Abstract
The role of alternative splicing (AS) in eliciting immune responses is poorly understood. We used quantitative AS microarray profiling to survey changes in AS during activation of Jurkat cells, a leukemia-derived T-cell line. Our results indicate that ∼10%–15% of the profiled alternative exons undergo a >10% change in inclusion level during activation. The majority of the genes displaying differential AS levels are distinct from the set of genes displaying differential transcript levels. These two gene sets also have overlapping yet distinct functional roles. For example, genes that show differential AS patterns during T-cell activation are often closely associated with cell-cycle regulation, whereas genes with differential transcript levels are highly enriched in functions associated more directly with immune defense and cytoskeletal architecture. Previously unknown AS events were detected in genes that have important roles in T-cell activation, and these AS level changes were also observed during the activation of normal human peripheral CD4+ and CD8+ lymphocytes. In summary, by using AS microarray profiling, we have discovered many new AS changes associated with T-cell activation. Our results suggest an extensive role for AS in the regulation of the mammalian immune response.
Keywords: alternative splicing, T-cell activation, T lymphocytes, microarray
INTRODUCTION
The adaptive immune system confers on an organism the ability to identify foreign antigen and to respond specifically and efficiently to each challenge. To accomplish this task, T cells, among other cell types, undergo numerous physical and biochemical changes upon interaction with antigen. Such functional changes, including cytoskeletal rearrangements, altered trafficking within the body, and induced expression and secretion of cytokines or cytotoxins, must occur at specific times during an immune response. Moreover, the induced cellular changes must be attenuated once the antigen is cleared, in order to prevent unregulated cell growth or a loss of tolerance to self. Not surprisingly, the cycle of functional changes that occur in response to antigen challenge of T cells requires the altered expression of a plethora of proteins (Miosge and Goodnow 2005), and much of this differential regulation is accomplished at the transcriptional level (Gerondakis et al. 1998; Moroy 2005). However, in addition to regulation of transcription, there is a growing awareness of the importance of regulated pre-mRNA splicing in controlling T-cell function (Lynch 2004).
Pre-mRNA splicing involves the precise removal of introns and joining of exons to produce a protein-coding mRNA. Importantly, the specificity of splicing can be regulated in a process known as alternative splicing (AS), such that a single pre-mRNA can give rise to multiple, independent mRNAs, each potentially encoding a functionally distinct protein isoform. Alternative splicing occurs in at least 74% of multiexon human genes and is a major mechanism responsible for the generation of protein diversity and gene regulation (Modrek et al. 2001; Johnson et al. 2003). In T cells, several genes are known to express multiple mRNA and protein isoforms via AS, including CD44, CD45, and CTLA4 (Lynch 2004). It has further been shown that the pattern of AS for these genes changes in response to antigen stimulation, resulting in important functional changes in protein expression (Trowbridge and Thomas 1994; Magistrelli et al. 1999; Lynch and Weiss 2000; Oaks et al. 2000; Ponta et al. 2003). Moreover, the metastatic spread of tumors and several immune-related diseases have been linked to the misregulation of splicing of these genes in humans cells, thereby demonstrating the functional importance of regulated AS (Lynch 2004, and references therein). However, despite the evidence that changes in AS are important during an immune response and can critically influence cellular function, there has been no systematic study to determine which genes are regulated at the level of AS in response to immunologic challenge.
During the past decade, microarray analyses have detected global transcriptional changes during T-cell activation both in vivo and in vitro (Marrack et al. 2000; Lin et al. 2003; Hess et al. 2004). These studies identified many genes that are differentially expressed in resting and activated T cells and have greatly enhanced our knowledge of the factors that are essential to elicit an immune response. More recently, custom-designed microarrays have been used to study the abundance of different isoforms generated through AS (Lee and Wang 2005; Blencowe 2006). Using a quantitative AS microarray platform, we have previously identified and characterized AS events in several mammalian tissues and cell lines (Pan et al. 2004, 2006). This technology thus has the potential to provide information about AS events that are pertinent to the regulation of the immune system.
Here we report the use of this microarray system to analyze global changes in AS during in vitro stimulation of the Jurkat T-cell lines. We find that AS and transcript level regulation affect different subsets of genes, with each mechanism, at specified detection thresholds, affecting at least 10% of the genes surveyed. Importantly, results from reverse transcriptase-polymerase chain reaction (RT-PCR) experiments indicate that >50% of the microarray-predicted AS events show changes when primary CD4+ or CD8+ human T cells are stimulated. Our results more than triple the number of known examples of activation-induced AS events in T lymphocytes and strongly suggest a widespread role for AS in controlling the mammalian immune response.
RESULTS
Monitoring changes in AS during Jurkat cell activation
In order to survey changes in AS during T-cell activation, we used our quantitative microarray platform in conjunction with a new microarray design implementing the Agilent 44K format that allows the monitoring of >5000 cassette-type AS events in human. The design of the system and the accompanying data analysis algorithm are described in detail by Pan et al. (2004) and Shai et al. (2006). Briefly, on each microarray, AS events mined from human cDNA and EST sequence databases are monitored by six different oligonucleotide probes: one probe for each exon body sequence, and one junction probe for each of the three splice-junction sequences generated by AS. The probe signals are analyzed by the generative model for the alternative splicing array platform (GenASAP) algorithm, which predicts the percent exclusion level (%ex) for each alternative exon and also provides a rank as a confidence score for each prediction (Pan et al. 2004; Shai et al. 2006). In order to gain a broader understanding of the immune system, additional probes were included on the microarray to monitor AS events that are associated with genes known to be involved in eliciting an immune response, as well as genes reported to have increased levels of expression during T-cell activation (Teague et al. 1999; Raghavan et al. 2002; Lin et al. 2003; Lynch 2004).
