Abstract
Carriers of premutation alleles (55–200 CGG repeats) of the fragile X mental retardation 1 (FMR1) gene have levels of FMR1 mRNA that are elevated by as much as 10-fold in peripheral blood leukocytes and CNS tissue. The excess expanded-repeat mRNA, per se, is now believed to result in forms of clinical involvement that are largely restricted to premutation carriers, including the neurodegenerative disorder, fragile X-associated tremor/ataxia syndrome (FXTAS). Although evidence to date suggests that the elevated mRNA is not due to increased stability, the basis for the increase is not known. In the current study, we have determined the relative transcriptional activities of premutation and normal FMR1 alleles using a highly sensitive nuclear run-on assay that involves immunocapture of digoxigenin-modified run-on transcripts followed by PCR amplification of the nascent transcripts. Using the nuclear run-on approach, we demonstrate that the rate of run-on synthesis of FMR1 transcripts is increased in premutation alleles. The current run-on assay should be broadly applicable to studies of other genes with promoters of weak to moderate strength. The fraction of capped FMR1 mRNA remains unaltered for premutation transcripts, indicating that elevated message levels are not due to premature escape from the cotranscriptional capping process. We also show that, in contrast to the situation with myotonic dystrophy, there is no net nuclear sequestration of premutation FMR1 mRNA. Finally, we have demonstrated that AGG interruptions within the CGG repeat element do not influence FMR1 mRNA levels.
Keywords: RNA toxicity, FXTAS, fragile X, nuclear run on, AGG, transcription rate
INTRODUCTION
Carriers of premutation alleles (55–200 CGG repeats) of the fragile X mental retardation 1 (FMR1) gene experience various forms of clinical involvement that appear to be unique to the premutation range, where the gene is active (Oostra and Willemsen 2003; Hagerman and Hagerman 2004). Such involvement includes premature ovarian failure (POF), seen in ∼20% of females who carry premutation alleles (Allingham-Hawkins et al. 1999; Marozzi et al. 2000), and fragile X-associated tremor/ataxia syndrome (FXTAS) (Hagerman et al. 2001, Jacquemont et al. 2003, 2004), a late-onset neurodegenerative disorder with core features of action tremor, gait ataxia, dementia, and parkinsonism. Larger CGG-repeat expansions (>200; full mutation) give rise to fragile X syndrome (OMIM +309550), the leading inherited form of mental impairment.
The presence of elevated levels of expanded-CGG repeat FMR1 mRNA in premutation carriers (Tassone et al. 2000a,c; Kenneson et al. 2001; Allen et al. 2004), coupled with the absence of FXTAS in older adults with fragile X syndrome, in whom the gene is silent, led us to propose that FXTAS is due to an RNA toxic gain of function (Hagerman et al. 2001; Greco et al. 2002, 2006; Hagerman and Hagerman 2004). Our model is analogous to the RNA gain-of-function model proposed for myotonic dystrophy, where CUG or CCUG repeat expansions (DMPK or ZNF9 genes, respectively), sequestered in the nuclei of affected cells, are believed to sequester one or more proteins to the expanded RNA repeat element (Finsterer 2002; Mankodi and Thornton 2002; Kanadia et al. 2003; Ho et al. 2004; Ranum and Day 2004). More recent studies of the expanded repeats in Drosophila (Jin et al. 2003), identification of the FMR1 mRNA in the intranuclear inclusions of FXTAS patients (Tassone et al. 2004b), and the recapitulation of CGG repeat-induced neural cell toxicity and inclusion formation in neural cell culture (Arocena et al. 2005) have provided additional support for an RNA-based pathogenesis of FXTAS (Hagerman and Hagerman 2004).
Although FMR1 mRNA levels are substantially elevated for alleles in the premutation range (Tassone et al. 2000a,c; Kenneson et al. 2001; Allen et al. 2004), the basis for the elevated mRNA levels is not known. Beilina et al. (2004) identified multiple FMR1 transcriptional initiation sites and demonstrated that the relative efficiencies among the start sites are modulated by the number of CGG repeats in the downstream 5′ untranslated region (5′ UTR) of the gene, suggesting that at least some of the influence of the CGG repeat is at the level of initiation. Studies of the decay of FMR1 mRNA levels in cell culture following treatment with actinomycin D suggested that there is no increase in the stability of the expanded-repeat mRNA (Tassone et al. 2000c); however, such studies are both indirect and subject to a large degree of uncertainty due to the relatively long half-life (∼8–10 h) of the FMR1 mRNA in lymphoid cells. Therefore, to address the basis for increased FMR1 mRNA levels associated with premutation alleles, we have performed a series of nuclear run-on experiments to measure directly the transcriptional activity of the FMR1 gene.
The nuclear run-on assay is based on the principle that genes in isolated nuclei will contain a number of stalled transcription complexes (due to loss of rNTPs during isolation of cell nuclei) in direct proportion to the transcriptional activity of the gene in vivo (Greenberg and Bender 1997). Thus, addition of labeled rUTP (and remaining rNTPs) to the isolated nuclei yields labeled run-on transcripts in proportion to the number of Pol II complexes that were stalled on the gene, since reinitiation does not occur in the isolated nuclei.
However, the standard nuclear run-on approach does not readily allow studies of low-abundance mRNAs such as the FMR1 mRNA (Zalfa et al. 2003) due to limitations in the sensitivity of detection of run-on transcripts. To overcome this intrinsic limitation of the method, we have developed a PCR-based approach to the nuclear run-on assay in which digoxygenin-labeled transcripts were affinity purified, followed by quantitative reverse transcriptase polymerase chain reaction (RT-PCR) amplification. We have used this assay to provide direct evidence that the increased FMR1 mRNA levels are due to increased transcriptional activity for both spliced and unspliced transcripts. In addition, using RNA in situ hybridization, we have demonstrated that FMR1 mRNA is principally located in the cytoplasm for both normal and premutation alleles, thus ruling out nuclear sequestration as a contributing factor in the elevated mRNA levels.
