Abstract
Most riboswitches are composed of a single metabolite-binding aptamer and a single expression platform that function together to regulate genes in response to changing metabolite concentrations. In rare instances, two aptamers or sometimes two complete riboswitches reside adjacent to each other in untranslated regions (UTRs) of mRNAs. We have examined an example of a tandem riboswitch in the Gram-positive bacterium Bacillus anthracis that includes two complete riboswitches for thiamine pyrophosphate (TPP). Unlike other complex riboswitch systems described recently, tandem TPP riboswitches do not exhibit cooperative ligand binding and do not detect two different types of metabolites. In contrast, both riboswitches respond independently to TPP and are predicted to function in concert to mimic the more “digital” gene control outcome observed when two aptamers bind ligands cooperatively. Our findings further demonstrate that simple gene control elements made only of RNA can be assembled in different architectures to yield more complex gene control outcomes.
Keywords: aptamer, metabolite-binding RNA, thiamine pyrophosphate, transcription termination
INTRODUCTION
Many bacteria make extensive use of ligand-binding RNAs called riboswitches (Nahvi et al. 2002) to sense the concentrations of important metabolites and to control genes that are important for metabolite biosynthesis and transport (Mandal and Breaker 2004; Soukup and Soukup 2004; Winkler and Breaker 2005). An exceptionally widespread class of riboswitches forms a highly conserved structure that selectively binds thiamine pyrophosphate (TPP) (Mironov et al. 2002; Winkler et al. 2002b). TPP-specific aptamers form two distinct pockets that separately bind the HMP (4-amino-5-hydroxymethyl-2-methylpyrimidine) and pyrophosphate moieties of the coenzyme (Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006; Thore et al. 2006). The thiazole moiety of TPP bridges these two domains of the aptamer, which aids in stabilizing the overall RNA fold. The structural changes brought about by ligand binding to the aptamer are harnessed by the adjoining expression platform to alter transcription or translation of the downstream open reading frame (ORF).
The structural requirements for ligand binding by RNA place a strong selective pressure for riboswitches to remain conserved in nucleotide sequence and structure throughout evolution. Most riboswitch aptamers remain so well conserved that their nucleotide sequences and secondary structure features are used as distinguishing characteristics for each riboswitch class. The conserved features can be used to create bioinformatics search algorithms that identify additional members of riboswitch classes from rapidly increasing collections of genomic DNA sequences (e.g., Vitreschak et al. 2002, 2003; Sudarsan et al. 2003a; Barrick et al. 2004; Nahvi et al. 2004; Corbino et al. 2005). As a result, numerous bacterial TPP riboswitches (Miranda-Rios et al. 2001; Rodionov et al. 2002; Winkler et al. 2002b) have been discovered, and several TPP riboswitch candidates also have been identified in archaeans and eukaryotes (Kubodera et al. 2003; Sudarsan et al. 2003a, 2005; Yamauchi et al. 2005).
It has been observed that some riboswitch aptamers occur in tandem within the same mRNA (Mandal et al. 2004; Sudarsan et al. 2006), including some TPP aptamers (Fig. 1A; Rodionov et al. 2004; Sudarsan et al. 2006). The juxtaposition of metabolite-binding RNAs can produce more complex ligand binding and gene control characteristics. Glycine-binding riboswitches carry the most common tandem aptamer arrangement known (Fig. 1B; Mandal et al. 2004; Barrick et al. 2004). The two aptamers bind glycine cooperatively and accomplish their gene control function through a single expression platform. This more sophisticated interplay between two aptamers allows the composite riboswitch to be more responsive to smaller changes in glycine concentration, which yields a genetic switch that is more digital (the gene is either maximally expressed or completely inhibited) (Mandal et al. 2003). In contrast, riboswitches that carry a single aptamer with only one ligand-binding site require nearly a 100-fold change in metabolite concentration to progress between the near-unsaturated and the near-saturated states (e.g., Nahvi et al. 2002; Winkler et al. 2002a,b; Mandal et al. 2003).
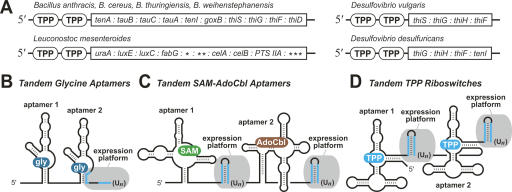
FIGURE 1.
Locations and architecture of tandem TPP riboswitches. (A) TPP aptamers (encircled TPP) reside adjacent to each other in several bacteria upstream of operons usually carrying genes for TPP metabolism. (*) Lmes02000088 ORF, (**) Lmes02000087 ORF, (***) Lmes02000083 ORF. In some organisms, tenI is named thiE, and goxB is named dadA. See Fig. 2 for description of gene functions. (B) Arrangement of tandem aptamers and single expression platform (gray) of some glycine riboswitches. (Un) Stretch of repetitive U residues that is characteristic of intrinsic transcription terminators (Gusarov and Nudler 1999; Yarnell and Roberts 1999). Base-pairing portions of terminator (blue lines). (C) Tandem SAM-AdoCbl riboswitch arrangement that permits control of gene expression by two different metabolites (Sudarsan et al. 2006). (D) Arrangement of tandem TPP riboswitches.