Two different clones of Jurkat cells, JSL1 (Lynch and Weiss 2000) and Trex-Jurkat (Invitrogen), were activated by incubation with the phorbol ester PMA for 7 h and 60 h, respectively. These stimulation periods were used to cover both the short-term and prolonged changes that happen after activation, and hereafter we will refer to these time points as “early” and “late” activation, respectively. Total RNA was collected from the different cell populations, and the activation status of the cells was verified by performing RT-PCR assays to monitor Galectin-9 and CD45, which are known to undergo AS changes after T-cell activation by PMA (data not shown; Lynch and Weiss 2000; Lahm et al. 2004). Poly A+ mRNA was extracted from the total RNA, reverse transcribed, labeled, and hybridized to the Agilent 44K array as described (Hughes et al. 2006; Pan et al. 2004). Signals from the microarray were input into GenASAP to generate %ex predictions. Transcript levels were also monitored for the same set of genes, using the averages of the signals from the probes specific for the constitutive exons flanking each cassette alternative exon represented on the microarray.
Based on our previous experience with the microarray platform, the most reliable predictions for %ex are from AS events that are ranked by GenASAP within the top third to top half of the data set, and which have constitutive exon probe values that are above the ∼95th percentile of the values from negative control probes included on the microarray. Similar criteria were used to generate the lists of AS events with higher confidence predictions for %ex for the analysis described below. In total, there are ∼1600 AS events belonging to genes that are expressed above background levels in either the resting and activated states. Importantly, we observed a high degree of correlation (∼0.9) between GenASAP %ex values and between transcript levels, at the corresponding time points for the two clones of Jurkat cells. For each of the time points, 35 events were chosen for RT-PCR validation based on biological relevance and a representative range of differences in %ex levels between resting and activated cells. In all cases, the magnitude of this difference was >10%. Representative RT-PCR reactions are shown in Figure 1. The correlation between the RT-PCR measurements and GenASAP predictions for %ex levels for events ranking in the top one-third portion of the data is comparable to those obtained in our previous validation experiments (e.g., ∼0.7) (data not shown; Pan et al. 2004; Shai et al. 2006).
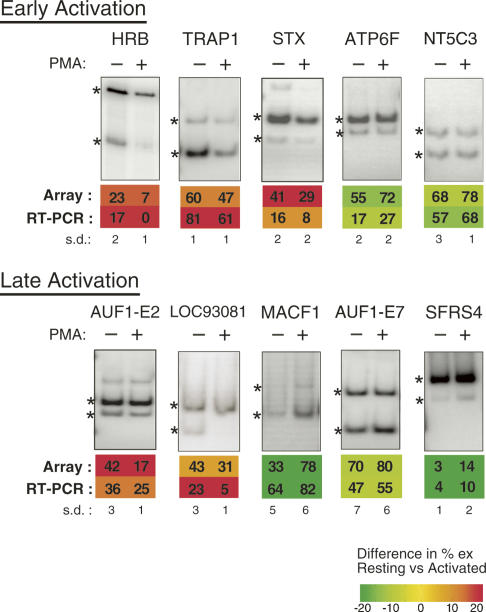
FIGURE 1.
Analysis of isoform expression in Jurkat cells of genes predicted by microarray to undergo changes in alternative splicing (AS) upon stimulation. Representative RT-PCR assays performed using Jurkat RNA isolated at either early or late time points after treatment with PMA, as described in Materials and Methods. Quantification of %ex and standard deviation (s.d.) from three independent experiments are shown, along with the corresponding prediction from the AS microarray profiling data. Numbers given indicate %exon skipping (i.e., exclusion). Asterisks indicate RT-PCR products derived from exon included and exon skipped isoforms.
Different sets of genes are controlled at the AS and transcriptional levels during Jurkat cell activation
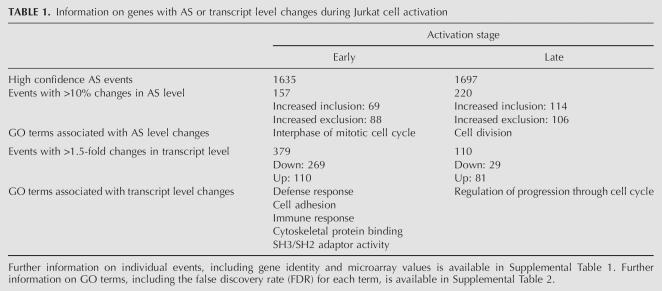
Among the profiled AS events with high confidence GenASAP %ex values, ∼10%–15% displayed >10% changes in splicing levels post-activation at each time point. More genes display altered splicing patterns at the late time point (220 AS events) compared with the early time point (157 AS changes) (Table 1; Supplemental Table 1). However, there are more genes with >1.5-fold changes in transcript levels at the early time point during activation (379 genes) than observed later in the activation response (110) (Table 1; Supplemental Table 1). Consistent with previous studies comparing gene expression changes in T cells during short and long periods of activation (Teague et al. 1999; Marrack et al. 2000; Hess et al. 2004), our results support the conclusion that dynamic changes in transcriptional regulation happen during the early phase of T-cell activation in order to generate a rapid immune response.
TABLE 1.
Information on genes with AS or transcript level changes during Jurkat cell activation
Within the group of genes that show changes in AS upon activation, approximately equal numbers of genes display increased exon inclusion and exclusion levels, and this is observed at both time points (Table 1). On the other hand, at the transcript level, a larger fraction of genes have decreased steady-state levels at the early time point, whereas the majority of genes have increased steady-state levels at the late time point. These results raised the question as to the extent of the overlap between genes regulated at the AS and transcript levels in the context of the rapid responses that occur in Jurkat cells following stimulation. In previous studies, we and others observed that different subsets of genes are regulated at the AS and transcript levels in differentiated mammalian tissues (Le et al. 2004; Pan et al. 2004). Consistent with these previous studies, at both early and late activation points, the majority of the genes that show a change at the level of AS do not show a change at the transcript level (Fig. 2). This implies that primarily different sets of genes are regulated at the AS and transcript levels in order to generate an immune response. Interestingly however, ∼47 genes with AS level changes of >10%ex also display >1.5-fold changes at the transcript level. It is possible that the AS events associated with this subset of genes are controlled by mechanisms involving coupling between transcription and splicing. Nevertheless, while communication between transcription and splicing may play a role in gene regulation (Kornblihtt et al. 2004; Kornblihtt 2005), our data do not allow us to distinguish whether these genes are regulated via mechanisms involving coupling, or rather are independently regulated at the AS and transcript levels.