RESULTS
Nuclear run-on transcription measurements demonstrate an increase in transcriptional activity for the premutation FMR1 allele
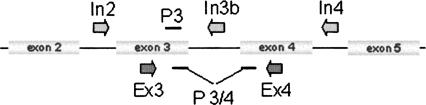
To obtain a direct measure of relative promoter activity for expanded (premutation) alleles of the FMR1 gene, we have developed a modification of the standard nuclear run-on assay (Greenberg and Bender 1997) in which the detection limit imposed by the filter-hybridization approach is avoided by using an RT-PCR-based method for quantification of run-on transcripts. Specifically, our method uses digoxigenin 11-UTP to label nascent RNA transcripts, which are then affinity purified with anti-digoxigenin-coated magnetic particles. Affinity purification of nascent, digoxygenin-labeled transcripts is followed by quantitative (real-time) RT-PCR specific for either mature FMR1 mRNA or incompletely spliced transcripts, depending on primer/probe selection (Fig. 1).
FIGURE 1.
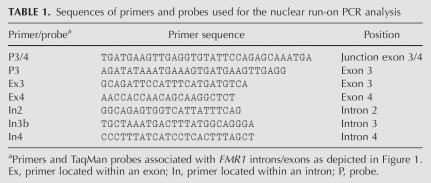
Diagram of the approximate locations of primers and probes used for the current RT-PCR analysis. Sequences of the primers and probes are given in Table 1.
In the current study, probe sets include primers and probes that target the intron adjacent to exon 3, to detect unprocessed or partially processed FMR1 transcripts, and a set that spans the intron/exon boundary between exons 3 and 4, to detect transcripts that are processed (Tassone et al. 2000c), at least with respect to the exon 3/4 junction. This step represents a second advantage relative to the standard, filter-hybridization approach, which does not distinguish between processed and unprocessed mRNA and is therefore subject to distortion by contamination of the processed RNA originally present in the preparation.
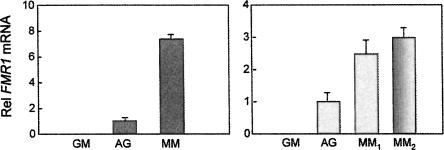
Quantitative RT-PCR, described by Tassone et al. (2000c), was performed on immunopurified, digoxygenin-labeled RNA transcripts (both nascent and mature), isolated from three lymphoblastoid cell lines harboring FMR1 alleles with differing CGG repeat numbers: a normal female (AG) with alleles of 16 and 29 CGG repeats, a premutation male (MM) with 160 repeats, and a full mutation male (GM) with ∼600 repeats. The results of our analysis (Fig. 2) demonstrate that the rate of transcription of the FMR1 gene is increased for premutation alleles. When compared to normal FMR1 alleles (AG), increased transcriptional activity was reflected by increased run-on production of both processed and unprocessed transcripts. Furthermore, the magnitude of the measured increase in transcriptional activity, determined in the run-on experiments for the premutation line (MM), were comparable to the magnitude of the increase in the steady-state level of FMR1 mRNA for the same premutation lymphoblastoid cell line (Primerano et al. 2002).
FIGURE 2.
Relative FMR1 mRNA and transcript levels in premutation versus normal lymphoblastoid cell lines. (A) Analysis of spliced message using primers located in exons 3 and 4, with the probe spanning the intron/exon junction. (B) Analysis of pre- or partially spliced transcripts using primers located in introns 2 and 3 (MM1) or in exon 3 and intron 4 (MM2). GM, hypermethylated, full mutation allele (∼600 CGG repeats); AG, normal allele (female; 16, 29 CGG repeats); MM, large premutation allele (160 CGG repeats). Error bars reflect measurements performed in triplicate.
Premutation alleles are not associated with nuclear sequestration
To determine whether nuclear sequestration contributes to the elevated mRNA levels for the expanded-repeat RNA, quantitative RT-PCR was performed on RNA isolated from separate cytoplasmic and nuclear fractions from lymphoblastoid cell lines derived from two individuals with moderate-to-large premutation alleles (PZ, 126 CGG; MM, 183 CGG), both presenting with high levels of FMR1 mRNA expression, and from an individual with a normal FMR1 allele (AG). Comparison of the cytoplasmic/nuclear mRNA ratios for the normal control (AG, ratio: 2.1 ± 0.07) and the two premutation cases (MM, ratio: 2.9 ± 0.10; PZ, 2.7 ± 0.12) demonstrates that there is no preferential nuclear sequestration of the mRNA derived from the two premutation cases. For all three cell lines, the majority of the mRNA is in the cytoplasm.
Thus, the elevated levels of FMR1 mRNA associated with premutation FMR1 alleles cannot be explained by a net redistribution of the FMR1 mRNA to the nucleus (e.g., nuclear sequestration) as a consequence of the expanded, noncoding CGG repeat. This result would, in turn, argue against a model for the lowered FMRP levels, observed in the upper premutation range (Tassone et al. 1999, 2000c; Kenneson et al. 2001), as being due to a relative deficiency of cytoplasmic mRNA.
