferase revealed no loss of silencing effect of siRNA in cultured cells. Additionally, one LNA at the 5′ end of the sense strand was shown to be fully compatible with silencing activity, while an LNA at the 5′ end of the antisense strand dramatically impaired the silencing effect. Since the strand that displays the weakest binding energy at its closing 5′ base pair is incorporated preferentially into the RISC (Khvorova et al. 2003; Schwarz et al. 2003), the conjugation of LNA at the 5′ sense strand might be used to alter strand bias in favor of incorporation of the antisense strand. It may help in reducing off-target effects and increasing potency. Additionally, the potency of siRNAs may be improved by simply lowering the number of RISCs loaded with the sense strand. LNA modifications of the 3′ or 5′ termini of the sense strand of siRNAs can abrogate their immunostimulatory activity. However, depending on the extent and location of LNA modifications, they can block not only the induction of IFNs but also the gene silencing activity of the siRNA (Hornung et al. 2005).
Bioconjugation
Bioconjugation of one or both strands of siRNAs with lipids and polymers is often desirable to (1) further increase their thermodynamic and nuclease stability, (2) improve the biodistribution and pharmacokinetic profiles of siRNAs, and (3) target them to specific cell types.
Lipid conjugation
Conjugation with lipids may enhance siRNA uptake via receptor-mediated endocytosis or by an increased membrane permeability of the otherwise negatively charged RNA. Conjugation of nucleic acids with cholesterol has been demonstrated to enhance cellular uptake in cell culture and hepatic deposition after systemic administration (Cheng et al. 2006). This is because cholesterol conjugation increases the hydrophobicity and cellular association of nucleic acids. Lorenz et al. (2004) have conjugated siRNAs with cholesterol derivatives like lithocholic and lauric acids at the 5′ end of the sense strand. Cholesterol conjugation was shown to increase the cellular uptake of siRNAs in human liver cells without use of any transfection reagent. Incubation of cells during 4 h with 50 nM of siRNAs with a modified sense strand down-regulated β-galactosidase expression to a higher extent than siRNAs with a modified antisense strand or two modified strands.
Apolipoprotein B (ApoB) is expressed predominantly in the liver and jejunum, and is an essential protein for the assembly and secretion of very-low-density lipoprotein (VLDL) and low-density lipoprotein (LDL), which are required for the transport and metabolism of cholesterol. Elevated ApoB and LDL levels are correlated with increased risk of coronary artery disease, and inadequate control of LDL-cholesterol after acute coronary syndromes results in increased risk of recurrent cardiac events or death. Soutschek et al. (2004) reported the conjugation of cholesterol to the 3′ end of the sense strand of an siRNA molecule targeting ApoB. The concentration of this protein in human blood samples correlates with those of cholesterol, and higher levels of both compounds are associated with an increased risk of coronary heart disease. Intravenous administration of radiolabeled siRNA-chol conjugates into rats at 50 mg/kg has shown improved pharmacokinetic properties compared to unconjugated siRNAs. siRNA-chol conjugates showed an elimination half-life (t 1/2) of 95 min and a plasma clearance (CL) of 0.5 mL/min, whereas unconjugated siRNAs have a t 1/2 of 6 min and a CL of 17.6 mL/min. Significant levels of this conjugate were found in the liver, heart, kidney, lung, and adipose tissue. Most importantly, siRNAs efficiently reduced the levels of ApoB mRNA by >50% in the liver and by 70% in the jejunum, which resulted in a lowering of the blood cholesterol levels comparable to that observed in mice in which the ApoB gene had been deleted.
Peptide conjugation
Protein transduction domains (PTDs) offer an alternative to the traditional methods of siRNA delivery. PTDs are short amino acid sequences that are able to interact with the plasma membrane in a way that leads to a highly efficient uptake into the cytoplasm. Only short stretches, mainly consisting of positively charged amino acids, such as arginine and lysine, are responsible for translocating the PTDs through the plasma membrane—hence the name cell-penetrating peptides (CPPs) or membrane permeant peptides (MPPs). Stretches of these amino acids help to maintain the helical structure of the PTD, which is the structural prerequisite for membrane penetration. Many natural and synthetic MPPs are known to contain this structural property of PTDs (Mahat et al. 1999; Schepers 2005).
Cellular uptake of MPPs takes place in a receptor-independent fashion. MPPs are mostly amphipathic molecules that interact with the negatively charged head groups of the plasma membrane via their positive amino acid residues. MPPs can transfect different cell types with very high efficiency and rapid uptake kinetics. Their great versatility with respect to cargo and cell type, as well as their high efficiency in delivering cargo molecules into cells, makes the MPPs a valuable tool for transfecting siRNAs into mammalian cells and even fully grown organisms. Some cargo molecules may only be coupled at one defined site, where the chemical modification does not alter functionality. This can be achieved by making use of single functional groups that naturally occur in the cargo molecule or may be introduced by the utilization of chemically modified building block in the synthesis of the MPPs. This system can be exploited for efficient delivery of siRNAs into cells and into animals. For coupling of the siRNA, both the MPP and the siRNA must be modified by functional groups (Hallbrink et al. 2001; Schepers 2005).
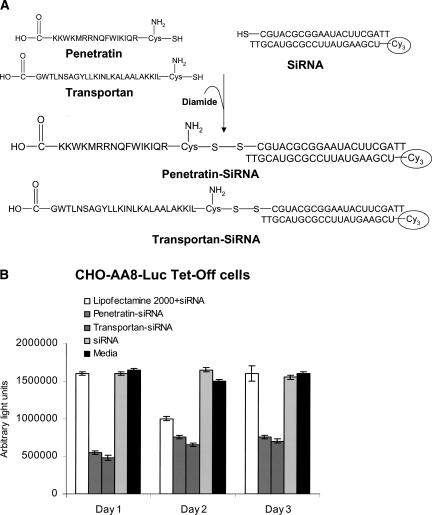
Muratovska and Eccles (2004) have shown that siRNAs can be delivered directly into the cytoplasm when conjugated with MPPs, such as penetratin or transportan (Fig. 5A). Constitutively expressed luciferase and GFP genes were successfully silenced in a high proportion of cells of different types with penetratin– or transportan–siRNA conjugates. For example, in CHO-AA8-Luc Tet Off cells, the percentage of luciferase activity silenced on each of the 3 d following incubation of the cells with MPP–siRNA conjugates was markedly superior to the silencing following transfection with Lipofectamine 2000 (Fig. 5B). The salient features of this system are as follows: (1) the peptide facilitates transport across the plasma membrane, and the MPP–siRNA conjugates are freely translocated into the cytoplasm; (2) the disulfide bond is reduced in the cytoplasm, releasing the bioactive siRNA to cause sequence-specific mRNA degradation; and (3) the uptake of MPP–siRNA conjugates is rapid and occurs directly through the membrane with no need for classical receptor-mediated uptake or endo- or pinocytosis.
FIGURE 5.
Gene silencing effect of siRNA conjugated with membrane permeant peptides (MPPs). (A) Conjugation of siRNA with two different MPPs: penetratin and transportan. MPPs and siRNAs were synthesized with a thiol group attached. The reaction between the thiol groups on the siRNAs and the MPPs was catalyzed by the oxidant diamide. (B) Silencing effect of siRNA on luciferase activity. CHO-AA8-Luc Tet-Off cells stably expressing luciferase were transfected with 25 nM siRNA conjugated with MPPs (penetratin and transportan) or complexed with Lipofectamine 2000. Luciferase activity was measured at days 1, 2, and 3 post-transfection. Reproduced with permission from Muratovska and Eccles (2004).
Simeoni et al. (2003) described a new peptide-based gene delivery system, MPG, which forms stable noncovalent complexes with nucleic acids. MPG is a bipartite amphipathic peptide derived from both the fusion peptide domain of HIV-1 gp41 protein and the nuclear localization signal (NLS) of Simian virus 40 large T antigen. The investigators showed that cell entry is independent of the endosomal pathway and that the NLS of MPG is involved in both electrostatic interactions with nucleic acid and nuclear targeting. MPG/nucleic acid particles interact with the nuclear import machinery; however, a mutation that affects the NLS of MPG disrupts these interactions and prevents nuclear delivery of nucleic acids. Therefore, this mutated-MPG system was used for siRNA delivery into the cytoplasm. The investigators demonstrated that this mutation yields a variant of MPG, which is a powerful tool for siRNA delivery into mammalian cells, enabling rapid release of the siRNA into the cytoplasm and promoting robust down-regulation of target mRNA.
Complex formation
One of the major challenges of transforming siRNAs from laboratory reagents to therapeutics is to develop an effective drug delivery system. Previous efforts to deliver siRNAs to the liver involved high-pressure tail-vein injections, resulting in silencing of a reporter gene in the liver (Morrissey et al. 2005a). However, this harsh treatment requires rapid injection of solutions two-and-a-half times the total blood volume of the animal. Although transfection efficiency of siRNAs into cultured cells is satisfactory for most in vitro applications, its therapeutic applications in vivo present an altogether more daunting challenge. Ideally, a delivery mechanism would (1) be capable of binding siRNAs in a reversible manner to ensure subsequent release of the siRNAs in target cells; (2) protect siRNAs from nuclease degradation during transit through the circulation; (3) escape from endosomal compartment; (4) be biocompatible; and (5) avoid rapid clearance by the liver and kidney.