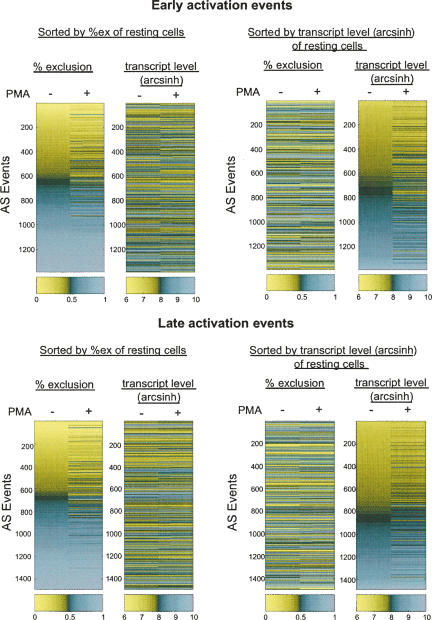
FIGURE 2.
Different subsets of genes are regulated at the AS and transcript levels during Jurkat cell activation. The %ex values and transcript level following treatment with PMA for 7 h (early activation) or 60 h (late activation) are plotted. In the left panel, AS events are sorted according to the %ex values in the resting state from low to high exclusion, and the corresponding transcript levels for the same genes are shown in the adjacent column. In the right panel, events are sorted according to transcript levels in the resting state from low to high expression, and the %ex levels of the AS events in the same genes are shown in the adjacent column.
Genes displaying differential AS and transcript level regulation during Jurkat cell activation are associated with different functions
Based on the above results, we next asked whether the different sets of genes that are controlled at the AS and transcript levels during T-cell activation belong to different functional categories. We used the online tool GOStat (http://gostat.wehi.edu.au/) to determine if these different subsets of genes are enriched in specific gene ontology (GO) terms (Beissbarth and Speed 2004). Among the genes expressed above background levels, the subset of genes displaying differential AS levels between resting and early activation periods are strongly enriched in the GO term “interphase of mitotic cell cycle.” Included among this list of GO term-enriched genes are CDK2, ANAPC5, and CCNB1, which function in the regulation of cell-cycle progression. Other GO terms associated with cell-cycle control were also enriched in the sets of AS-regulated genes that are differentially regulated between resting and both activation periods, although at relatively modest levels (false discovery rate [FDR] >0.1; refer to Materials and Methods). Interestingly, a different set of GO terms are highly enriched among the genes that are differentially regulated at the transcript level at the early time-point post-activation. These GO terms are associated with immune defense and cytoskeletal functions (Table 1; Supplemental Table 2). However, genes that are regulated at the transcript level at the later time point are enriched with genes involved in cell-cycle regulation, consistent with results of previous studies (Teague et al. 1999). Together, these findings agree with the aforementioned observation that different sets of genes are regulated at the AS and transcript levels during activation of the Jurkat cell line, and are also consistent with changes that would be expected during activation of T cells. For example, in addition to the expected enrichment of functions associated with immune defense, enrichment of genes that are associated functionally with the cytoskeleton among the genes with transcript level changes likely reflects the extensive rearrangements that are known to occur in the cytoskeleton during T-cell activation (Poenie et al. 2004; Sechi and Wehland 2004). These results therefore support the functional validity of changes we have detected at the AS and transcript levels using our microarray system and analysis methods.
In addition to the enrichment of the specific GO terms mentioned above, we also observe many AS-regulated and transcriptionally regulated sets of genes that are associated with RNA metabolism. In order to more systematically investigate the extent of regulation in this functional class, we analyzed data from a set of additional probes included on the microarray that monitor the expression of known and putative splicing factors. Data from these probes, which were analyzed separately from the data generated from the probes for monitoring genes with AS events, revealed changes in transcript levels for ∼22 defined splicing factors following activation, and the majority of these changes resulted in increased transcript levels (Supplemental Table 3). These results are consistent with previous studies demonstrating roles for splicing factors in T-cell biology (Wang et al. 2001; Rothrock et al. 2005; Heyd et al. 2006), and that a number of splicing factors have altered expression levels after T-cell activation (Screaton et al. 1995; Lemaire et al. 1999; ten Dam et al. 2000). In summary, we have observed that distinct yet overlapping sets of genes and functional categories are differentially regulated at the AS and transcriptional levels during activation of the Jurkat cells. The detection of a specific set of AS-regulated genes with GO term enrichment suggests that these AS events may be coordinated in a manner that is functionally relevant to T-cell activation.
AS events identified during activation of Jurkat cells are also detected in normal peripheral blood lymphocytes
The Jurkat cell lines used for the microarray profiling experiments are known to mimic many features of naive peripheral human T cells, such as the increased expression of IL-2 and CD69 upon stimulation through the CD3 subunit of the T-cell receptor or with pharmacological agents (Abraham and Weiss 2004). However, there are aspects of gene regulation and cellular function that differ between T-cell-derived lines and primary T cells (Lin et al. 2003). For example, Jurkat cells are defective in the expression of two lipid phosphatases, PTEN and SHIP, leading to the constitutive activation of the kinases PI3K and ITK (Abraham and Weiss 2004). Since the consequences of these differences in the context of T-cell biology are not well understood (Abraham and Weiss 2004), we assessed the relevance of our microarray profiling data to normal T-cell physiology by validating a subset of the %ex values in purified human peripheral T cells. Naive (CD45RA+) CD4+CD3+ or CD8+CD3+ cells were purified from pooled human blood by negative selection (see Materials and Methods). Flow cytometry determined the purity of the CD4+CD3+ population to be >90%, while the CD8+CD3+ population was ∼80% pure (data not shown). In both cases the contaminating cells were CD3− and were not further characterized. The purified cell populations were then cultured with or without the lectin PHA to mimic antigen stimulation. Total RNA was isolated from each of the cell populations and analyzed by a low-cycle RT-PCR assay that we have previously shown to provide a quantitative measure of changes in RNA isoform expression (Lynch and Weiss 2000).