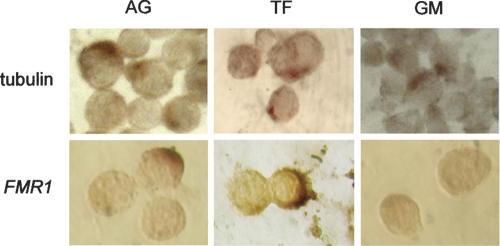
To further characterize the localization of the FMR1 transcript within the cells, we have performed RNA in situ hybridization using digoxygenin-labeled riboprobes that are specific for FMR1 and for tubulin (control) mRNAs. Sense and antisense probes were used for both targets. As demonstrated in Figure 3, both FMR1 and tubulin mRNAs are principally located in the cytoplasm for both normal and premutation alleles. As a control, sense riboprobes for both FMR1 and tubulin RNA did not reveal any signal, thus demonstrating specificity of the in situ hybridization reaction (data not shown). These observations provide further evidence against nuclear sequestration as a major contributing factor for the elevated expanded FMR1 mRNA levels.
FIGURE 3.
In situ hybridization with β-tubulin and FMR1 antisense strand in lymphoblastoid cell line derived from a normal individual (AG), from a male premutation carrier (TF, 190 CGG repeats), and from an individual with a hypermethylated full mutation (GM, ∼600 CGG repeats).
Expanded FMR1 mRNAs are not subject to preferential escape from cotranscriptional capping
We have also investigated the possibility that the elevated mRNA levels associated with premutation alleles may reflect at least partial escape from the cotranscriptional capping that normally occurs with the transition from transcriptional initiation to elongation (Coppola et al. 1983). Were the expanded CGG repeat to facilitate such an escape, one would expect that decreased levels of capping of the mature mRNA would accompany the increases in transcript level. Reduced capping efficiency would predict reduced efficiency of translation of FMRP, consistent with the reduced FMRP levels observed in the upper premutation range (Tassone et al. 1999, 2000c; Kenneson et al. 2001); however, reduced capping efficiency would also lead to the expectation of lowered mRNA levels as a result of reduced stability of the uncapped message, which is not observed.
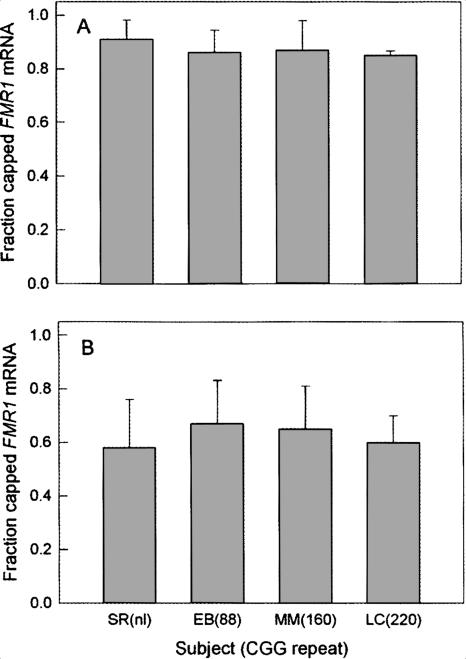
To address this question directly, we have performed a capping fractionation assay on lymphoblastoid cell lines derived from four individuals with FMR1 alleles of differing CGG repeat number (SR, 25 CGG repeats; EB, 88 repeats; MM, 160 repeats; LC, 220 CGG repeats, ∼85% methylated). Capped and uncapped RNAs were reverse transcribed, and quantitative RT-PCR was performed as described above. Both GUS and GAPDH were used as reference genes. These cap-fractionation assays demonstrate the lack of a significant difference in capping efficiency with increasing CGG repeat number (Fig. 4). In particular, the percent of capped FMR1 mRNA did not differ in the total RNA pools for normal and premutation CGG repeat ranges using either GUS or GAPDH as the controls. Interestingly, the apparent capping efficiency overall seemed to be decreased for the GAPDH control; however, this is likely due to errors associated with the lower levels of GAPDH mRNA relative to FMR1 mRNA. In any case, the ratios do not change with increasing CGG repeat number.
FIGURE 4.
Expansion of the CGG repeat in the FMR1 mRNA does not affect the fraction of message that possesses a 5′ cap. Capped and uncapped mRNAs were fractionated by affinity chromatography (see Materials and Methods); capped and uncapped mRNAs were quantified by TaqMan RT-PCR, using either GUS (A) or GAPDH (B) mRNAs as reference genes.
AGG interruptions within the CGG repeat tract do not influence transcript levels within the premutation range
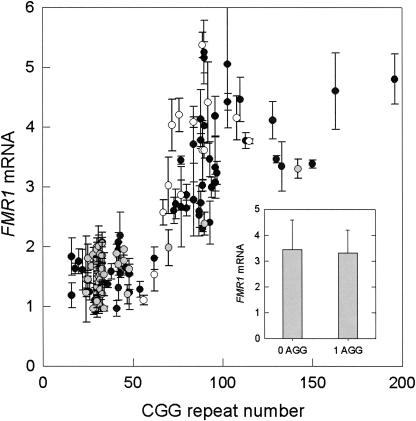
One of the distinguishing features of expanded (CGG) repeat FMR1 alleles is the loss of the two to three AGG interruptions that occur every ∼9–10 CGG repeats within the repeat element (Eichler et al. 1994; Kunst and Warren 1994; Snow et al. 1994; Zhong et al. 1994). Such elements are generally regarded as exerting a stabilizing effect on repeat size during transmission (Dombrowski et al. 2002) and have been demonstrated to affect the structure of both DNA and RNA (Pearson et al. 1998; Napierala et al. 2005); however, there are currently no data that address a possible role of the AGG repeat on either transcriptional activity or mRNA stability. To address the possibility that the AGG interruptions have an influence on FMR1 mRNA levels, we have performed a preliminary series of ANCOVAs on separate sample data sets (mRNA level, CGG repeat, AGG repeat number) in each of the three CGG repeat domains: normal (<45 CGG repeats), gray zone (45–54 CGG repeats), and premutation (55–200 CGG repeats). The breakdown in AGG repeat number and number of subjects was as follows (number of AGG interruptions [number of subjects]): Normal range: 0 (1), 1 (20), 2 (55); gray zone: 0 (0), 1 (5), 2 (5); premutation: 0 (21), 1 (36), 2 (2).