Cationic lipids
Since the introduction in 1987 of the transfection reagent Lipofectin, an equimolar mixture of the cationic lipid N-[1-(2,3-dioleyloxy)propyl]-N,N,N-trimethylammonium chloride (DOTMA) and the colipid dioleoylphosphatidylethanolamine (DOPE), many cationic liposomes have been tested for in vitro and in vivo transfection of nucleic acids. The flexibility in the design of cationic lipid structure and liposome composition combined with the diversity of methods for their preparation and in vivo efficiency have promoted the notion that cationic lipids can be efficiently used for human gene transfer (Mahato et al. 1997). The physicochemical properties of lipid/nucleic acid complexes are strongly influenced by the (1) relative proportions of each component; (2) structure of the headgroup of the cationic lipids; (3) cationic lipid: colipid molar and charge ratio; (4) particle size of lipid/nucleic complexes; and (5) liposome size (Mahato et al. 1998; Spagnou et al. 2004). Many effective cationic lipids contain protonable polyamines linked to dialkyl or cholesterol anchors. The cationic lipids in the liposomal formulation form complexes with DNA or RNA, leading to their enhanced cellular association. In general, cationic lipids with multivalent headgroups have been shown to be more effective in gene transfer compared to their monovalent counterparts (Mahato et al. 1998, 1999; Keller 2005).
siRNAs interact electrostatically with cationic liposomes to form particles that are able to transfer them into cells. Since the nature of cationic lipid interactions with DNA and RNA is identical, it seems obvious to adapt pDNA formulations for siRNA delivery. However, pDNA and siRNA are otherwise very different from each other in molecular weight and topography with potentially important consequences. In contrast to pDNA, siRNA cannot condense into particles of nanometric dimensions, being already a small subnanometric nucleic acid (Spagnou et al. 2004). Additionally, electrostatic interactions between siRNA and cationic liposomes pose two potential problems: (1) a relatively uncontrolled interaction process leading to lipid/siRNA complexes of excessive size and poor stability and (2) incomplete encapsulation of siRNA molecules, which thereby exposes siRNA to potential enzymatic or physical degradation prior to delivery to cells. Such considerations should make clear the fact that pDNA and siRNA are completely different kinds of nucleic acids and that a lipid/siRNA complex should be regarded as different from lipid/pDNA complex formulations (Spagnou et al. 2004; Keller 2005).
Torchilin and colleagues (Zhang et al. 2006) successfully delivered siRNA into lung tumor cells by loading siRNA into liposomes bearing arginine octamer (R8), a type of MPP, attached to the liposome surface. siRNA-loaded R8 liposomes were formulated using 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP)/siRNA with the addition of the PEG-phosphatidylethanolamine. The R8 liposomes loaded with HDM2–siRNA demonstrated a high stability and protected the incorporated siRNA from degradation by blood serum even after 24 h of incubation. This siRNA formulation showed very high transfection efficiency in lung tumor cells. The R8-liposome-mediated transfection takes place even in the presence of plasma proteins.
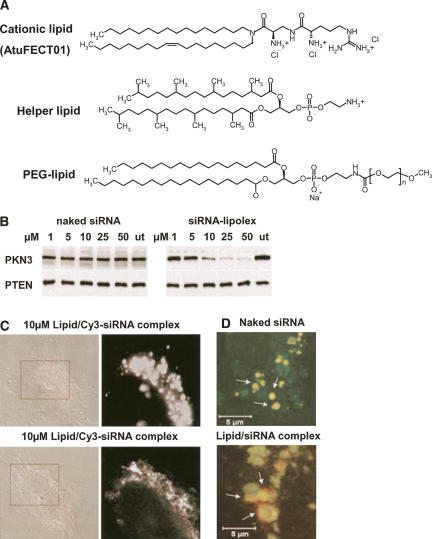
Cationic liposomes are known to interact with serum proteins, lipoproteins, heparin, and glycosaminoglycans in the extracellular matrix, leading to aggregation or release of nucleic acids from the complexes even before reaching the target cells. Cationic lipids can also activate the complement system, leading to their rapid clearance by macrophages of the reticuloendothelial systems (Mahato et al. 1997). To circumvent these problems, many liposomal carriers have been coated with PEG to minimize their interaction with serum proteins or complement system and to improve the circulation time. In addition, PEGylation may help to stabilize the lipid/nucleic acid complexes, leading to the reduction in macrophage clearance. Santel et al. (2006) developed a new liposomal system for siRNA delivery. This system consists of 50 mol% of β-L-arginyl-2,3-L-diaminopropionic acid-N-palmityl-N-oleyl-amide trihydrochloride (AtuFECT01), 49 mol% of neutral/helper lipid 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine (DPhyPE), and 1 mol% PEGylated lipid N-(carbonyl-methoxypolyethyleneglycol-2000)-1,2-distearoyl-sn-glycero-3-phosphoethanol-amine sodium salt (DSPE-PEG) (Fig. 6A). The new liposomal system was able to deliver Cy3–siRNA into different cells lines (HeLa, HUVEC) in a very low concentration (10 nM). siRNA delivery by this liposome was able to silence protein kinase N3 (PKN3) expression in a concentration-dependent manner as shown in Figure 6B. The liposomal system also showed improved siRNA cellular uptake and, more importantly, escape from the endocytotic/endosomal pathway into the cytoplasm (Fig. 6C,D). Data from IL-12 ELISA demonstrated that 1 mol% of DSPE-PEG in the liposome/siRNA complex was sufficient to reduce unspecific toxic side effects in vivo. The researchers also injected 1.88 mg/kg of complexed Cy3–siRNA intravenously in mice to evaluate the effect of the liposome on siRNA biodistribution. Complexed siRNAs were taken up by the vasculature endothelium in different organs, mainly liver, heart, lung, spleen, and kidney, showing a profound delay in the clearance rate.
FIGURE 6.
In vitro evaluation of lipid/siRNA complexes. (A) Chemical structures of lipids used for preparation of cationic liposomes and complex formation with siRNAs. (AtuFECT01) β-L-Arginyl-2,3-L-diaminopropionic acid-N-palmityl-N-oleyl-amide trihydrochoride; (helper lipid) 1,2-diphytanoyl-sn-glycero-3-phosphoethanolamine; (PEG-lipid) distearoyl-sn-glycero-3-phospho-ethanolamine sodium salt. (B) Concentration-dependent inhibition of PKN3 protein expression with lipid/siRNA complex, but not with naked siRNA in HeLa cells as assessed by immunoblot. PTEN served as the loading control. (C) Intracellular distribution of lipid/Cy3–siRNA complexes in HeLa cells analyzed by confocal microscopy. (D) Merged high-magnification pictures of a cytoplasmic area from HUVECs showing endosomal/lysosomal localization of siRNAs. Naked siRNAs remain trapped in the endosomal pathway (upper panel), whereas formulated siRNAs escape the late endosomal/lysosomal vesicles (lower panel). Reprinted by permission from Macmillan Publishers Ltd.: [Gene Therapy] (Santel et al.) © (2006).
Cationic polymers
Due to high cytotoxicity on repeated use and potent anti-inflammatory activity in vivo (Mahato et al. 1998), interest in polymeric gene carriers is growing. Like cationic lipids, cationic polymers spontaneously form complexes with nucleic acids because of electrostatic interactions between positively charged amine groups of the polycations and negatively charged phosphate groups of the nucleic acids. The interaction between cationic polymers/nucleic acid complex and negatively charged cell membranes can enhance its uptake by cells and thus increase transfection efficiency (Han et al. 2000). Various cationic polymers have been studied as potential vectors for gene delivery. These include proteins such as histones and cationized human serum albumin, polypeptides such as poly-L-lysine (PLL) and poly-L-ornithine, and polyamines such as PEI and starburst polyamidoamine (pAMAM) dendrimers. Copolymers containing hydrophilic segments such as PEG and dextran have also been synthesized.
pAMAM dendrimers are a class of highly branched spherical polymers whose surface charge and diameter are determined by the number of synthetic steps. The three-dimensional spherical structure of dendrimers offers synthesis control in terms of degree and generation of branching. This control allows the production of polymer particles with a very low degree of polydispersity, which is a significant advantage over other polymers such as PLL. The primary amino groups present on the surface of pAMAM dendrimers are positively charged at physiological pH. These surface amino groups provide useful moieties for the functional modification of dendrimers since they allow covalent coupling reactions under mild conditions and in a controlled fashion (Mahato et al. 1999). Although pAMAM dendrimers have been largely applied for delivery of plasmids and ODNs (Santhakumaran 2004; Hollins 2004; Choi 2005), this strategy remains to be explored for siRNA delivery. Generally, it is expected that the advantages observed for ODNs may be adapted for siRNA. However, Kang et al. (2005) reported that pAMAM dendrimers were not effective for delivery of siRNA against multidrug resistance 1 protein (MDR-1) in NIH 3T3 MDR cells.
Among the cationic polymers employed for gene delivery, PEI has been the most widely used polymer for siRNA delivery. PEIs with various molecular weights, degrees of branching, and other modifications have been largely used for transfection of siRNAs in different cell lines and live animals (Urban-Klein et al. 2005; Grzelinski et al. 2006). The high transfection efficiency of PEI can be attributed to its buffering effect or the “proton sponge effect” due to its secondary and tertiary amines. The cytotoxicity and transfection efficiency of PEI are directly proportional to its molecular weight. PEI is often grafted with PEG or cholesterol to reduce its cytotoxicity (Wang et al. 2002; Patil et al. 2005). Using atomic force microscopy, Grzelinski et al. (2006) showed that complexation of unmodified siRNAs with PEI leads to the formation of complexes that condense and completely cover siRNAs. The investigators showed that the delivery of siRNAs against growth factor pleiotrophin complexed with PEI was able to generate antitumoral effects in an orthotropic mouse glioblastoma model with U87 cells growing intracranially. Urban-Klein et al. (2005) showed that noncovalent complexation of synthetic siRNAs with low-molecular-weight PEI efficiently stabilizes siRNAs and delivers siRNAs into cells where they display full bioactivity at nontoxic concentrations. In a subcutaneous mouse tumor model, intraperitoneal administration of PEI/siRNA complexes, but not of naked siRNAs, was delivered into the tumors, resulting in a marked reduction of tumor growth through siRNA-mediated HER-2 receptor down-regulation.