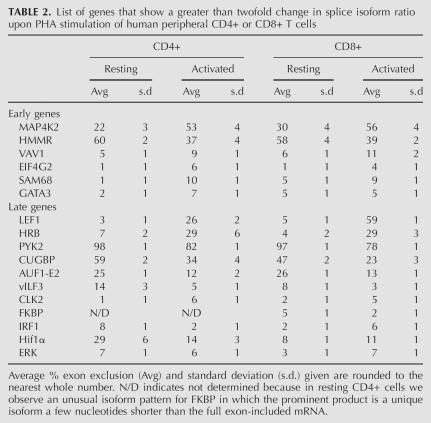
We initially chose to analyze the AS of transcripts from 28 genes. Twenty-five of these genes were selected from among the high confidence microarray predictions of genes that undergo activation-induced AS, while an additional three (Pyk2, CUGBP, and Hif1α) were selected based on their biological interest and strong %ex change. Three of the 28 genes were not expressed at sufficient levels in CD4+ peripheral T cells to allow quantification, but we were able to quantify isoform levels for the other 25. Of 11 genes predicted by the microarray data to have an AS level change at the early time-point post-stimulation, six showed a change in isoform ratio of at least twofold (refer to Materials and Methods) in human CD4+ T cells after 7 h of stimulation with PHA (Table 2; Fig. 3), while a seventh (ILF3) showed a significant change in isoform expression at 72 h post-PHA treatment. Five of the six rapidly responding genes were further observed to undergo a change in AS in the purified CD8+ human T cells, while the sixth (GATA3) did not show a significant change in AS level in CD8+ cells (Table 2; Fig. 3).
TABLE 2.
List of genes that show a greater than twofold change in splice isoform ratio upon PHA stimulation of human peripheral CD4+ or CD8+ T cells
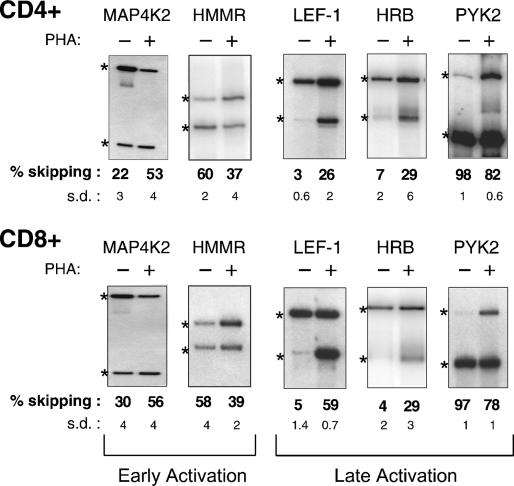
FIGURE 3.
Analysis of isoform expression in human CD4+ and CD8+ cells of genes predicted to undergo changes in AS upon stimulation. Representative RT-PCR assays performed using RNA isolated from CD4+ or CD8+ cells at either early or late time points after treatment with PHA, as described in Materials and Methods. Quantification and standard deviation (s.d.) from three to four experiments is shown. Numbers given indicate %exon skipping (i.e., exclusion). Asterisks indicate RT-PCR products derived from exon included and exon skipped isoforms.
Alternative splicing of the additional 14 genes was analyzed in RNA from resting cells versus cells harvested 72 h post-stimulation with PHA. In marked agreement with the microarray data from the cell lines, 11 of these 14 showed a greater than twofold change in isoform expression upon stimulation in at least one cell type (Table 2; Fig. 3; see Materials and Methods), while a 12th (PPP1CA) showed a more modest change (1.6-fold). Seven of the 11 strong responders showed activation-induced changes in splicing in both the CD4+ and CD8+ cells, whereas two showed evidence of regulated AS in only one of the two cell types (Hif1A and Erk) and another two showed a different pattern (FKBP) or direction (IRF1) of splicing in the CD4+ versus CD8+ cells (Table 2; Fig. 3).
The above results demonstrate that a significant number of genes predicted by the microarray results to undergo AS changes in the cell lines also have activation-induced AS changes in normal human T cells (i.e., six of 11 early events and 11 of 14 late events). However, we do note differences in the absolute levels and/or the direction of the changes in AS levels when comparing the splicing patterns in the Jurkat lines and CD4+ or CD8+ cells. This is not surprising given the differences between transformed and primary cell lines, as well as the differences between the latter two cell populations (e.g., the differences in directionality and/or extent of regulation between purified CD4+ and CD8+ cells for AS events such as observed for IRF1, Hif1α, etc.). However, to further test the prognostic ability of the microarray and to confirm that our high rate of validation was not simply reflective of an overall abundance of activation-induced AS in immune cells, we assayed the splicing of 14 genes predicted by the microarray to not have altered AS levels upon stimulation. These genes were analyzed at multiple time points following stimulation of CD4+ cells with PHA. As shown in Supplemental Table 4, only one of these 14 genes (COPS3) was observed to undergo changes in splicing pattern in response to stimulation, despite the fact that at least nine of the 14 genes did express two isoforms such that we would have readily detected a change in isoform ratio. Thus, we conclude that at least 50% of the microarray-predicted AS changes in the Jurkat cell lines likely correspond to AS events that are also differentially regulated upon antigen-stimulation of normal human T cells.
Using rpsblast (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), we performed a search on all translated sequences of the activation-induced AS events that occurred in primary blood lymphocytes (PBLs) to determine if these events have the potential to affect conserved protein domains. Among 17 of the alternative exons, nine encode regions overlapping conserved domains in the Conserved Domain Database (CDD) (Supplemental Table 5). Interestingly, three of these activation-induced AS changes affect the catalytic domains of serine/threonine protein kinases. Future studies on these exons and the conserved domains they encode may shed light on the roles of the AS events we have identified in T-cell activation and thereby accelerate our understanding of T-cell biology.
DISCUSSION
An essential hallmark of the adaptive immune system is its ability to rapidly change protein expression and cellular function in response to specific stimuli. Both experimental and bioinformatic approaches have demonstrated that a notable proportion of genes expressed in the immune system contain alternative exons, and these observations suggest that regulation of splicing could play a critical role in altering lymphocyte function (Lynch, 2004). However, there has been no systematic effort to identify AS events that are regulated during lymphocyte activation, or any attempt to understand the contributions of transcription and splicing as potentially complementary mechanisms to regulate the functions of the immune system.