The central result of this analysis is as follows: whereas the dependence of mRNA levels on CGG repeat is highly significant in the premutation range (p = 0.0036) (Fig. 5), there is no dependence of the mRNA level on the number of AGG repeats (mainly, 0 or 1 AGGs; p = 0.87). This is true whether the premutation allele derives from individuals with or without FXTAS. Thus, the number of CGG interruptions does not seem to play a role in either transcriptional activity or in the pathogenesis of FXTAS. In the normal range, there is no dependence of the mRNA level on AGG repeat number (mainly, 1 or 2 AGGs), CGG repeat number, or gender. We also did not see any effect of the number of AGG interruptions on the FMR1 protein (FMRP) measure (% FMRP-positive lymphocytes) in the premutation range, although there is a negative correlation (p = 0.017) between CGG repeat number and %FMRP-positive lymphocytes in the premutation range, consistent with previous observations in peripheral blood leukocytes (Tassone et al. 2000c) and lymphoblastoid lines (Kenneson et al. 2001).
FIGURE 5.
Graph of the relationship between relative FMR1 mRNA levels, the number of CGG repeats, and the number of AGG interruptions, for a cohort of 128 adult male premutation carriers and controls. Gray circles, 2 AGG interruptions; open circles, 1 AGG; black circles, 0 AGGs. (Inset) Histogram representing the average FMR1 mRNA levels for premutation alleles with 0 AGG interruptions or with 1 AGG interruption (includes two cases with 2 AGGs); error bars represent SEMs.
DISCUSSION
We have previously reported that FMR1 mRNA levels are elevated by as much as 10-fold in peripheral blood leukocytes of carriers of premutation alleles of the FMR1 gene (Tassone et al. 2000a,c); with the magnitude of the elevation displaying a strong positive correlation with the length of the CGG repeat in both leukocytes (Tassone et al. 2000a,c; Allen et al. 2004) and transformed lymphoblastoid cells (Kenneson et al. 2001); elevated levels extend well into the full mutation range for unmethylated alleles (Tassone et al. 2000b). Such increases do not appear to be a result of increased message stability (Tassone et al. 2000c), since the rate of decay of FMR1 mRNA does not appear to be altered in premutation cells, relative to controls, following treatment with the Pol II inhibitor, actinomycin D. However, such measurements provide only indirect evidence for increased transcriptional activity, with the added caveat that the interpretation of actinomycin results is much less certain for messages with intrinsically long half-lives, which is the situation with FMR1 mRNA.
Increased message levels were also observed when luciferase reporter constructs harboring the FMR1 5′ UTR with premutation CGG repeat expansions were transfected into either SK or 293 cells (Chen et al. 2003). This observation suggests that the expanded CGG repeat itself, not lowered FMRP levels (Tassone et al. 2000c; Kenneson et al. 2001), is driving increased mRNA production; however, the increased mRNA levels observed in the Chen et al. (2003) study were somewhat more modest than the elevations observed for the same alleles in peripheral blood leukocytes.
To more directly approach the question of the basis for the elevated mRNA levels, we used a sensitive, PCR-based nuclear transcription run-on approach to determine whether expanded-repeat FMR1 alleles are associated with increased transcriptional activity. Using probes for both spliced and unspliced transcripts, we demonstrated that premutation alleles are associated with increased run-on transcription; thus, our current results support increased transcriptional activity as the basis for most, if not all, of the elevated levels of FMR1 expanded alleles.
We also addressed the possibility that nuclear sequestration plays a contributing role, perhaps through stabilization of the retained mRNA; significant sequestration does occur for the expanded (CUG) repeat in the 3′ UTR of the myotonic dystrophy protein kinase (DMPK) gene (Mankodi and Thornton 2002) associated with myotonic dystrophy (DM1; MIM 160,900); lowered cytoplasmic levels of DMPK mRNA are thought to contribute to lowered levels of DMPK protein (MIM 605,377). Using two separate methods, RT-PCR of RNA isolated from nuclear and cytoplasmic fractions and in situ hybridization of FMR1 mRNA, we found no evidence for a CGG-repeat effect on nuclear retention. Thus, increased stability through selective retention of expanded-repeat mRNA cannot explain the elevated mRNA levels. Moreover, these results rule out nuclear sequestration as an explanation for the lowered FMRP levels observed in leukocytes obtained from carriers of premutation alleles, a conclusion that is in agreement with other evidence supporting a model in which the expanded CGG repeat in the 5′ UTR acts as a direct impediment to translation (Primerano et al. 2002). It should be noted that whereas neither cytoplasmic/nuclear fractionation nor in situ labeling would be capable of detecting small shifts in message distribution, the former because nuclear and cytoplasmic fractions are never fully separated and the latter because of its intrinsically qualitative nature, the methods are clearly capable of ruling out the large redistribution that would be required to account for retention-coupled mRNA stabilization as the basis for the increase in mRNA levels.
It is not clear how the expanded CGG repeat leads to increased transcription of the FMR1 gene; none of the trans-acting factors that presumably transduce the downstream signal presented by CGG repeat has been identified. However, it is known that the CGG repeat does influence the choice of transcription start site (Beilina et al. 2004), with most premutation transcripts initiating from an initiator element (Inr) that is ∼50 nucleotides (nt) upstream of the principal Inr element for transcription of normal alleles. Further studies are clearly necessary to elucidate the mechanisms that lead to transcriptional dysregulation of the expanded-repeat FMR1 gene. Excess of expanded-repeat mRNA is now believed to give rise to the neurodegenerative disorder FXTAS. In addition, the Drosophila melanogaster model, with an expanded CGG transcript of ∼90 CGG repeats, has shown that the rCGG alone can lead to the development of neurogenerative changes in the eye, accompanied by the formation of inclusions (Jin et al. 2003). Thus, detailed knowledge of the mechanism of transcriptional initiation should lead to targeted therapeutic approaches for the treatment of FXTAS.