The presence of readily accessible amine groups on the cationic polymers provides a convenient method for introducing ligands for receptor-mediated gene delivery. A common approach is to modify the amine using the N-hydroxyl succinimide (NHS) moiety, which forms amide and imide bonds with primary and secondary amines, respectively. The carboxyl groups of PEG can be coupled to the amine in the presence of NHS and carbodiimide reagents. In many cases, a hetero-bifunctional linker is used for conjugating the ligand to the polymer. One such linker that is widely used is N-succinimidyl 3-(2-pyridyldithio) proprionate (SPDP), containing an NHS moiety on one end and a 2-pyridyldithiol group on the other end, which is specific toward sulfhydryl groups. Use of sulfhydryl groups is especially convenient when conjugating peptides, because this functional group can be easily included during peptide synthesis in the form of cysteine. Alternatively, a sulfhydryl group can be introduced on the polymer by using reagents such as dithiobis(succicimidyl propionate). Hetero-bifunctional PEG containing an NHS moiety on one end and a sulfhydryl-reactive maleimide or vinyl sulfone group has also been used (Mahato et al. 1999; Hughes and Rao 2005; Ikeda and Taira 2006).
Encapsulation into lipid particles
Recently, Zimmermann et al. (2006) demonstrated that siRNAs, when delivered systemically in a liposomal formulation, can silence ApoB in nonhuman primates. ApoB-specific siRNAs were encapsulated in stable nucleic acid lipid particles (SNALPs) and administered by intravenous injection to cynomolgus monkeys at doses of 1 or 2.5 mg/kg. SNALPs consist of a lipid bilayer containing a mixture of cationic and fusogenic lipids that enables the cellular uptake and endosomal release of the particle's nucleic acid payload. SNALPs are also coated with a diffusible poly(ethylene glycol)-lipid (PEGylated lipid) conjugate that provides a neutral hydrophilic exterior and stabilizes the particle during formulation. The surface coating also shields the cationic colayer in vivo, preventing rapid systemic clearance. Upon administration, PEGylated lipid dissociates from the SNALP, transforming the particle into a transfection-competent entity. A single siRNA injection resulted in dose-dependent silencing of ApoB mRNA expression in the liver 48 h after administration, with maximal silencing of >90%. Significant reductions in ApoB protein, serum cholesterol, and low-density lipoprotein levels were observed as early as 24 h after treatment and lasted for 11 d at the highest siRNA dose, thus demonstrating the potent biological effect of siRNA treatment. Similar results were also obtained after intraveneous injection of siRNA encapsulated SNALPs in mice. In contrast, no detectable reduction was observed with a SNALP-formulated mismatched siRNA (siApoB-MM) or empty SNALP vesicles, indicating that silencing is specific to the siRNA and is not caused by the liposomal carrier. This silencing effect of SNALP-formulated siRNA represents >100-fold improvement in potency compared with systemic administration of cholesterol-conjugated siRNAs targeted against ApoB. These findings show clinically relevant RNAi-mediated gene silencing in nonhuman primates, supporting RNAi therapeutics as a potential new class of drugs.
Morrissey et al. (2005b) chemically modified siRNAs targeted against hepatitis B virus (HBV) by substituting all 2′-OH residues on the RNA with 2′-F, 2′-O-Me, or 2′-H residues. These chemically modified siRNAs were incorporated into SNALPs and administered intravenously into mice carrying replicating HBV. There was improved efficacy of siRNA–SNALP compared to unformulated siRNA, which correlated with a longer half-life of siRNA encapsulated in SNALPs in plasma and liver. Three daily intravenous injections of 3 mg/kg/d reduced serum HBV DNA >1.0 log10. This reduction in HBV DNA was specific, dose dependent, and lasted for up to 7 d after dosing. Furthermore, reductions were seen in serum HBV DNA for up to 6 wk with weekly dosing. This persistent in vivo activity is a major advance relative to previous reports showing silencing activity for only 18 h or 24 h. The daily loading doses demonstrating efficacy in the current study are >30-fold lower than those reported previously (1 versus ≥30 mg/kg). Moreover, the single maintenance dose given each week is approximately two logs lower than doses used previously. Substantial quantities of SNALP accumulated in liver (28% ± 1.7%) and spleen (8.2% ± 2.8%), but there was little accumulation in the lung (0.3% ± 0.1%) by 24 h after intravenous injection. Intense staining was observed at 2 h after intravenous injection of Cy3-labeled siRNA–SNALP into female mice. Regions of fluorescence were seen in hepatocytes, although staining intensity decreased slightly at 8 h. An intense signal was observed in some Kupffer cells and sinusoidal endothelial cells and in blood vessels up to 72 h after dosing. Administration of an siRNA that had been chemically stabilized improved the elimination half-life (T 1/2 = 0.8 h or 49 min) compared to the unstabilized siRNA. However, the most dramatic improvement to the pharmacokinetics resulted from formulation of the siRNA with SNALP (T 1/2 = 6.5 h). Unmodified siRNA–SNALP formulations strongly induced inflammatory cytokines, such as IFN-α, TNF-α, and IL-6 in the serum of injected mice. In contrast, administration of modified siRNA–SNALP formulations induced no detectable increase in these inflammatory cytokines. These data indicate that the modified siRNA not only increases siRNA in vivo stability but also abrogates undesirable immunostimulatory properties of unmodified siRNA.
Ebola virus (EBOV) infection causes a frequently fatal hemorrhagic fever that is refractory to treatment with currently available antiviral therapeutics. Geisbert et al. (2006) formulated four siRNAs targeted to the polymerase gene of the Zaire species of EBOV (ZEBOV) in SNALPs or formed complexes with polyethylenimine (PEI). When guinea pigs were treated with PEI/siRNA complexes shortly before the ZEBOV challenge, there were reductions in plasma viremia levels and partial protection of the animals from death. In contrast, siRNA–SNALP particles were found to be more efficacious, as these formulations completely protected guinea pigs against viremia and death when administered shortly after the ZEBOV challenge.
Target specificity
Ligands that recognize cell-specific receptors expressed by the target cell can be conjugated to polymers and cationic lipids in order to promote specific cellular uptake via receptor-mediated endocytosis. For systemic application, targeted siRNA delivery systems should have the following characteristics: (1) stability in blood with a prolonged half-life; (2) transport across capillary endothelial walls; (3) specific binding to cell plasma membranes; (4) active endocytosis in the cell; and (5) siRNA activity in the target cells with the desired biological end point. The preferential accumulation of siRNAs in specific cells in the body is dependent on the interplay between these processes.
Table 3 displays some ligands used to achieve site-specific gene delivery via receptor-mediated endocytosis. Several groups have exploited the asialoglycoprotein receptors (ASGPR) for gene delivery to hepatocytes, using polymeric carriers with ligands such as galactose and lactose (Kim et al. 2005b). The ASGPR are expressed on the surface of hepatocytes. The receptor mediates endocytosis and subsequent internalization of proteins that are conjugated to galactose-terminated oligosaccharide moieties. Dendritic cells form part of the mammalian immune system and express mannose receptors and mannose-related receptors (Ikeda and Taira 2006). On the other hand, folate receptor is expressed on many human tumor cells (Kim et al. 2006). Tumor cells often overexpress transferrin receptors on their surface. Another group of receptors that has received significant attention are the integrins. Integrins are heterodimeric cell-adhesion receptors that are composed of α and β subunits. The arginine-glycine-aspartic acid (RGD) motif has often been used for targeted delivery of drugs and genes because of its ability to bind to integrins that are expressed on the activated endothelial cells found in tumor vasculature (Schiffelers et al. 2004; Ikeda and Taira 2006).
TABLE 3.
Ligands used in target-specific delivery of siRNA and other gene therapies
Combinations of PEG-block-polycation/ODN and PEG-block-ODN (or PEG-ODN conjugate)/polycation are feasible for polyion complex (PIC) micelle formation with a segregated PIC core surrounded by a palisade of flexible and hydrophilic PEG layers to increase biocompatibility and enzymatic tolerability. Ligands may also be conjugated on the periphery of the PEG palisade of the PIC micelles to increase the uptake into the target cells through a receptor-mediated endocytotic pathway. Based on this strategy, Oishi et al. (2005) conjugated siRNAs with lactosylated PEG through acid-labile linkage of β-thiopropionate (Lac-PEG–siRNA), followed by complex formation with PLL (Fig. 7A). The β-thiopropionate linkage is readily cleaved at acidic pH of the endosomal compartment. Both the Lac-PEG–siRNA conjugate and the PLL micelles were tested in human hepatoma HuH-7 cells possessing ASGPR. Significant silencing of firefly luciferase gene expression was observed for both systems. However, when Lac-PEG–siRNA conjugate was delivered after formation of PLL micelles, the 50% inhibitory concentration (IC50) drastically decreased from 91.4 nM to 1.3 nM, that is, an almost 100-fold increase in RNAi activity due to PLL micellization of Lac-PEG–siRNA (Fig. 7B, right panel). The ASGP receptor-mediated endocytosis was confirmed using asialofetuin, the inhibitor for ASGP receptor. The investigators also demonstrated that the acid-labile linkage in the conjugate contributes to endosomal release (Fig. 7B, left panel). Moreover, several important factors are likely to be synergistically involved in the pronounced RNAi activity of the PLL micelles, such as: (1) improvement of the stability against enzymatic degradation; (2) minimal interaction with serum proteins; (3) enhancement of the cellular uptake through the ASGP receptor-mediated endocytosis; and (4) effective transport of the free siRNA from endosome to the cytoplasm.
FIGURE 7.