In this study, we have utilized a quantitative microarray system to analyze both splicing and transcript level changes in ∼4000 genes upon stimulation of two T-cell-derived Jurkat cell lines. Our results indicate that stimulation-induced changes in transcript and AS levels occur with similar frequency in these cells and, in each case, involve ∼10%–15% of the microarray-profiled genes. However, relatively few genes are affected by both mechanisms. The GO terms associated with genes that are differentially regulated in activated Jurkat cells at the transcript versus AS levels are also largely distinct. At the early time point, genes regulated at the transcriptional levels are significantly enriched in GO terms associated with immune defense and cytoskeletal functions, whereas genes regulated at the AS level are enriched for GO terms associated with cell-cycle regulation. Independent roles for AS and transcription in the regulation of gene expression programs are consistent with previous results (Le et al. 2004; Pan et al. 2004, 2006). Collectively, these observations suggest that separate mechanisms acting at the AS and transcriptional levels are typically used to achieve distinct functional outcomes in a diverse range of physiological contexts. Moreover, we found that there are more genes regulated at the transcript level at the early time point during cell activation compared to the later time point. This agrees with previous observations that T-cell activation is a reversible process. After prolonged activation periods, the transcriptional program will gradually resume to a resting state in order to prevent overactivation and self-intolerance (Georgopoulos et al. 1997; Lenardo 2003; Miosge and Goodnow 2005).
Widespread changes in transcription in response to T-cell activation have long been known to occur and have been extensively studied by others (Marrack et al. 2000; Lin et al. 2003; Hess et al. 2004). However, surprisingly, our data also reveal extensive regulation of AS upon stimulation of T cells. Given the lack of precedence for such abundant signal-responsive regulation of splicing, we sought to determine whether this mode of regulation is also frequent in normal peripheral human T lymphocytes. Strikingly, >50% of the genes predicted by the microarray data to have an altered splicing pattern upon stimulation of the Jurkat T-cell lines also show regulated AS upon PHA activation of naive CD4+ and/or CD8+ peripheral human T cells. These results confirm the utility of the microarray approach for predicting new AS events associated with T-cell activation and also reveal the physiologic pervasiveness of regulated AS in the immune system.
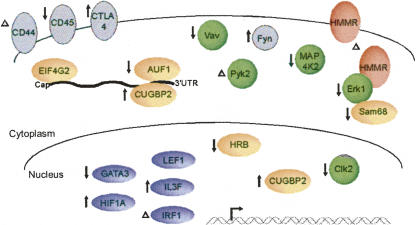
Importantly, our studies confirm at least 17 novel examples of activation-induced AS in primary T lymphocytes, thereby more than tripling the number of genes known to be regulated by AS in this branch of the mammalian immune system. In addition to cell-cycle-regulated genes, many of the splicing-regulated genes encode RNA binding proteins, transcription factors, and other proteins that have important functions in T cells (Fig. 4; Supplemental Table 5). Some of the splicing changes we observed are quite pronounced (i.e., a ∼20%ex change or greater), such as in the genes encoding MAP4K2, HMMR, LEF-1, PYK2, and CUGBP2, whereas other AS events appear to have a relatively modest effect on the levels of the predominant isoforms. However, in many instances (such as SAM68, VAV, and ERK1; see Supplemental Table 5), the minor isoform corresponds to a precise deletion of a single functional domain, which could result in a dominant-negative activity. In such cases, an increase in the inhibitory isoform could have significant functional consequences, even if it remains a minor percentage of the total message. Consistent with this possibility, relatively small (i.e., ∼10%) changes in the relative ratios of the spliced isoforms have been linked to neurological diseases such as Alzheimer's (Glatz et al. 2006). Moreover, in our analysis we did not sort cellular populations following activation, so we cannot distinguish between genes that undergo modest changes in splicing in a large percentage of cells versus those that undergo large changes in splicing in a small subpopulation of cells. We note that the activities of some of the genes we have analyzed, such as GATA3 and IRF1, are known to correlate with particular cell fates following T-cell engagement with antigen, suggesting that regulation may differ between distinct subpopulations (Farrar et al. 2001; Romeo et al. 2002). Finally, even if some of the changes in AS upon T-cell activation are only modest, the fact that many of the regulated genes are engaged in similar functions strongly suggests that activation-induced AS is of physiological relevance. It is interesting to consider, for example, that these changes in AS could operate as a rheostat, in which multiple transitions in the expression of a cascade of functionally linked splice variants serve to fine tune the cellular response to antigen (see Fig. 4).
FIGURE 4.
Model for the functions of AS during activation of human T cells. Schematic diagram of proteins affected by AS in T cells, as predicted by AS microarray profiling and confirmed by RT-PCR analysis of RNA from human CD4+ or CD8+ cells, and the predicted consequences. Color coding of proteins is as follows: green for kinases and signal adaptor molecules, orange for proteins involved in RNA metabolism, blue for transcription factors, pink for receptors, and gray for proteins shown in previous studies to be regulated by AS in T cells. The location of each proteins is shown according to its known functional role. These classifications are not precise as some proteins fit into more than one functional category and others are known to function in more than one location in the cell. Up and down arrows and Δ, respectively, indicate putative increase, decrease, or altered function of a protein as a consequence of AS upon T cell activation. See Supplemental Table 5 for further information.
Interestingly, the genes confirmed to undergo changes in AS rapidly after stimulation correspond broadly to genes that encode immediate T-cell effector functions such as signaling (e.g., VAV, SAM68 [this study]), migration (e.g., HMMR [this study], CD44 [Konig et al. 1998]), proliferation, and initiating fate decisions (GATA3 [this study]). In contrast, many genes that only display a switch in AS pattern following prolonged activation encode proteins important for homeostasis or development of immunologic memory (e.g., CD45 [Majeti et al. 2000; Rothrock et al. 2003] CTLA4 [Magistrelli et al. 1999], LEF1, and AUF1 [this study]). Regulation of AS at early versus late time points likely occurs by distinct pathways and proteins, as has been shown for the CD44 and CD45 genes that, respectively, undergo AS changes at 7 h and 48 h following T-cell stimulation (Konig et al. 1998; Lynch and Weiss 2000). However, genes regulated at a similar time point may be under the control of coordinated mechanisms, as has been suggested for CD45 and CTLA4 (Rothrock et al. 2003). Such coordination of AS regulation has been observed recently in the mammalian nervous system (Ule et al. 2005) and is due at least in part to the activity of neuronal-specific splicing factors such as Nova, which regulate the AS of multiple functionally linked genes (Ule et al. 2005).