We have also explored the possibility that the increased rate of transcript synthesis in vivo of the expanded-repeat mRNA may reflect at least partial escape from cotranscriptional capping of the nascent transcript (Coppola et al. 1983). Such an escape process would predict lowered levels of capped mRNA for the expanded CGG alleles; however, our data show that total RNA pools for both normal and premutation CGG repeat ranges contain FMR1 mRNA that have equivalent extents of capping. These results are significant for two reasons. First, the fact that there is no difference between normal and premutation alleles suggests that the possibility raised above is unlikely to be responsible for the increase in mRNA levels in the premutation range. Second, as a practical consideration, these results indicate that the FMR1 mRNA within our samples is largely intact at the 5′ end.
Within the CGG repeat element in the 5′ UTR region of the FMR1 gene, two to three AGG repeats are generally associated with normal alleles, whereas in the gray zone and premutation range, one or both of the AGG interruptions is lost. These AGG interruptions are thought to impart increased genetic (DNA) stability during replication and repair by precluding the formation of secondary structures (e.g., CGG hairpins) (Kunst and Warren 1994; Pearson et al. 1998; Dombrowski et al. 2002) within the repeat region. However, there has been no study of the influence of the AGG repeat on mRNA levels. As an initial approach to this issue, we have determined whether there is an association between the presence/absence of the AGG interruptions and mRNA level. The results of our analysis indicate that the AGG element does not have a significant effect on mRNA levels within either normal or premutation ranges.
Finally, the current study has important ramifications for potential therapeutic approaches to treat FXTAS, which now appears to be due to a toxic gain of function of the expanded (CGG) repeat FMR1 mRNA (Hagerman et al. 2001; Greco et al. 2002; Jacquemont et al. 2003; Jin et al. 2003; for review, see Hagerman and Hagerman 2004). In particular, targeted reduction of the mRNA levels would be expected to reduce the ongoing pathogenic process. Since the elevated mRNA levels appear to be driven primarily by increased transcription, both the mRNA per se and dysregulation of the FMR1 promoter represent potential targets.
MATERIALS AND METHODS
Nuclei were isolated from lymphoblastoid cell lines following the protocol of Greenberg and Bender (1997). Briefly, 5 × 107 cells were pelleted in a conical tube and washed twice with ice-cold phosphate saline buffer (PBS). Cells were then lysed on ice for 5 min in lysis buffer containing 0.5% (v/v) NP-40, 10 mM Tris-HCl at pH 7.4, 10 mM NaCl, and 3 mM MgCl2. After centrifugation (15 min at 500 relative centrifuge force [rcf]), the supernatant was removed and used to isolate cytoplasmic RNA by standard methods (Trizol, Invitrogen, Life technologies). The nuclear pellet was resuspended and subjected to a repeat (5 min) lysis to remove any remaining intact cells. Following centrifugation, the supernatant was discarded and the nuclei were resuspended in 200 μL of glycerol storage buffer (50 mM Tris-HCl at pH 8.3, 5 mM MgCl2, 0.1 mM EDTA, 40% [v/v] glycerol); cells were stored in liquid nitrogen until further use.
Transcriptional elongation was carried out as described by Greenberg and Bender (1997) with several modifications. Frozen nuclei, isolated as described above, were thawed and resuspended in one volume (200 μL) of reaction buffer (10 mM Tris-HCl at pH 8.0, 5 mM MgCl2, and 0.3 M KCl, 0.1 mM of each ATP, GTP, CTP), and were incubated for 30 min at 37°C in the presence of 0.2 μM of digoxigenin 11-UTP (Roche). Nuclei were then treated with DNase I and Proteinase K for 15 min at 37°C. Labeled transcripts were then isolated and purified using the Trizol method (Invitrogen, Life technologies). Finally, nascent digoxygenin-labeled RNA transcripts were affinity purified using anti-digoxygenin-coated magnetic particles according to the manufacturer's protocol (Roche). Specifically, the purified (digoxigenin-labeled) total RNA was incubated with anti-digoxigenin beads for 30 min at room temperature in TEN buffer (10 mM Tri-HCl, 1 mM EDTA, 100 mM NaCl at pH 7.5), with gentle and occasional inversion. Particles were washed twice in washing buffer (10 mM Tri-HCl, 1 mM EDTA, 1 M NaCl at pH 7.5) and equilibrated in 100 μL of the same buffer with guanidine HCl added to a final concentration of 6 M. Concentrations of the eluted mRNAs were determined spectrophotometrically. Aliquots of 500 ng of isolated RNA were used for quantitative (TaqMan) RT-PCR assays as described by Tassone et al. (2000c).
Quantitative (fluorescence) RT-PCR was used to obtain precise estimates of FMR1 transcript levels, using β-glucoronidase (GUS) as the control RNA. Mature mRNA levels were measured in both cytoplasmic and nuclear compartments of lymphoblastoid cell lines derived from a normal individual (AG), from an individual with a full mutation (GM), and from a premutation carrier (MM). Quantitative RT-PCR was also used to measure nascent transcript from the FMR1 gene using the digoxygenin labeling strategy described above. Details of the RT-PCR method and its application to studies of FMR1 mRNA levels are described by Tassone et al. (2000c). For mature mRNA, the 81-base-pair (bp) amplicon specific for the reference gene β-glucoronidase (GUS) (GenBank NM000181) spans exons 11 and 12 of the GUS gene, while the amplicon specific for the FMR1 gene (GenBank NM000181) spans the boundary between exons 3 and 4. In addition, sets of primers and probes spanning intron 2 and exon 4 and lying between exon 2 and intron 4 were used to measure the levels of spliced or partially spliced FMR1 mRNA. All RT-PCR analyses were performed in duplicate and for three different RNA concentrations, and standards were included for each assay to compensate for any changes in reaction efficiency, as previously described (Tassone et al. 2000c). Sequences of the primers and probes used for the current study are listed in Table 1.