Receptor-mediated delivery of lactosylated PEG siRNA conjugate (Lac-PEG–siRNA) complexed with poly-L-lysine (PLL). (A) Chemical structure of Lac-PEG–siRNA. (B) RNAi activities against firefly luciferase gene generated in human hepatoma HuH-7 cells. A significant decrease in the firefly luciferase expression was observed after transfection with Lac-PEG–siRNA (conjugate alone) and with Lac-PEG–siRNA complexed with PLL (polyion complex, PIC micelle) (right panel). Cellular uptake via asialoglycoprotein (ASGP) receptor-mediated endocytosis was confirmed using asialofetuin (ASF), which is the inhibitor for ASGP receptor. The inhibitor of endosomal acidification, nigericin, was used to confirm that the acid-labile linkage in the conjugate contributes to RNAi activity (left panel). Reproduced from Oishi et al. (2005). Copyright (2006) American Chemical Society.
To develop a tumor-selective delivery system, Schiffelers et al. (2004) complexed siRNA with PEGylated PEI. RGD peptide ligand was attached to the distal end of PEG as a means to target tumor neovasculature-expressing integrins. The PEG-PEI-RGD system was used for delivery of siRNAs inhibiting VEGF receptor 2 (VEGF-R2). siRNA complexation with PEG-PEI-RGD presented improved serum stability over 12 h compared to unformulated siRNA. Tail-vein injection of 40 μg of FITC-labeled VEGF-R2–siRNA into tumor-bearing mice showed selective tumor uptake, siRNA sequence-specific inhibition of protein expression within the tumor, and inhibition of both tumor angiogenesis and growth rate. Additionally, the modular chemical design of the PEG-PEI-RGD allows substitution with other ligands, or combinations of ligands, to selectively target other tissues. Alternatively, Kim et al. (2006) conjugated GFP targeting siRNAs with PEI and PEI-graft-poly(ethylene glycol)-folate (PEI-PEG-FOL). The PEI-PEG-FOL/siRNA complex showed the most dose-effective and fastest gene silencing effect for folate receptor overexpressing KB cells, probably due to the fact that siRNA could be directly delivered, via folate receptor-mediated endocytosis, into the cytoplasm compartment where the RNAi process takes place.
Hu-Lieskovan et al. (2005) demonstrated that a cyclodextrin-based polycation delivery system can be used to target metastatic tumors. This multicomponent delivery system includes short polycations containing cyclodextrins that enable assembly with other components linked to targeting ligands. The cyclodextrin-containing polycations self-assembled with siRNAs and formed colloidal particles ∼50 nm in diameter, and their terminal imidazole groups assisted in the intracellular trafficking and release of the nucleic acid. Some of the PEG chains used for stabilization in biologic fluid are linked to transferring as a targeting ligand specific for tumor cells. When introduced systemically into mice by tail-vein injection, siRNA targeting EWS-FLI1 at a 2.5 mg/kg dose was able to achieve transient reduction of tumor growth in a murine model of metastatic Ewing's sarcoma. The tumor growth inhibition was correlated with a sequence-specific down-regulation of EWS-FLI1 expression, which is a transcriptional activator of tumorigenesis. The long-term systemic administration of the cyclodextrin-based polycation delivery system did not elicit detectable immune response or any changes in mouse physiology.
The use of monoclonal antibodies to direct the delivery of siRNAs carried inside liposomes to specific cell types has been demonstrated in vivo. Pirollo et al. (2006) employed a cationic immunoliposome made of DOTAP:DOPE bearing an anti-transferrin receptor single-chain antibody fragment. 6-FAM-labeled siRNA (9 mg/kg) was intravenously injected into mice in which pancreatic, primary, and metastatic tumors were previously induced. The immunoliposome was able to efficiently and specifically deliver siRNAs to both primary and metastatic tumors, including prostrate, pancreatic, and breast tumors after systemic administration.
Song et al. (2005) designed a protamine-antibody fusion protein to deliver siRNAs to HIV-infected or envelope-transfected cells. The fusion protein (F105-P) was designed with the protamine coding sequence linked to the C terminus of the heavy-chain Fab fragment of an HIV-1 envelope antibody. The siRNA–antibody complexes induced silencing only in cells expressing HIV-1 envelope. Additionally, siRNAs targeted against the HIV-1 capsid gene gag inhibited HIV replication in HIV-infected primary T-cells, which are highly resistant to transfection. Intravenous or intratumoral injection of F105-P-complexed siRNAs could deliver siRNA to env-expressing (env+) B16 melanoma cells, but not into normal tissues or env-negative tumors. Moreover, administration of F105-P-complexed siRNAs against c-myc, MDM2, and VEGF inhibited envelope-expressing subcutaneous B16 tumors. Furthermore, an ErbB2 single-chain antibody fused with protamine delivered siRNAs specifically into ErbB2-expressing cancer cells.
Aptamers can be used for site-specific delivery of siRNAs, as they possess high affinity and specificity for their targets (Hicke and Stephens 2000; Pestourie et al. 2005). Prostate-specific membrane antigen (PSMA) is a cell-surface receptor overexpressed in prostate cancer cells and in tumor vascular endothelium, and thus PSMA-specific aptamers can be internalized into PSMA-expressing cells (Farokhzad et al. 2004). McNamara et al. (2006) developed anti-PSMA aptamer compounds, which can specifically deliver siRNAs to PSMA-expressing cells. The aptamer portion of this system (A10) binds to PSMA, while siRNA silences two anti-apoptotic genes, such as polo-like kinase 1 (PLK1) and BCL2, which are overexpressed in most human tumors. When applied to LNCaP cells that express PSMA, siRNAs were internalized and processed by Dicer, resulting in depletion of target proteins and cell death. In contrast, these molecules did not bind to or function in cells that do not express PSMA. When administered intratumorally at 200 pmol/75 μL injection, these reagents also specifically inhibited tumor growth and mediated tumor regression in a xenograft model of prostate cancer.
In another strategy, anti-PSMA aptamer was coupled to siRNAs via biotin–streptavidin binding (Chu et al. 2006). Sense strands of siRNAs were first biotinylated and then noncovalently joined one to another with streptavidin. When transfected into LNCaP cells at 45 nM, siRNAs silenced lamin A/C gene expression. The inhibition of gene expression was dependent on conjugation to the aptamer and was faster than that produced by lipid-based siRNA delivery. Cell exposure to siRNA–aptamer conjugates for up to 72 h did not result in interferon response. The use of aptamers has some advantages over other methods for targeted delivery of siRNAs. Aptamers have low immunogenicity, can be easily synthesized in large quantities at relatively low cost, and are amenable to a variety of chemical modifications that confer both resistance to degradation in physiologic fluids and improved pharmacokinetics. Additionally, the smaller size of aptamers compared to that of antibodies (<15 kDa versus 150 kDa) facilitates their in vivo delivery (Hicke and Stephens 2000; Pestourie et al. 2005).
CONCLUDING REMARKS
Given the ability of silencing essentially any gene of interest, RNAi technologies have generated intense research activity. While silencing of specific genes by siRNA is showing promise in laboratories, there are several obstacles that need to be overcome before its therapeutic application can be realized. Gene silencing is greatly dependent on siRNA size, sequence, and structure, and varies from cell type to cell type. The selection of mRNA target sequences lacking putative immunostimulatory motifs can generate siRNA duplexes that mediate RNAi with minimal immunoactivation.
A proper balance between effectiveness and specificity is necessary for turning siRNA into therapeutics. Highly effective molecules that are not specific can result in off-target effects, complicating data interpretation. An effective siRNA sequence must be chosen that should not cause any unintended effect. Also, one must decide whether to use siRNA or shRNA, and whether or not it should be chemically synthesized or expressed endogenously from a range of vectors, as these decisions will affect the therapeutic effectiveness of these RNAi technologies.
In conclusion, RNAi offers great hope for treating different chronic and acquired diseases. In fact, the results from Phase I clinical trials of siRNA targeting AMD are quite promising.
ABBREVIATIONS
AAV, Adeno-associated virus; Ago2, Argonaute 2; AMD, age-related macular degeneration; ApoB, apolipoprotein B; ASGPR, asialoglycoprotein receptor; Chol, cholesterol; DSPE, N-(carbonyl-methoxypolyethyleneglycol-2000)-1,2-distearoyl-sn-glycero-3-phosphoethanol-amine sodium salt; disRNA, Dicer substrate double-stranded RNA; DNAzyme, deoxyribozyme; DOPE, dioleoylphosphatidylethanolamine; DOTAP, 1,2-dioleoyl-3-trimethylammonium-propane; DOTMA, N[1-(2,3-dioleyloxy)propyl]-N,N,N-trimeyhylammonium chloride; dsRNA, double-stranded RNA; EGFP, enhanced green fluorescent protein; EIF-2α, eukaryotic translation initiation factor-2 alpha; GFP, green fluorescent protein; hCav, human caveolin-1; HEK, human embryonic kidney; HTF, human tissue factor; IFN, interferon; IL, interleukin; IRF, interferon regulatory factor; LDL, low density lipoprotein; LNA, locked nucleic acid; Luc, luciferase; MPP, membrane permeant peptide; miRNA, microRNA; MoMLV, Moloney murine leukemia virus; NF-κB, nuclear factor kappa B; NHS, N-hydroxyl succinimide; NLS, nuclear localization signal; 2′5′-OAS, 2′-5′-oligoadenylate synthetase; ODN, oligodeoxynucleotide; pAMAM, starburst polyamidoamine; PDC, plasmacytoid dendritic cell; PEG, poly(ethylene glycol); PEI, polyethyleneimine; PIC, polyion complex; PKN3, protein kinase N3; PKR, protein kinase R; PLK1, polo-like kinase 1; PLL, poly-L-lysine; Pol II, RNA polymerase II; Pol III, RNA polymerase III; PS, phosphorothioate; PSMA, prostate-specific membrane antigen; PTD, protein transduction domain; re-hDicer, recombinant human Dicer; RGD, glycine-aspartic acid motif; RIG-I, retinoic acid-inducible gene I; RISC, RNA-induced silencing complex; RNAi, RNA interference; RNPC, Ribonucleoprotein complex; SEC, siRNA expression cassette; shRNA, short hairpin RNA; siRNA, small interfering RNA; SNALP, Stable nucleic acid lipid particle; SPDP, N-succinimidyl 3-(2-pyridyldithio) proprionate; ssRNA, single-stranded RNA; tetO, tetracycline operon sequence; tetR, tetracycline repressor; TLR, Toll-like receptor; TNF-α, tumor nechosis factor alpha; UTR, untranslated region; VEGF, vascular endothelial growth factor; VLDL, very-low-density lipoprotein.