Our data suggest that, far from being a minor phenomenon, regulation of AS is a pervasive mechanism for altering gene expression in response to T-cell stimulation and may be at least as common as regulated transcription. The identity of the genes that undergo activation-responsive AS and the predicted functional consequences of the changes in AS that we observe suggest that regulation of AS in response to T-cell stimulation has the potential to significantly alter cellular activity. Ultimately, identification of the functions of individual alternative exons as well as of the factors that regulate AS, particularly those that influence coordinated groups of AS events in T cells, will be required to determine the specific functional consequences of this abundant mode of gene regulation in shaping the activity of mammalian immune system.
MATERIALS AND METHODS
Cell culture, T-cell activation, and RNA purification
The JSL1 and Trex Jurkat (Invitrogen) cell lines were maintained in RPMI+ 10% heat-inactivated fetal calf serum (FCS), as described in detail previously (Lynch and Weiss 2000), and stimulated by the addition of 20 ng/mL PMA (Sigma) for the times indicated. Isolation of naive CD4+ or CD8+ T cells from human peripheral blood was done by negative selection as described previously (Lynch and Weiss 2000). Briefly, PBLs were obtained from ∼150 mL of human blood by Ficoll gradient. PBLs were then incubated for 30 min at 4°C with antibodies specific for CD14, CD16, CD19, CD45R0, CD11b, CD235a, and either CD8 (for CD4+ purification) or CD4 (for CD8+ purification). All antibodies were obtained from Clontech and used at a concentration of 1 μL of antibody per 1 million PBLs. Cells that reacted with antibody were then removed by two successive rounds of incubation and depletion with magnetic beads coated with sheep anti-mouse IgG antibodies (Dynal Inc.). Remaining cells were counted and cultured at a concentration of 0.3 million/mL in RPMI + 10% FCS in the absence or presence of 0.5 mg/mL PHA for the times indicated. An aliquot of the resulting cell population was also analyzed by flow cytometry for expression of CD4, CD8, CD3, and CD45 RA to estimate purity. Collection and use of blood was approved under IRB no. 072006–032 to K.W.L. RNA was isolated from the cell using Trizol (Invitrogen) following the manufacturer's protocol. Extraction of poly A+ RNA was performed as described (Pan et al. 2006).
Identification of AS events in human transcripts
AS events represented on the human 44K microarray were identified and selected as described previously (Pan et al. 2004, 2005, 2006), using cDNA/EST sequences data from UniGene (ftp://ftp.ncbi.nih.gov/repository/UniGene; Build no. 158) and human genome sequence data (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens).
Microarray hybridization, image processing, and data analysis
Microarray design, hybridization, and data analysis for 5183 nonredundant human AS events (represented on a 44K Agilent microarray) were performed as described previously (Pan et al. 2004; Hughes et al. 2006). Percent exon exclusion values and associated rankings for these values were generated using the GenASAP algorithm, as described previously (Pan et al. 2004; Shai et al. 2006). The analysis of AS levels used AS events with %ex values in the top third-ranking portion of the data and with transcript levels above the 96th percentile of the negative control probes on the microarray. Analysis of transcript levels of alternatively spliced genes was performed using the averages of signal intensities from the probes specific for the pairs of constitutive exons flanking each monitored alternative exon (Pan et al. 2006).
GO enrichment analysis was performed using the online tool GOStat (http://gostat.wehi.edu.au/). Genes with differential AS levels analyzed for GO term enrichment displayed AS level changes of at least 10% upon Jurkat activation, and were ranked in the top one-third portion of the data set by GenASAP. This group of genes was compared with all other alternatively spliced and GO annotated genes represented on the array that are expressed above background levels, as defined by the 96th percentile of the negative control probes. For the GO term enrichment analysis of genes with transcript level changes, the genes with at least 1.5-fold changes in transcript levels between resting and activated states were compared against all genes with an above background level of expression in at least one of the two time points being compared. FDR was used as the correction for multiple hypothesis testing.
RT-PCR
RT-PCR assays on Jurkat cell RNA were carried out using the One-Step RT-PCR Kit (Qiagen) as described previously (Pan et al. 2006). RNA from the human CD4+ and CD8+ cells was analyzed by low-cycle RT-PCR as described in detail previously (Lynch and Weiss 2000), except that for each target gene the PCR reaction was terminated at four different cycle numbers in order to determine empirically that quantification was within the linear range. Fold Change in isoform ratio is calculated as (%isoform1/%isoform2)resting/(%isoform1/%isoform2)activated. We have previously determined a fold change of >2 as being a valid cutoff for significant changes in AS detected by RT-PCR (45). A complete listing of primers and annealing temperatures is given in online Supplemental Table 6.
SUPPLEMENTAL DATA
All supplemental material can be found at http://www.utoronto.ca/intron/T-cell-AS/.
ACKNOWLEDGMENTS
We thank Ofer Shai and Matthew Fagnani for assistance with data analysis, and John Calarco for helpful comments on the manuscript. Our research was supported by Operating Grants from the National Cancer Institute of Canada, Canadian Institutes for Health Research, and Genome Canada to B.J.B. and by NIH grant R01 GM067719 and NSF grant MCB0347104 to K.W.L.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.457207.