TABLE 1.
Sequences of primers and probes used for the nuclear run-on PCR analysis
Lymphoblastoid cell lines were maintained in RPMI-1640 supplemented with 15% fetal bovine serum (heat inactivated) (Gibco BRL), glutamine, and penicillin/streptomycin (Gibco BRL) at 37°C in a humidified atmosphere of 5% CO2. Cultures were grown until they reached a density of 1 × 106 cells/mL, when they were used for the isolation of nuclei and cytoplasmic fractions.
CGG repeat size was determined by both Southern blot and by PCR analysis. Southern blot was performed on DNA isolated from lymphoblastoid cell lines using the FMR1 genomic probe StB12.3, labeled with digoxygenin-11-dUTP by PCR (PCR dig synthesis Kit, Roche Diagnostics), as previously described (Tassone et al. 2004a). Genomic DNA was also amplified by PCR using primers c and f (Fu et al. 1991) and performed using the Expand Long Template PCR System (Roche Diagnostics) in conjunction with the osmolite betaine (B0300, Sigma-Aldrich), as described by Saluto et al. (2005). Analysis and calculation of the repeat size for both Southern blot and PCR analysis were carried out using an Alpha Innotech FluorChem 8800 Image Detection System.
Samples of 1.8 × 106 lymphoblastoid cells were cytospin fixed at room temperature for 30 min with 4% paraformaldehyde in 1× PBS, followed by washing in 1× PBS. Cells were permeabilized at 37°C for 5 min with PRONASE at a final concentration of 10 μg/mL (Roche Applied Science), followed by prehybridization for 2 h at 55°C in 50% formamide, 5× SSC, 5× Denhardt's solution, 0.2% SDS, 100 μg/mL heparin, 250 μg/mL yeast tRNA, and 250 μg/mL salmon sperm DNA. Hybridization was performed overnight at 55°C in the same solution, with addition of 300–800 ng/mL of the in vitro synthesized FMR1 digoxigenin-labeled riboprobe complementary to nucleotides 1891–3583 of FMR1 mRNA sequence (L23971). Beta-tubulin riboprobe (nucleotides 453–1052 of gene mRNA sequence; M13441) was used as the control. Washes were performed first at room temperature in 1× SSC plus 0.1% SDS, then at 50°C in 0.2× SSC plus 0.1% SDS. RNase treatment was for 30 min at 37°C (20 μg/mL). The hybridized probe was detected immunocytochemically using a sheep anti-digoxigenin antibody (1:500 dilution) coupled to alkaline phosphatase for 5 h at room temperature; nitro blue tetrazolium and 5-bromo-4-chloro-3-indolyl phosphate were used as a substrate according to the manufacturer's protocol (Roche Applied Sciences). Lymphoblastoid cell nuclei were stained with 1 μg /mL DAPI at room temperature for 10 min. Cells were photographed on a Nikon Eclipse TE200 microscope.
The number of AGG interruptions was determined for a total of 128 subjects, distributed across three categories on the basis of CGG repeat size, as follows: normal alleles (<45 CGG repeats; n = 70), gray-zone alleles (45–54 CGG repeats; n = 9), and premutation alleles (55–200 CGG repeats; n = 49). For each of the 129 male subjects, the number of AGG interruptions was determined by restriction enzyme digestion of PCR products that were generated according to the protocol of Saluto et al. (2005) using primers c and f (Fu et al. 1991). PCR products were purified using the MinElute purification kit (Qiagen), followed by digestion with the restriction enzyme, EciI, which specifically targets the CGG sequence containing the AGG trinucleotide interruption (5′-GCGGAN11). Eci cleavage allows one to determine the number and approximate locations of AGG interruptions, based on the size and the number of bands of the digested PCR product on a 3% agarose gel. In a normal allele, three bands are usually observed, a 30-bp fragment and two larger fragments (one of ∼160 bp and one of ∼120 bp).
Fractionation of total cell RNA into capped and uncapped RNA populations was performed by selective binding of the capped RNA to recombinant GST-eIF4E (McCracken et al. 1997). Briefly, 10 μg of total RNA from each sample, along with ∼25 ng of 32P-labeled in vitro transcribed/capped c-myc transcript (as pulldown capped control), were incubated with 20 μg of GST-eIF4E (purified as described by McCracken et al. 1997) for 1 h at 4°C in binding buffer (McCracken et al. 1997). The mRNA:eIF4E complex was then incubated in 50 μL of glutathione sepharose-bead slurry for an additional hour, and the resulting fractionated RNA was extracted with phenol/chloroform, EtOH precipitated, and resuspended in 50 μL of H2O. To determine cap fractionation efficiency, 10 μL of each fraction was run on an 8% acrylamide gel and analyzed by autoradiography of 32P-labeled capped-myc.
ACKNOWLEDGMENTS
We thank Dr. D. Zorio for help with the cap fractionation assay and Boris Eleuteri for the in situ hybridization. This research was supported by grants from the National Institutes of Health (PH40661, P.J.H.; GM58613, D.B.), by the UC Davis Health Systems Research Award Program (F.T.), and by Ministero della Salute and Istituto Superiore di Sanità (C.B.).