ACKNOWLEDGMENTS
We thank the National Institutes of Health for financial support (R01 DK069968 and R01 DK064633), USPH Grant 47379. D.D.P. was a recipient of Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES, Brazil).
Footnotes
Abbreviations: See list at end of paper.
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.459807.
REFERENCES
- Allerson, C.R., Sioufi, N., Jarres, R., Prakash, T.P., Naik, N., Berdeja, A., Wanders, L., Griffey, R.H., Swayze, E.E., Bhat, B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- Amarasinghe, A.K., Calin-Jageman, I., Harmouch, A., Sun, W., Nicholson, A.W. Escherichia coli ribonuclease III: Affinity purification of hexahistidine-tagged enzyme and assays for substrate binding and cleavage. Methods Enzymol. 2001;342:143–158. doi: 10.1016/s0076-6879(01)42542-0. [DOI] [PubMed] [Google Scholar]
- Amarzguioui, M., Holen, T., Babaie, E., Prydz, H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003;31:589–595. doi: 10.1093/nar/gkg147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarzguioui, M., Rossi, J.J., Kim, D. Approaches for chemically synthesized siRNA and vector-mediated RNAi. FEBS Lett. 2005;579:5974–5981. doi: 10.1016/j.febslet.2005.08.070. [DOI] [PubMed] [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bernstein, E., Caudy, A.A., Hammond, S.M., Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- Bertrand, J.R., Pottier, M., Vekris, A., Opolon, P., Maksimenko, A., Malvy, C. Comparison of antisense oligonucleotides and siRNAs in cell culture and in vivo. Biochem. Biophys. Res. Commun. 2002;296:1000–1004. doi: 10.1016/s0006-291x(02)02013-2. [DOI] [PubMed] [Google Scholar]
- Birmingham, A., Anderson, E.M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. doi: 10.1038/nmeth854. [DOI] [PubMed] [Google Scholar]
- Bondensgaard, K., Petersen, M., Singh, S.K., Rajwanshi, V.K., Kumar, R., Wengel, J., Jacobsen, J.P. Structural studies of LNA:RNA duplexes by NMR: Conformations and implications for RNase H activity. Chemistry (Easton) 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A., Corey, D.R. Locked nucleic acid (LNA): Fine-tuning the recognition of DNA and RNA. Chem. Biol. 2001;8:1–7. doi: 10.1016/s1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A., Jensen, S., Liu, Y., Kaur, K., Arar, K., White, M.A., Corey, D.R. RNA interference in mammalian cells by chemically-modified RNA. Biochemistry. 2003;42:7967–7975. doi: 10.1021/bi0343774. [DOI] [PubMed] [Google Scholar]
- Braasch, D.A., Paroo, Z., Constantinescu, A., Ren, G., Oz, O.K., Mason, R.P., Corey, D.R. Biodistribution of phosphodiester and phosphorothioate siRNA. Bioorg. Med. Chem. Lett. 2004;14:1139–1143. doi: 10.1016/j.bmcl.2003.12.074. [DOI] [PubMed] [Google Scholar]
- Breaker, R.R., Joyce, G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Bridge, A.J., Pebernard, S., Ducraux, A., Nicoulaz, A.L., Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003;34:263–264. doi: 10.1038/ng1173. [DOI] [PubMed] [Google Scholar]
- Brummelkamp, T.R., Bernards, R., Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- Castanotto, D., Li, H., Rossi, J.J. Functional siRNA expression from transfected PCR products. RNA. 2002;8:1454–1460. doi: 10.1017/s1355838202021362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.T. An economic and efficient method of RNAi vector constructions. Anal. Biochem. 2004;334:199–200. doi: 10.1016/j.ab.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Cheng, K., Ye, Z., Guntaka, R.V., Mahato, R.I. Enhanced hepatic uptake and bioactivity of type α1(I) collagen gene promoter-specific triplex-forming oligonucleotides after conjugation with cholesterol. J. Pharmacol. Exp. Ther. 2006;317:797–805. doi: 10.1124/jpet.105.100347. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L., Rana, T.M. RNAi in human cells: Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. doi: 10.1016/s1097-2765(02)00652-4. [DOI] [PubMed] [Google Scholar]
- Chiu, Y.L., Rana, T.M. siRNA function in RNAi: A chemical modification analysis. RNA. 2003;9:1034–1048. doi: 10.1261/rna.5103703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi, Y.S., Thomas, T., Kotlyar, A., Islam, M.T., Baker, J.R., Jr. Synthesis and functional evalution of DNA-assembled polyamidoamine dendrimer clusters for cancer cell-specific targeting. Chem. Biol. 2005;12:35–43. doi: 10.1016/j.chembiol.2004.10.016. [DOI] [PubMed] [Google Scholar]
- Chu, T.C., Twu, K.Y., Ellington, A.D., Levy, M. Aptamer mediated siRNA delivery. Nucleic Acids Res. 2006;34:e73. doi: 10.1093/nar/gkl388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna, F., Fechtner, M., Dames, S., Aygun, H., Klippel, A., Pronk, G.J., Giese, K., Kaufmann, J. Structural variations and stabilising modifications of synthetic siRNAs in mammalian cells. Nucleic Acids Res. 2003a;31:2705–2716. doi: 10.1093/nar/gkg393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czauderna, F., Santel, A., Hinz, M., Fechtner, M., Durieux, B., Fisch, G., Leenders, F., Arnold, W., Giese, K., Klippel, A., et al. Inducible shRNA expression for application in a prostate cancer mouse model. Nucleic Acids Res. 2003b;31:e127. doi: 10.1093/nar/gng127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, O., Picard, D. RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res. 2002;30:e46. doi: 10.1093/nar/30.10.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Lieberman, J. RUNNING INTERFERENCE: Prospects and obstacles to using small interfering RNAs as small molecule drugs. Annu. Rev. Biomed. Eng. 2006;8:377–402. doi: 10.1146/annurev.bioeng.8.061505.095848. [DOI] [PubMed] [Google Scholar]
- Dykxhoorn, D.M., Novina, C.D., Sharp, P.A. Killing the messenger: Short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. doi: 10.1038/nrm1129. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001a;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- Elbashir, S.M., Lendeckel, W., Tuschl, T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001b;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Martinez, J., Patkaniowska, A., Lendeckel, W., Tuschl, T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001c;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir, S.M., Harborth, J., Weber, K., Tuschl, T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- Elmen, J., Thonberg, H., Ljungberg, K., Frieden, M., Westergaard, M., Xu, Y., Wahren, B., Liang, Z., Orum, H., Koch, T., et al. Locked nucleic acid (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farokhzad, O.C., Jon, S., Khademhosseini, A., Tran, T.N., Lavan, D.A., Langer, R. Nanoparticle-aptamer bioconjugates: A new approach for targeting prostate cancer cells. Cancer Res. 2004;64:7668–7672. doi: 10.1158/0008-5472.CAN-04-2550. [DOI] [PubMed] [Google Scholar]
- Feldman, A.R., Sen, D. A new and efficient DNA enzyme for the sequence-specific cleavage of RNA. J. Mol. Biol. 2001;313:283–294. doi: 10.1006/jmbi.2001.5058. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans . Nature. 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- Geisbert, T.W., Hensley, L.E., Kagan, E., Yu, E.Z., Geisbert, J.B., Daddario-DiCaprio, K., Fritz, E.A., Jahrling, P.B., McClintock, K., Phelps, J.R., et al. Postexposure protection of guinea pigs against a lethal ebola virus challenge is conferred by RNA interference. J. Infect. Dis. 2006;193:1650–1657. doi: 10.1086/504267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm, D., Kay, M.A. Therapeutic short hairpin RNA expression in the liver: Viral targets and vectors. Gene Ther. 2006;13:563–575. doi: 10.1038/sj.gt.3302727. [DOI] [PubMed] [Google Scholar]
- Grimm, D., Streetz, K.L., Jopling, C.L., Storm, T.A., Pandey, K., Davis, C.R., Marion, P., Salazar, F., Kay, M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- Grzelinski, M., Urban-Klein, B., Martens, T., Lamszus, K., Bakowsky, U., Hobel, S., Czubayko, F., Aigner, A. RNA interference-mediated gene silencing of pleiotrophin through polyethylenimine-complexed small interfering RNAs in vivo exerts antitumoral effects in glioblastoma xenografts. Hum. Gene Ther. 2006;17:751–766. doi: 10.1089/hum.2006.17.751. [DOI] [PubMed] [Google Scholar]
- Gupta, S., Schoer, R.A., Egan, J.E., Hannon, G.J., Mittal, V. Inducible, reversible, and stable RNA interference in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:1927–1932. doi: 10.1073/pnas.0306111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall, A.H., Wan, J., Shaughnessy, E.E., Ramsay Shaw, B., Alexander, K.A. RNA interference using boranophosphate siRNAs: Structure–activity relationships. Nucleic Acids Res. 2004;32:5991–6000. doi: 10.1093/nar/gkh936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallbrink, M., Floren, A., Elmquist, A., Pooga, M., Bartfai, T., Langel, U. Cargo delivery kinetics of cell-penetrating peptides. Biochim. Biophys. Acta. 2001;1515:101–109. doi: 10.1016/s0005-2736(01)00398-4. [DOI] [PubMed] [Google Scholar]