REFERENCES
- Abraham, R.T., Weiss, A. Jurkat T cells and development of the T-cell receptor signalling paradigm. Nat. Rev. Immunol. 2004;4:301–308. doi: 10.1038/nri1330. [DOI] [PubMed] [Google Scholar]
- Beissbarth, T., Speed, T.P. GOstat: Find statistically overrepresented gene ontologies within a group of genes. Bioinformatics. 2004;20:1464–1465. doi: 10.1093/bioinformatics/bth088. [DOI] [PubMed] [Google Scholar]
- Blencowe, B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Farrar, J.D., Ouyang, W., Lohning, M., Assenmacher, M., Radbruch, A., Kanagawa, O., Murphy, K.M. An instructive component in T helper cell type 2 (Th2) development mediated by GATA-3. J. Exp. Med. 2001;193:643–650. doi: 10.1084/jem.193.5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos, K., Winandy, S., Avitahl, N. The role of the Ikaros gene in lymphocyte development and homeostasis. Annu. Rev. Immunol. 1997;15:155–176. doi: 10.1146/annurev.immunol.15.1.155. [DOI] [PubMed] [Google Scholar]
- Gerondakis, S., Grumont, R., Rourke, I., Grossmann, M. The regulation and roles of Rel/NF-κB transcription factors during lymphocyte activation. Curr. Opin. Immunol. 1998;10:353–359. doi: 10.1016/s0952-7915(98)80175-1. [DOI] [PubMed] [Google Scholar]
- Glatz, D.C., Rujescu, D., Tang, Y., Berendt, F.J., Hartmann, A.M., Faltraco, F., Rosenberg, C., Hulette, C., Jellinger, K., Hampel, H., et al. The alternative splicing of tau exon 10 and its regulatory proteins CLK2 and TRA2-BETA1 changes in sporadic Alzheimer's disease. J. Neurochem. 2006;96:635–644. doi: 10.1111/j.1471-4159.2005.03552.x. [DOI] [PubMed] [Google Scholar]
- Hess, K., Yang, Y., Golech, S., Sharov, A., Becker, K.G., Weng, N.P. Kinetic assessment of general gene expression changes during human naive CD4+ T cell activation. Int. Immunol. 2004;16:1711–1721. doi: 10.1093/intimm/dxh172. [DOI] [PubMed] [Google Scholar]
- Heyd, F., ten Dam, G., Moroy, T. Auxiliary splice factor U2AF26 and transcription factor Gfi1 cooperate directly in regulating CD45 alternative splicing. Nat. Immunol. 2006;7:859–867. doi: 10.1038/ni1361. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., Hiley, S.L., Saltzman, A.L., Babak, T., Blencowe, B.J. Microarray analysis of RNA processing and modification. Methods Enzymol. 2006;410:300–316. doi: 10.1016/S0076-6879(06)10014-2. [DOI] [PubMed] [Google Scholar]
- Johnson, J.M., Castle, J., Garrett-Engele, P., Kan, Z., Loerch, P.M., Armour, C.D., Santos, R., Schadt, E.E., Stoughton, R., Shoemaker, D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Konig, H., Ponta, H., Herrlich, P. Coupling of signal transduction to alternative pre-mRNA splicing by a composite splice regulator. EMBO J. 1998;17:2904–2913. doi: 10.1093/emboj/17.10.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblihtt, A.R. Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 2005;17:262–268. doi: 10.1016/j.ceb.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Kornblihtt, A.R., de la Mata, M., Fededa, J.P., Munoz, M.J., Nogues, G. Multiple links between transcription and splicing. RNA. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahm, H., Andre, S., Hoeflich, A., Kaltner, H., Siebert, H.C., Sordat, B., von der Lieth, C.W., Wolf, E., Gabius, H.J. Tumor galectinology: Insights into the complex network of a family of endogenous lectins. Glycoconj. J. 2004;20:227–238. doi: 10.1023/B:GLYC.0000025817.24297.17. [DOI] [PubMed] [Google Scholar]
- Le, K., Mitsouras, K., Roy, M., Wang, Q., Xu, Q., Nelson, S.F., Lee, C. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2004;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C., Wang, Q. Bioinformatics analysis of alternative splicing. Brief. Bioinform. 2005;6:23–33. doi: 10.1093/bib/6.1.23. [DOI] [PubMed] [Google Scholar]
- Lemaire, R., Winne, A., Sarkissian, M., Lafyatis, R. SF2 and SRp55 regulation of CD45 exon 4 skipping during T-cell activation. Eur. J. Immunol. 1999;29:823–837. doi: 10.1002/(SICI)1521-4141(199903)29:03<823::AID-IMMU823>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Lenardo, M.J. Molecular regulation of T-lymphocyte homeostasis in the healthy and diseased immune system. Immunol. Res. 2003;27:387–398. doi: 10.1385/IR:27:2-3:387. [DOI] [PubMed] [Google Scholar]
- Lin, Z., Fillmore, G.C., Um, T.H., Elenitoba-Johnson, K.S., Lim, M.S. Comparative microarray analysis of gene expression during activation of human peripheral blood T cells and leukemic Jurkat T cells. Lab. Invest. 2003;83:765–776. doi: 10.1097/01.lab.0000073130.58435.e5. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W. Consequences of regulated pre-mRNA splicing in the immune system. Nat. Rev. Immunol. 2004;4:931–940. doi: 10.1038/nri1497. [DOI] [PubMed] [Google Scholar]
- Lynch, K.W., Weiss, A. A model system for the activation-induced alternative-splicing of CD45 implicates protein kinase C and Ras. Mol. Cell. Biol. 2000;20:70–80. doi: 10.1128/mcb.20.1.70-80.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magistrelli, G., Jeannin, P., Herbault, N., Benoit De Coignac, A., Gauchat, J.F., Bonnefoy, J.Y., Delneste, Y. A soluble form of CTLA-4 generated by alternative splicing is expressed by nonstimulated human T cells. Eur. J. Immunol. 1999;29:3596–3602. doi: 10.1002/(SICI)1521-4141(199911)29:11<3596::AID-IMMU3596>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Majeti, R., Xu, Z., Parslow, T.G., Olson, J.L., Daikh, D.I., Killeen, N., Weiss, A. An inactivation point mutation in the inhibitory wedge of CD45 causes lymphoproliferation and autoimmunity. Cell. 2000;103:1059–1070. doi: 10.1016/s0092-8674(00)00209-9. [DOI] [PubMed] [Google Scholar]