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.280807.
REFERENCES
- Allen, E.G., He, W., Yadav-Shah, M., Sherman, S.L. A study of the distributional characteristics of FMR1 transcript levels in 238 individuals. Hum. Genet. 2004;114:439–447. doi: 10.1007/s00439-004-1086-x. [DOI] [PubMed] [Google Scholar]
- Allingham-Hawkins, D.J., Babul-Hirji, R., Chitayat, D., Holden, J.J., Yang, K.T., Lee, C., Hudson, R., Gorwill, H., Nolin, S.L., Glicksman, A., et al. Fragile X premutation is a significant risk factor for premature ovarian failure: The International Collaborative POF in Fragile X study–preliminary data. Am. J. Med. Genet. 1999;83:322–325. [PMC free article] [PubMed] [Google Scholar]
- Arocena, D.G., Iwahashi, C.K., Won, N., Beilina, A., Ludwig, A.L., Tassone, F., Schwartz, P.H., Hagerman, P.J. Induction of inclusion formation and disruption of lamin A/C structure by premutation CGG-repeat RNA in human cultured neural cells. Hum. Mol. Genet. 2005;14:3661–3671. doi: 10.1093/hmg/ddi394. [DOI] [PubMed] [Google Scholar]
- Beilina, A., Tassone, F., Schwartz, P.H., Sahota, P., Hagerman, P.J. Redistribution of transcription start sites within the FMR1 promoter region with expansion of the downstream CGG-repeat element. Hum. Mol. Genet. 2004;13:543–549. doi: 10.1093/hmg/ddh053. [DOI] [PubMed] [Google Scholar]
- Chen, L.S., Tassone, F., Sahota, P., Hagerman, P.J. The (CGG)n repeat element within the 5′ untranslated region of the FMR1 message provides both positive and negative cis effects on in vivo translation of a downstream reporter. Hum. Mol. Genet. 2003;12:3067–3074. doi: 10.1093/hmg/ddg331. [DOI] [PubMed] [Google Scholar]
- Coppola, J.A., Field, A.S., Luse, D.S. Promoter-proximal pausing by RNA polymerase II in vitro: Transcripts shorter than 20 nucleotides are not capped. Proc. Natl. Acad. Sci. 1983;80:1251–1255. doi: 10.1073/pnas.80.5.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrowski, C., Levesque, S., Morel, M.L., Rouillard, P., Morgan, K., Rousseau, F. Premutation and intermediate-size FMR1 alleles in 10,572 males from the general population: Loss of an AGG interruption is a late event in the generation of fragile X syndrome alleles. Hum. Mol. Genet. 2002;11:371–378. doi: 10.1093/hmg/11.4.371. [DOI] [PubMed] [Google Scholar]
- Eichler, E.E., Holden, J.J., Popovich, B.W., Reiss, A.L., Snow, K., Thibodeau, S.N., Richards, C.S., Ward, P.A., Nelson, D.L. Length of uninterrupted CGG repeats determines instability in the FMR1 gene. Nat. Genet. 1994;8:88–94. doi: 10.1038/ng0994-88. [DOI] [PubMed] [Google Scholar]
- Finsterer, J. Myotonic dystrophy type 2. Eur. J. Neurol. 2002;9:441–447. doi: 10.1046/j.1468-1331.2002.00453.x. [DOI] [PubMed] [Google Scholar]
- Fu, Y.H., Kuhl, D.P., Pizzuti, A., Pieretti, M., Sutcliffe, J.S., Richards, S., Verkerk, A.J., Holden, J.J., Fenwick R.G., Jr., Warren, S.T., et al. Variation of the CGG repeat at the fragile X site results in genetic instability: Resolution of the Sherman paradox. Cell. 1991;67:1047–1058. doi: 10.1016/0092-8674(91)90283-5. [DOI] [PubMed] [Google Scholar]
- Greco, C.M., Hagerman, R.J., Tassone, F., Chudley, A.E., Del Bigio, M.R., Jacquemont, S., Leehey, M., Hagerman, P.J. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- Greco, C.M., Berman, R.F., Martin, R.M., Tassone, F., Schwartz, P.H., Chang, A., Trapp, B.D., Iwahashi, C., Brunberg, J., Grigsby, J., et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- Greenberg, M., Bender, T. Nuclear runoff transcription in mammalian cells. Curr. Prot. Molec. Bio. Supp. 1997;37:4.10.11–14.10.11. [Google Scholar]
- Hagerman, P.J., Hagerman, R.J. The fragile-X premutation: A maturing perspective. Am. J. Hum. Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman, R.J., Leehey, M., Heinrichs, W., Tassone, F., Wilson, R., Hills, J., Grigsby, J., Gage, B., Hagerman, P.J. Intention tremor, parkinsonism, and generalized brain atrophy in male carriers of fragile X. Neurology. 2001;57:127–130. doi: 10.1212/wnl.57.1.127. [DOI] [PubMed] [Google Scholar]
- Ho, T.H., Charlet, B.N., Poulos, M.G., Singh, G., Swanson, M.S., Cooper, T.A. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont, S., Hagerman, R.J., Leehey, M., Grigsby, J., Zhang, L., Brunberg, J.A., Greco, C., Des Portes, V., Jardini, T., Levine, R., et al. Fragile X premutation tremor/ataxia syndrome: Molecular, clinical, and neuroimaging correlates. Am. J. Hum. Genet. 2003;72:869–878. doi: 10.1086/374321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquemont, S., Hagerman, R.J., Leehey, M.A., Hall, D.A., Levine, R.A., Brunberg, J.A., Zhang, L., Jardini, T., Gane, L.W., Harris, S.W., et al. Penetrance of the fragile X-associated tremor/ataxia syndrome in a premutation carrier population. JAMA. 2004;291:460–469. doi: 10.1001/jama.291.4.460. [DOI] [PubMed] [Google Scholar]