- Han, S., Mahato, R.I., Sung, Y.K., Kim, S.W. Development of biomaterials for gene therapy. Mol. Ther. 2000;2:302–317. doi: 10.1006/mthe.2000.0142. [DOI] [PubMed] [Google Scholar]
- Hannon, G.J. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- Heidel, J.D., Hu, S., Liu, X.F., Triche, T.J., Davis, M.E. Lack of interferon response in animals to naked siRNAs. Nat. Biotechnol. 2004;22:1579–1582. doi: 10.1038/nbt1038. [DOI] [PubMed] [Google Scholar]
- Heil, F., Hemmi, H., Hochrein, H., Ampenberger, F., Kirschning, C., Akira, S., Lipford, G., Wagner, H., Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303:1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- Hicke, B.J., Stephens, A.W. Escort aptamers: A delivery service for diagnosis and therapy. J. Clin. Invest. 2000;106:923–928. doi: 10.1172/JCI11324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holen, T., Amarzguioui, M., Wiiger, M.T., Babaie, E., Prydz, H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757–1766. doi: 10.1093/nar/30.8.1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollins, A.J., Benboubetra, M., Omidi, Y., Zinselmeyer, B.H., Schatzlein, A.G., Uchegbu, I.F., Akhtar, S. Evaluation of generation 2 and 3 poly(propylenimine) dendrimers for the potential cellular delivery of antisense oligonucleotides targeting the epidermal growth factor receptor. Pharm Res. 2004;21:458–466. doi: 10.1023/B:PHAM.0000019300.04836.51. [DOI] [PubMed] [Google Scholar]
- Hornung, V., Guenthner-Biller, M., Bourquin, C., Ablasser, A., Schlee, M., Uematsu, S., Noronha, A., Manoharan, M., Akira, S., de Fougerolles, A., et al. Sequence-specific potent induction of IFN-α by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005;11:263–270. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]
- Huang, B., Kochanek, S. Adenovirus-mediated silencing of huntingtin expression by shRNA. Hum. Gene Ther. 2005;16:618–626. doi: 10.1089/hum.2005.16.618. [DOI] [PubMed] [Google Scholar]
- Hughes, J.A., Rao, G.A. Targeted polymers for gene delivery. Expert Opin. Drug Deliv. 2005;2:145–157. doi: 10.1517/17425247.2.1.145. [DOI] [PubMed] [Google Scholar]
- Hu-Lieskovan, S., Heidel, J.D., Bartlett, D.W., Davis, M.E., Triche, T.J. Sequence-specific knockdown of EWS-FLI1 by targeted, nonviral delivery of small interfering RNA inhibits tumor growth in a murine model of metastatic Ewing's sarcoma. Cancer Res. 2005;65:8984–8992. doi: 10.1158/0008-5472.CAN-05-0565. [DOI] [PubMed] [Google Scholar]
- Ikeda, Y., Taira, K. Ligand-targeted delivery of therapeutic siRNA. Pharm. Res. 2006;23:1631–1640. doi: 10.1007/s11095-006-9001-x. [DOI] [PubMed] [Google Scholar]
- Inoue, H., Hayase, Y., Imura, A., Iwai, S., Miura, K., Ohtsuka, E. Synthesis and hybridization studies on two complementary nona(2′-O-methyl)ribonucleotides. Nucleic Acids Res. 1987;15:6131–6148. doi: 10.1093/nar/15.15.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson, A.L., Bartz, S.R., Schelter, J., Kobayashi, S.V., Burchard, J., Mao, M., Li, B., Cavet, G., Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003;21:635–637. doi: 10.1038/nbt831. [DOI] [PubMed] [Google Scholar]
- Jackson, A.L., Burchard, J., Leake, D., Reynolds, A., Schelter, J., Guo, J., Johnson, J.M., Lim, L., Karpilow, J., Nichols, K., et al. Position-specific chemical modification of siRNAs reduces “off-target” transcript silencing. RNA. 2006;12:1197–1205. doi: 10.1261/rna.30706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, J., Wernli, M., Klimkait, T., Erb, P. Enhanced gene silencing by the application of multiple specific small interfering RNAs. FEBS Lett. 2003;552:247–252. doi: 10.1016/s0014-5793(03)00893-7. [DOI] [PubMed] [Google Scholar]
- Judge, A.D., Sood, V., Shaw, J.R., Fang, D., McClintock, K., MacLachlan, I. Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat. Biotechnol. 2005;23:457–462. doi: 10.1038/nbt1081. [DOI] [PubMed] [Google Scholar]
- Kang, H., Delong, R., Fisher, M.H., Juliano, R.L. Tat-conjugated PAMAM dendrimers as delivery agents for antisense and siRNA oligonucleotides. Pharm. Res. 2005;22:2099–2106. doi: 10.1007/s11095-005-8330-5. [DOI] [PubMed] [Google Scholar]
- Kariko, K., Bhuyan, P., Capodici, J., Weissman, D. Small interfering RNAs mediate sequence-independent gene suppression and induce immune activation by signaling through toll-like receptor 3. J. Immunol. 2004;172:6545–6549. doi: 10.4049/jimmunol.172.11.6545. [DOI] [PubMed] [Google Scholar]
- Kawakami, S., Hattori, Y., Lu, Y., Higuchi, Y., Yamashita, F., Hashida, M. Effect of cationic charge on receptor-mediated transfection using mannosylated cationic liposome/plasmid DNA complexes following the intravenous administration in mice. Pharmazie. 2004;59:405–408. [PubMed] [Google Scholar]
- Kawasaki, H., Taira, K. Short hairpin type of dsRNAs that are controlled by tRNAVal promoter significantly induce RNAi-mediated gene silencing in the cytoplasm of human cells. Nucleic Acids Res. 2003;31:700–707. doi: 10.1093/nar/gkg158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki, H., Suyama, E., Iyo, M., Taira, K. siRNAs generated by recombinant human Dicer induce specific and significant but target site-independent gene silencing in human cells. Nucleic Acids Res. 2003;31:981–987. doi: 10.1093/nar/gkg184. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Keller, M. Lipidic carriers of RNA/DNA oligonucleotides and polynucleotides: What a difference a formulation makes! J. Control. Release. 2005;103:537–540. doi: 10.1016/j.jconrel.2004.12.024. [DOI] [PubMed] [Google Scholar]
- Khvorova, A., Reynolds, A., Jayasena, S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Longo, M., Han, Y., Lundberg, P., Cantin, E., Rossi, J.J. Interferon induction by siRNAs and ssRNAs synthesized by phage polymerase. Nat. Biotechnol. 2004;22:321–325. doi: 10.1038/nbt940. [DOI] [PubMed] [Google Scholar]
- Kim, D.H., Behlke, M.A., Rose, S.D., Chang, M.S., Choi, S., Rossi, J.J. Synthetic dsRNA Dicer substrates enhance RNAi potency and efficacy. Nat. Biotechnol. 2005a;23:222–226. doi: 10.1038/nbt1051. [DOI] [PubMed] [Google Scholar]
- Kim, E.M., Jeong, H.J., Park, I.K., Cho, C.S., Moon, H.B., Yu, D.Y., Bom, H.S., Sohn, M.H., Oh, I.J. Asialoglycoprotein receptor targeted gene delivery using galactosylated polyethylenimine-graft-poly(ethylene glycol): In vitro and in vivo studies. J. Control. Release. 2005b;108:557–567. doi: 10.1016/j.jconrel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Kim, S.H., Mok, H., Jeong, J.H., Kim, S.W., Park, T.G. Comparative evaluation of target-specific GFP gene silencing efficiencies for antisense ODN, synthetic siRNA, and siRNA plasmid complexed with PEI-PEG-FOL conjugate. Bioconjug. Chem. 2006;17:241–244. doi: 10.1021/bc050289f. [DOI] [PubMed] [Google Scholar]
- Kraynack, B.A., Baker, B.F. Small interfering RNAs containing full 2′-O-methylribonucleotide-modified sense strands display Argonaute2/eIF2C2-dependent activity. RNA. 2006;12:163–176. doi: 10.1261/rna.2150806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layzer, J.M., McCaffrey, A.P., Tanner, A.K., Huang, Z., Kay, M.A., Sullenger, B.A. In vivo activity of nuclease-resistant siRNAs. RNA. 2004;10:766–771. doi: 10.1261/rna.5239604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y.S., Nakahara, K., Pham, J.W., Kim, K., He, Z., Sontheimer, E.J., Carthew, R.W. Distinct roles for Drosophila Dicer-1 and Dicer-2 in the siRNA/miRNA silencing pathways. Cell. 2004;117:69–81. doi: 10.1016/s0092-8674(04)00261-2. [DOI] [PubMed] [Google Scholar]
- Lesnik, E.A., Guinosso, C.J., Kawasaki, A.M., Sasmor, H., Zounes, M., Cummins, L.L., Ecker, D.J., Cook, P.D., Freier, S.M. Oligodeoxynucleotides containing 2′-O-modified adenosine: Synthesis and effects on stability of DNA:RNA duplexes. Biochemistry. 1993;32:7832–7838. doi: 10.1021/bi00081a031. [DOI] [PubMed] [Google Scholar]
- Leuschner, P.J., Ameres, S.L., Kueng, S., Martinez, J. Cleavage of the siRNA passenger strand during RISC assembly in human cells. EMBO Rep. 2006;7:314–320. doi: 10.1038/sj.embor.7400637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis, D.L., Hagstrom, J.E., Loomis, A.G., Wolff, J.A., Herweijer, H. Efficient delivery of siRNA for inhibition of gene expression in postnatal mice. Nat. Genet. 2002;32:107–108. doi: 10.1038/ng944. [DOI] [PubMed] [Google Scholar]