- Marrack, P., Mitchell, T., Hildeman, D., Kedl, R., Teague, T.K., Bender, J., Rees, W., Schaefer, B.C., Kappler, J. Genomic-scale analysis of gene expression in resting and activated T cells. Curr. Opin. Immunol. 2000;12:206–209. doi: 10.1016/s0952-7915(99)00075-8. [DOI] [PubMed] [Google Scholar]
- Miosge, L.A., Goodnow, C.C. Genes, pathways and checkpoints in lymphocyte development and homeostasis. Immunol. Cell Biol. 2005;83:318–335. doi: 10.1111/j.1440-1711.2005.01353.x. [DOI] [PubMed] [Google Scholar]
- Modrek, B., Resch, A., Grasso, C., Lee, C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroy, T. The zinc finger transcription factor Growth factor independence 1 (Gfi1) Int. J. Biochem. Cell Biol. 2005;37:541–546. doi: 10.1016/j.biocel.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Oaks, M.K., Hallett, K.M., Penwell, R.T., Stauber, E.C., Warren, S.J., Tector, A.J. A native soluble form of CTLA-4. Cell. Immunol. 2000;201:144–153. doi: 10.1006/cimm.2000.1649. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Shai, O., Misquitta, C., Zhang, W., Saltzman, A.L., Mohammad, N., Babak, T., Siu, H., Hughes, T.R., Morris, Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Bakowski, M.A., Morris, Q., Zhang, W., Frey, B.J., Hughes, T.R., Blencowe, B.J. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan, Q., Saltzman, A.L., Kim, Y.K., Misquitta, C., Shai, O., Maquat, L.E., Frey, B.J., Blencowe, B.J. Quantitative microarray profiling provides evidence against widespread coupling of alternative splicing with nonsense-mediated mRNA decay to control gene expression. Genes & Dev. 2006;20:153–158. doi: 10.1101/gad.1382806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poenie, M., Kuhn, J., Combs, J. Real-time visualization of the cytoskeleton and effector functions in T cells. Curr. Opin. Immunol. 2004;16:428–438. doi: 10.1016/j.coi.2004.05.016. [DOI] [PubMed] [Google Scholar]
- Ponta, H., Sherman, L., Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Immunol. 2003;4:33–45. doi: 10.1038/nrm1004. [DOI] [PubMed] [Google Scholar]
- Raghavan, A., Ogilvie, R.L., Reilly, C., Abelson, M.L., Raghavan, S., Vasdewani, J., Krathwohl, M., Bohjanen, P.R. Genome-wide analysis of mRNA decay in resting and activated primary human T lymphocytes. Nucleic Acids Res. 2002;30:5529–5538. doi: 10.1093/nar/gkf682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romeo, G., Fiorucci, G., Chiantore, M.V., Percario, Z.A., Vannucchi, S., Affabris, E. IRF-1 as a negative regulator of cell proliferation. J. Interferon Cytokine Res. 2002;22:39–47. doi: 10.1089/107999002753452647. [DOI] [PubMed] [Google Scholar]
- Rothrock, C., Cannon, B., Hahm, B., Lynch, K.W. A conserved signal-responsive sequence mediates activation-induced alternative splicing of CD45. Mol. Cell. 2003;12:1317–1324. doi: 10.1016/s1097-2765(03)00434-9. [DOI] [PubMed] [Google Scholar]
- Rothrock, C.R., House, A.E., Lynch, K.W. HnRNP L represses exon splicing via a regulated exonic splicing silencer. EMBO J. 2005;24:2792–2802. doi: 10.1038/sj.emboj.7600745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton, G.R., Caceres, J.F., Mayeda, A., Bell, M.V., Plebanski, M., Jackson, D.G., Bell, J.I., Krainer, A.R. Identification and characterization of three members of the human SR family of pre-mRNA splicing factors. EMBO J. 1995;14:4336–4349. doi: 10.1002/j.1460-2075.1995.tb00108.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechi, A.S., Wehland, J. Interplay between TCR signalling and actin cytoskeleton dynamics. Trends Immunol. 2004;25:257–265. doi: 10.1016/j.it.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Shai, O., Morris, Q.D., Blencowe, B.J., Frey, B.J. Inferring global levels of alternative splicing isoforms using a generative model of microarray data. Bioinformatics. 2006;22:606–613. doi: 10.1093/bioinformatics/btk028. [DOI] [PubMed] [Google Scholar]
- Teague, T.K., Hildeman, D., Kedl, R.M., Mitchell, T., Rees, W., Schaefer, B.C., Bender, J., Kappler, J., Marrack, P. Activation changes the spectrum but not the diversity of genes expressed by T cells. Proc. Natl. Acad. Sci. 1999;96:12691–12696. doi: 10.1073/pnas.96.22.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dam, G.B., Zilch, C.F., Wallace, D., Wieringa, B., Beverley, P.C.L., Poels, L.G., Screaton, G.R. Regulation of alternative splicing of CD45 by antagonistic effects of SR protein splicing factors. J. Immunol. 2000;164:5287–5295. doi: 10.4049/jimmunol.164.10.5287. [DOI] [PubMed] [Google Scholar]
- Trowbridge, I.S., Thomas, M.L. CD45: An emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu. Rev. Immunol. 1994;12:85–116. doi: 10.1146/annurev.iy.12.040194.000505. [DOI] [PubMed] [Google Scholar]
- Ule, J., Ule, A., Spencer, J., Williams, A., Hu, J.S., Cline, M., Wang, H., Clark, T., Fraser, C., Ruggiu, M., et al. Nova regulates brain-specific splicing to shape the synapse. Nat. Genet. 2005;37:844–852. doi: 10.1038/ng1610. [DOI] [PubMed] [Google Scholar]
- Wang, H.-Y., Xu, X., Ding, J.-H., Bermingham, J.R., Fu, X.-D. SC35 plays a role in T cell development and alternative splicing of CD45. Mol. Cell. 2001;7:331–342. doi: 10.1016/s1097-2765(01)00181-2. [DOI] [PubMed] [Google Scholar]