- Jin, P., Zarnescu, D.C., Zhang, F., Pearson, C.E., Lucchesi, J.C., Moses, K., Warren, S.T. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila . Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- Kanadia, R.N., Johnstone, K.A., Mankodi, A., Lungu, C., Thornton, C.A., Esson, D., Timmers, A.M., Hauswirth, W.W., Swanson, M.S. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Kenneson, A., Zhang, F., Hagedorn, C.H., Warren, S.T. Reduced FMRP and increased FMR1 transcription is proportionally associated with CGG repeat number in intermediate-length and premutation carriers. Hum. Mol. Genet. 2001;10:1449–1454. doi: 10.1093/hmg/10.14.1449. [DOI] [PubMed] [Google Scholar]
- Kunst, C.B., Warren, S.T. Cryptic and polar variation of the fragile X repeat could result in predisposing normal alleles. Cell. 1994;77:853–861. doi: 10.1016/0092-8674(94)90134-1. [DOI] [PubMed] [Google Scholar]
- Mankodi, A., Thornton, C.A. Myotonic syndromes. Curr. Opin.Neurol. 2002;15:545–552. doi: 10.1097/00019052-200210000-00005. [DOI] [PubMed] [Google Scholar]
- Marozzi, A., Vegetti, W., Manfredini, E., Tibiletti, M.G., Testa, G., Crosignani, P.G., Ginelli, E., Meneveri, R., Dalpra, L. Association between idiopathic premature ovarian failure and fragile X premutation. Hum. Reprod. 2000;15:197–202. doi: 10.1093/humrep/15.1.197. [DOI] [PubMed] [Google Scholar]
- McCracken, S., Fong, N., Rosonina, E., Yankulov, K., Brothers, G., Siderovski, D., Hessel, A., Foster, S., Shuman, S., Bentley, D.L. 5′-capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes & Dev. 1997;11:3306–3318. doi: 10.1101/gad.11.24.3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napierala, M., Bacolla, A., Wells, R.D. Increased negative superhelical density in vivo enhances the genetic instability of triplet repeat sequences. J. Biol. Chem. 2005;280:37366–37376. doi: 10.1074/jbc.M508065200. [DOI] [PubMed] [Google Scholar]
- Oostra, B.A., Willemsen, R. A fragile balance: FMR1 expression levels. Hum. Mol. Genet. 2003;12:R249–R257. doi: 10.1093/hmg/ddg298. [DOI] [PubMed] [Google Scholar]
- Pearson, C.E., Eichler, E.E., Lorenzetti, D., Kramer, S.F., Zoghbi, H.Y., Nelson, D.L., Sinden, R.R. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37:2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- Primerano, B., Tassone, F., Hagerman, R.J., Hagerman, P., Amaldi, F., Bagni, C. Reduced FMR1 mRNA translation efficiency in fragile X patients with premutations. RNA. 2002;8:1482–1488. [PMC free article] [PubMed] [Google Scholar]
- Ranum, L.P., Day, J.W. Myotonic dystrophy: RNA pathogenesis comes into focus. Am. J. Hum. Genet. 2004;74:793–804. doi: 10.1086/383590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saluto, A., Brussino, A., Tassone, F., Arduino, C., Cagnoli, C., Pappi, P., Hagerman, P., Migone, N., Brusco, A. An enhanced polymerase chain reaction assay to detect pre- and full mutation alleles of the fragile X mental retardation 1 gene. J. Mol. Diagn. 2005;7:605–612. doi: 10.1016/S1525-1578(10)60594-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow, K., Tester, D.J., Kruckeberg, K.E., Schaid, D.J., Thibodeau, S.N. Sequence analysis of the fragile X trinucleotide repeat: Implications for the origin of the fragile X mutation. Hum. Mol. Genet. 1994;3:1543–1551. doi: 10.1093/hmg/3.9.1543. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Iklé, D.N., Dyer, P.N., Lampe, M., Willemsen, R., Oostra, B.A., Taylor, A.K. FMRP expression as a potential prognostic indicator in fragile X syndrome. Am. J. Med. Genet. 1999;84:250–261. [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Chamberlain, W.D., Hagerman, P.J. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000a;97:195–203. doi: 10.1002/1096-8628(200023)97:3<195::AID-AJMG1037>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Loesch, D.Z., Lachiewicz, A., Taylor, A.K., Hagerman, P.J. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels of FMR1 messenger RNA. Am. J. Med. Genet. 2000b;94:232–236. doi: 10.1002/1096-8628(20000918)94:3<232::aid-ajmg9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Taylor, A.K., Gane, L.W., Godfrey, T.E., Hagerman, P.J. Elevated levels of FMR1 mRNA in carrier males: A new mechanism of involvement in the fragile-X syndrome. Am. J. Hum. Genet. 2000c;66:6–15. doi: 10.1086/302720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone, F., Hagerman, R.J., Garcia-Arocena, D., Khandjian, E.W., Greco, C.M., Hagerman, P.J. Intranuclear inclusions in neural cells with premutation alleles in fragile X associated tremor/ataxia syndrome. J. Med. Genet. 2004a;41:e43. doi: 10.1136/jmg.2003.012518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone, F., Iwahashi, C., Hagerman, P.J. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004b;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B., Bagni, C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- Zhong, N., Ye, L., Dobkin, C., Brown, W.T. Fragile X founder chromosome effects: Linkage disequilibrium or microsatellite heterogeneity? Am. J. Med. Genet. 1994;51:405–411. doi: 10.1002/ajmg.1320510421. [DOI] [PubMed] [Google Scholar]