- Li, M.J., Bauer, G., Michienzi, A., Yee, J.K., Lee, N.S., Kim, J., Li, S., Castanotto, D., Zaia, J., Rossi, J.J. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol. Ther. 2003;8:196–206. doi: 10.1016/s1525-0016(03)00165-5. [DOI] [PubMed] [Google Scholar]
- Liu, C.M., Liu, D.P., Dong, W.J., Liang, C.C. Retrovirus vector-mediated stable gene silencing in human cell. Biochem. Biophys. Res. Commun. 2004a;313:716–720. doi: 10.1016/j.bbrc.2003.11.174. [DOI] [PubMed] [Google Scholar]
- Liu, J., Carmell, M.A., Rivas, F.V., Marsden, C.G., Thomson, J.M., Song, J.J., Hammond, S.M., Joshua-Tor, L., Hannon, G.J. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004b;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- Lorenz, C., Hadwiger, P., John, M., Vornlocher, H.P., Unverzagt, C. Steroid and lipid conjugates of siRNAs to enhance cellular uptake and gene silencing in liver cells. Bioorg. Med. Chem. Lett. 2004;14:4975–4977. doi: 10.1016/j.bmcl.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Mahat, R.I., Monera, O.D., Smith, L.C., Rolland, A. Peptide-based gene delivery. Curr. Opin. Mol. Ther. 1999;1:226–243. [PubMed] [Google Scholar]
- Mahato, R.I., Rolland, A., Tomlinson, E. Cationic lipid-based gene delivery systems: Pharmaceutical perspectives. Pharm. Res. 1997;14:853–859. doi: 10.1023/a:1012187414126. [DOI] [PubMed] [Google Scholar]
- Mahato, R.I., Anwer, K., Tagliaferri, F., Meaney, C., Leonard, P., Wadhwa, M.S., Logan, M., French, M., Rolland, A. Biodistribution and gene expression of lipid/plasmid complexes after systemic administration. Hum. Gene Ther. 1998;9:2083–2099. doi: 10.1089/hum.1998.9.14-2083. [DOI] [PubMed] [Google Scholar]
- Mahato, R.I., Smith, L.C., Rolland, A. Pharmaceutical perspectives of nonviral gene therapy. Adv. Genet. 1999;41:95–156. doi: 10.1016/s0065-2660(08)60152-2. [DOI] [PubMed] [Google Scholar]
- Mahato, R.I., Cheng, K., Guntaka, R.V. Modulation of gene expression by antisense and antigene oligodeoxynucleotides and small interfering RNA. Expert Opin. Drug Deliv. 2005;2:3–28. doi: 10.1517/17425247.2.1.3. [DOI] [PubMed] [Google Scholar]
- Manoharan, M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- Marques, J.T., Williams, B.R. Activation of the mammalian immune system by siRNAs. Nat. Biotechnol. 2005;23:1399–1405. doi: 10.1038/nbt1161. [DOI] [PubMed] [Google Scholar]
- Marques, J.T., Devosse, T., Wang, D., Zamanian-Daryoush, M., Serbinowski, P., Hartmann, R., Fujita, T., Behlke, M.A., Williams, B.R. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat. Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- Marshall, W.S., Kaiser, R.J. Recent advances in the high-speed solid phase synthesis of RNA. Curr. Opin. Chem. Biol. 2004;8:222–229. doi: 10.1016/j.cbpa.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Matranga, C., Tomari, Y., Shin, C., Bartel, D.P., Zamore, P.D. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607–620. doi: 10.1016/j.cell.2005.08.044. [DOI] [PubMed] [Google Scholar]
- Matsukura, S., Jones, P.A., Takai, D. Establishment of conditional vectors for hairpin siRNA knockdowns. Nucleic Acids Res. 2003;31:e77. doi: 10.1093/nar/gng077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey, A.P., Meuse, L., Pham, T.T., Conklin, D.S., Hannon, G.J., Kay, M.A. RNA interference in adult mice. Nature. 2002;418:38–39. doi: 10.1038/418038a. [DOI] [PubMed] [Google Scholar]
- McNamara, J.O., II, Andrechek, E.R., Wang, Y., Viles, K.D., Rempel, R.E., Gilboa, E., Sullenger, B.A., Giangrande, P.H. Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat. Biotechnol. 2006;24:1005–1015. doi: 10.1038/nbt1223. [DOI] [PubMed] [Google Scholar]
- Meister, G., Tuschl, T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- Meister, G., Landthaler, M., Patkaniowska, A., Dorsett, Y., Teng, G., Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- Merdan, T., Callahan, J., Petersen, H., Kunath, K., Bakowsky, U., Kopeckova, P., Kissel, T., Kopecek, J. Pegylated polyethylenimine-Fab′ antibody fragment conjugates for targeted gene delivery to human ovarian carcinoma cells. Bioconjug. Chem. 2003;14:989–996. doi: 10.1021/bc0340767. [DOI] [PubMed] [Google Scholar]
- Milligan, J.F., Uhlenbeck, O.C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Morrissey, D.V., Blanchard, K., Shaw, L., Jensen, K., Lockridge, J.A., Dickinson, B., McSwiggen, J.A., Vargeese, C., Bowman, K., Shaffer, C.S., et al. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005a;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- Morrissey, D.V., Lockridge, J.A., Shaw, L., Blanchard, K., Jensen, K., Breen, W., Hartsough, K., Machemer, L., Radka, S., Jadhav, V., et al. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005b;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- Muratovska, A., Eccles, M.R. Conjugate for efficient delivery of short interfering RNA (siRNA) into mammalian cells. FEBS Lett. 2004;558:63–68. doi: 10.1016/S0014-5793(03)01505-9. [DOI] [PubMed] [Google Scholar]
- Myers, J.W., Jones, J.T., Meyer, T., Ferrell, J.E., Jr. Recombinant Dicer efficiently converts large dsRNAs into siRNAs suitable for gene silencing. Nat. Biotechnol. 2003;21:324–328. doi: 10.1038/nbt792. [DOI] [PubMed] [Google Scholar]
- Naito, Y., Yamada, T., Ui-Tei, K., Morishita, S., Saigo, K. siDirect: Highly effective, target-specific siRNA design software for mammalian RNA interference. Nucleic Acids Res. 2004;32:W124–W129. doi: 10.1093/nar/gkh442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishitsuji, H., Kohara, M., Kannagi, M., Masuda, T. Effective suppression of human immunodeficiency virus type 1 through a combination of short- or long-hairpin RNAs targeting essential sequences for retroviral integration. J. Virol. 2006;80:7658–7666. doi: 10.1128/JVI.00078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- No, D., Yao, T.P., Evans, R.M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc. Natl. Acad. Sci. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nykanen, A., Haley, B., Zamore, P.D. ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell. 2001;107:309–321. doi: 10.1016/s0092-8674(01)00547-5. [DOI] [PubMed] [Google Scholar]
- Ohkawa, J., Taira, K. Control of the functional activity of an antisense RNA by a tetracycline-responsive derivative of the human U6 snRNA promoter. Hum. Gene Ther. 2000;11:577–585. doi: 10.1089/10430340050015761. [DOI] [PubMed] [Google Scholar]
- Oishi, M., Nagasaki, Y., Itaka, K., Nishiyama, N., Kataoka, K. Lactosylated poly(ethylene glycol)-siRNA conjugate through acid-labile β-thiopropionate linkage to construct pH-sensitive polyion complex micelles achieving enhanced gene silencing in hepatoma cells. J. Am. Chem. Soc. 2005;127:1624–1625. doi: 10.1021/ja044941d. [DOI] [PubMed] [Google Scholar]
- Paddison, P.J., Caudy, A.A., Bernstein, E., Hannon, G.J., Conklin, D.S. Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes & Dev. 2002;16:948–958. doi: 10.1101/gad.981002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil, S.D., Rhodes, D.G., Burgess, D.J. DNA-based therapeutics and DNA delivery systems: A comprehensive review. AAPS J. 2005;7:E61–E77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul, C.P., Good, P.D., Winer, I., Engelke, D.R. Effective expression of small interfering RNA in human cells. Nat. Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- Persengiev, S.P., Zhu, X., Green, M.R. Nonspecific, concentration-dependent stimulation and repression of mammalian gene expression by small interfering RNAs (siRNAs) RNA. 2004;10:12–18. doi: 10.1261/rna5160904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestourie, C., Tavitian, B., Duconge, F. Aptamers against extracellular targets for in vivo applications. Biochimie. 2005;87:921–930. doi: 10.1016/j.biochi.2005.04.013. [DOI] [PubMed] [Google Scholar]
- Pirollo, K.F., Zon, G., Rait, A., Zhou, Q., Yu, W., Hogrefe, R., Chang, E.H. Tumor-targeting nanoimmunoliposome complex for short interfering RNA delivery. Hum. Gene Ther. 2006;17:117–124. doi: 10.1089/hum.2006.17.117. [DOI] [PubMed] [Google Scholar]
- Reynolds, A., Leake, D., Boese, Q., Scaringe, S., Marshall, W.S., Khvorova, A. Rational siRNA design for RNA interference. Nat. Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- Robbins, M.A., Li, M., Leung, I., Li, H., Boyer, D.V., Song, Y., Behlke, M.A., Rossi, J.J. Stable expression of shRNAs in human CD34+ progenitor cells can avoid induction of interferon responses to siRNAs in vitro. Nat. Biotechnol. 2006;24:566–571. doi: 10.1038/nbt1206. [DOI] [PubMed] [Google Scholar]
- Rubinson, D.A., Dillon, C.P., Kwiatkowski, A.V., Sievers, C., Yang, L., Kopinja, J., Rooney, D.L., Ihrig, M.M., McManus, M.T., Gertler, F.B., et al. A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat. Genet. 2003;33:401–406. doi: 10.1038/ng1117. [DOI] [PubMed] [Google Scholar]
- Santel, A., Aleku, M., Keil, O., Endruschat, J., Esche, V., Fisch, G., Dames, S., Loffler, K., Fechtner, M., Arnold, W., et al. A novel siRNA-lipoplex technology for RNA interference in the mouse vascular endothelium. Gene Ther. 2006;13:1222–1234. doi: 10.1038/sj.gt.3302777. [DOI] [PubMed] [Google Scholar]
- Santhakumaran, L.M., Thomas, T., Thomas, T.J. Enhanced cellular uptake of a triplex-forming oligonucleotide by nanoparticle formation in the presence of polypropylenimine dendrimers. Nucleic Acids Res. 2004;32:2102–2112. doi: 10.1093/nar/gkh526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders, L.R., Barber, G.N. The dsRNA binding protein family: Critical roles, diverse cellular functions. FASEB J. 2003;17:961–983. doi: 10.1096/fj.02-0958rev. [DOI] [PubMed] [Google Scholar]
- Schepers, U. Wiley-VCH; New York: 2005. RNA interference in practice: Principles, basics, and methods for gene silencing in C. elegans, Drosophila, and mammals . [Google Scholar]
- Schiffelers, R.M., Ansari, A., Xu, J., Zhou, Q., Tang, Q., Storm, G., Molema, G., Lu, P.Y., Scaria, P.V., Woodle, M.C. Cancer siRNA therapy by tumor selective delivery with ligand-targeted sterically stabilized nanoparticle. Nucleic Acids Res. 2004;32:e149. doi: 10.1093/nar/gnh140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schomber, T., Kalberer, C.P., Wodnar-Filipowicz, A., Skoda, R.C. Gene silencing by lentivirus-mediated delivery of siRNA in human CD34+ cells. Blood. 2004;103:4511–4513. doi: 10.1182/blood-2003-07-2397. [DOI] [PubMed] [Google Scholar]
- Schwarz, D.S., Hutvagner, G., Du, T., Xu, Z., Aronin, N., Zamore, P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Semizarov, D., Frost, L., Sarthy, A., Kroeger, P., Halbert, D.N., Fesik, S.W. Specificity of short interfering RNA determined through gene expression signatures. Proc. Natl. Acad. Sci. 2003;100:6347–6352. doi: 10.1073/pnas.1131959100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeoni, F., Morris, M.C., Heitz, F., Divita, G. Insight into the mechanism of the peptide-based gene delivery system MPG: Implications for delivery of siRNA into mammalian cells. Nucleic Acids Res. 2003;31:2717–2724. doi: 10.1093/nar/gkg385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siolas, D., Lerner, C., Burchard, J., Ge, W., Linsley, P.S., Paddison, P.J., Hannon, G.J., Cleary, M.A. Synthetic shRNAs as potent RNAi triggers. Nat. Biotechnol. 2005;23:227–231. doi: 10.1038/nbt1052. [DOI] [PubMed] [Google Scholar]
- Sioud, M. Therapeutic siRNAs. Trends Pharmacol. Sci. 2004;25:22–28. doi: 10.1016/j.tips.2003.11.006. [DOI] [PubMed] [Google Scholar]
- Sioud, M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- Sioud, M. Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2′-hydroxyl uridines in immune responses. Eur. J. Immunol. 2006;36:1222–1230. doi: 10.1002/eji.200535708. [DOI] [PubMed] [Google Scholar]
- Sohail, M., Doran, G., Riedemann, J., Macaulay, V., Southern, E.M. A simple and cost-effective method for producing small interfering RNAs with high efficacy. Nucleic Acids Res. 2003;31:e38. doi: 10.1093/nar/gng038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, E., Lee, S.K., Dykxhoorn, D.M., Novina, C., Zhang, D., Crawford, K., Cerny, J., Sharp, P.A., Lieberman, J., Manjunath, N., et al. Sustained small interfering RNA-mediated human immunodeficiency virus type 1 inhibition in primary macrophages. J. Virol. 2003a;77:7174–7181. doi: 10.1128/JVI.77.13.7174-7181.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, E., Lee, S.K., Wang, J., Ince, N., Ouyang, N., Min, J., Chen, J., Shankar, P., Lieberman, J. RNA interference targeting Fas protects mice from fulminant hepatitis. Nat. Med. 2003b;9:347–351. doi: 10.1038/nm828. [DOI] [PubMed] [Google Scholar]
- Song, E., Zhu, P., Lee, S.K., Chowdhury, D., Kussman, S., Dykxhoorn, D.M., Feng, Y., Palliser, D., Weiner, D.B., Shankar, P., et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 2005;23:709–717. doi: 10.1038/nbt1101. [DOI] [PubMed] [Google Scholar]
- Soutschek, J., Akinc, A., Bramlage, B., Charisse, K., Constien, R., Donoghue, M., Elbashir, S., Geick, A., Hadwiger, P., Harborth, J., et al. Therapeutic silencing of an endogenous gene by systemic administration of modified siRNAs. Nature. 2004;432:173–178. doi: 10.1038/nature03121. [DOI] [PubMed] [Google Scholar]
- Spagnou, S., Miller, A.D., Keller, M. Lipidic carriers of siRNA: Differences in the formulation, cellular uptake, and delivery with plasmid DNA. Biochemistry. 2004;43:13348–13356. doi: 10.1021/bi048950a. [DOI] [PubMed] [Google Scholar]
- Tiscornia, G., Singer, O., Ikawa, M., Verma, I.M. A general method for gene knockdown in mice by using lentiviral vectors expressing small interfering RNA. Proc. Natl. Acad. Sci. 2003;100:1844–1848. doi: 10.1073/pnas.0437912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ui-Tei, K., Naito, Y., Takahashi, F., Haraguchi, T., Ohki-Hamazaki, H., Juni, A., Ueda, R., Saigo, K. Guidelines for the selection of highly effective siRNA sequences for mammalian and chick RNA interference. Nucleic Acids Res. 2004;32:936–948. doi: 10.1093/nar/gkh247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urban-Klein, B., Werth, S., Abuharbeid, S., Czubayko, F., Aigner, A. RNAi-mediated gene-targeting through systemic application of polyethylenimine (PEI)-complexed siRNA in vivo. Gene Ther. 2005;12:461–466. doi: 10.1038/sj.gt.3302425. [DOI] [PubMed] [Google Scholar]
- Van Craenenbroeck, K., Vanhoenacker, P., Leysen, J.E., Haegeman, G. Evaluation of the tetracycline- and ecdysone-inducible systems for expression of neurotransmitter receptors in mammalian cells. Eur. J. Neurosci. 2001;14:968–976. doi: 10.1046/j.0953-816x.2001.01719.x. [DOI] [PubMed] [Google Scholar]
- Wang, D.A., Narang, A.S., Kotb, M., Gaber, A.O., Miller, D.D., Kim, S.W., Mahato, R.I. Novel branched poly(ethylenimine)-cholesterol water-soluble lipopolymers for gene delivery. Biomacromolecules. 2002;3:1197–1207. doi: 10.1021/bm025563c. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz, M., Trono, D. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J. Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, L.F., Ralph, G.S., Walmsley, L.E., Bienemann, A.S., Parham, S., Kingsman, S.M., Uney, J.B., Mazarakis, N.D. Lentiviral-mediated delivery of Bcl-2 or GDNF protects against excitotoxicity in the rat hippocampus. Mol. Ther. 2005;11:89–95. doi: 10.1016/j.ymthe.2004.08.026. [DOI] [PubMed] [Google Scholar]
- Wooddell, C.I., Van Hout, C.V., Reppen, T., Lewis, D.L., Herweijer, H. Long-term RNA interference from optimized siRNA expression constructs in adult mice. Biochem. Biophys. Res. Commun. 2005;334:117–127. doi: 10.1016/j.bbrc.2005.06.066. [DOI] [PubMed] [Google Scholar]
- Wu, X., Li, Y., Crise, B., Burgess, S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- Xia, H., Mao, Q., Paulson, H.L., Davidson, B.L. siRNA-mediated gene silencing in vitro and in vivo. Nat. Biotechnol. 2002;20:1006–1010. doi: 10.1038/nbt739. [DOI] [PubMed] [Google Scholar]
- Yang, D., Buchholz, F., Huang, Z., Goga, A., Chen, C.Y., Brodsky, F.M., Bishop, J.M. Short RNA duplexes produced by hydrolysis with Escherichia coli RNase III mediate effective RNA interference in mammalian cells. Proc. Natl. Acad. Sci. 2002;99:9942–9947. doi: 10.1073/pnas.152327299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye, Z., Cheng, K., Guntaka, R.V., Mahato, R.I. Targeted delivery of a triplex-forming oligonucleotide to hepatic stellate cells. Biochemistry. 2005;44:4466–4476. doi: 10.1021/bi047529j. [DOI] [PubMed] [Google Scholar]
- Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., Fujita, T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- Yuan, B., Latek, R., Hossbach, M., Tuschl, T., Lewitter, F. siRNA Selection Server: An automated siRNA oligonucleotide prediction server. Nucleic Acids Res. 2004;32:W130–W134. doi: 10.1093/nar/gkh366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore, P.D., Haley, B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Tang, N., Liu, X., Liang, W., Xu, W., Torchilin, V.P. siRNA-containing liposomes modified with polyarginine effectively silence the targeted gene. J. Control. Release. 2006;112:229–239. doi: 10.1016/j.jconrel.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann, T.S., Lee, A.C., Akinc, A., Bramlage, B., Bumcrot, D., Fedoruk, M.N., Harborth, J., Heyes, J.A., Jeffs, L.B., John, M., et al. RNAi-mediated gene silencing in non-human primates. Nature. 2006;441:111–114. doi: 10.1038/nature04688. [DOI] [PubMed] [Google Scholar]