Another tandem arrangement of aptamers occurs in the 5′ UTR of the Bacillus clausii metE mRNA (Fig. 1C). This mRNA carries an aptamer for S-adenosylmethionine (SAM) followed by another for adenosylcobalamin (AdoCbl or coenzyme B12) (Sudarsan et al. 2006). In this and related examples, each aptamer is followed by an expression platform, which suggests that each aptamer is part of a distinct riboswitch that functions independently of the other. Indeed, biochemical and genetic data demonstrate that this mixed tandem riboswitch architecture functions as a two-input Boolean NOR logic gate wherein binding of either SAM or AdoCbl causes gene repression (Sudarsan et al. 2006).
In the current study, we have examined the characteristics of another tandem arrangement of aptamers that is exhibited by some TPP riboswitches (Rodionov et al. 2004; Sudarsan et al. 2006), some candidate riboswitches (Sudarsan et al. 2006), and numerous T-box RNAs (Gutierrez-Preciado et al. 2005). The tandem architecture of these RNAs suggests that the two aptamers sense the same compound and that each aptamer is associated with its own expression platform (Fig. 1D). In this report, we describe the biochemical and genetic characteristics of a tandem TPP riboswitch system from Bacillus anthracis. Our findings indicate that both riboswitches function independently, but the composite arrangement of two complete riboswitches in a single mRNA most likely provides an alternative mechanism for generating a more digital response to changing ligand concentrations.
RESULTS
Molecular components of tandem TPP riboswitches from B. anthracis
We have used various bioinformatics strategies including filtered covariance model searches (Corbino et al. 2005; Weinberg and Ruzzo 2006) to identify numerous new representatives of known riboswitch classes (e.g., Sudarsan et al. 2003a; Barrick et al. 2004; Nahvi et al. 2004; Corbino et al. 2005). These algorithms rank each “hit” by assessing its similarity to a consensus sequence and structure for riboswitch aptamers established by comparative analysis of known representatives. Although an expression platform also is important for riboswitch function, this portion of a riboswitch is usually far less conserved and is not used in the search.
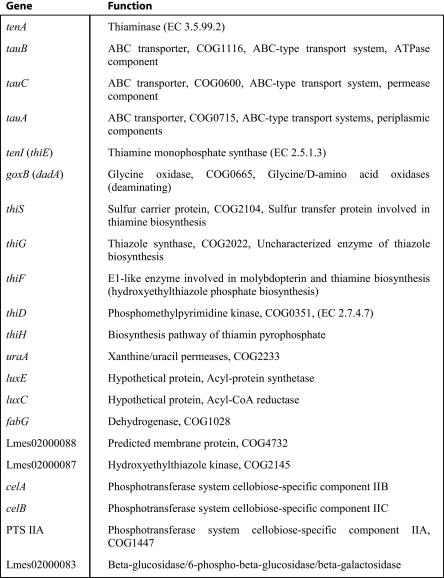
By scanning the output of these computer-aided searches, we have identified a number of instances in bacteria where two TPP aptamers appear in tandem (Fig. 1A). Additional tandem examples in bacteria were reported previously (Rodionov et al. 2004). A total of seven tandem TPP aptamer arrangements have been identified, and each resides immediately upstream of a multigene operon that codes for proteins involved in TPP biosynthesis. For example, the arrangement identified in B. anthracis resides upstream of an operon containing the tenA gene (Fig. 1A), which is one of 10 successive ORFs in a putative operon carrying TPP metabolic genes (Fig. 2).
FIGURE 2.
Description of genes associated with tandem TPP riboswitches from seven species of bacteria. Gene names and descriptions were adapted from that reported previously (Marchler-Bauer et al. 2005).
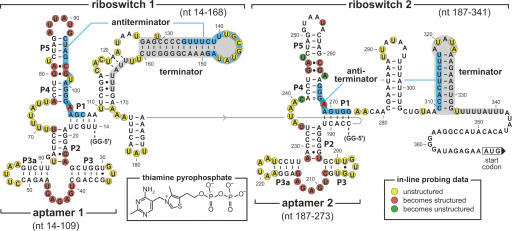
A closer examination of the tandem representative from B. anthracis (Fig. 3) indicates that both aptamers are followed by putative intrinsic transcription terminators (strongly base-paired stem followed by a run of U residues) (Gusarov and Nudler 1999; Yarnell and Roberts 1999). For each aptamer, there is the potential for the formation of an alternative base-paired region between the adjacent terminator stem and the core of the aptamer. These putative anti-terminator structures include nucleotides that are known to be essential for TPP binding (Winkler et al. 2002b; Edwards and Ferré-D'Amaré 2006; Serganov et al. 2006; Thore et al. 2006). Therefore, both aptamers most likely function as TPP-responsive genetic OFF switches because TPP binding and anti-terminator formation should be mutually exclusive.
FIGURE 3.
Sequence and secondary structure architecture of the tandem TPP riboswitches from the 5′ UTR of the tenA operon of B. anthracis. Aptamer models are based on phylogenetic (Mironov et al. 2002; Winkler et al. 2002b) and X-ray structure (Serganov et al. 2006; Thore et al. 2006) data. Ranges of nucleotides identify the various constructs used for biochemical analyses. Putative antiterminator structures (blue-shaded nucleotides), putative terminator structures (gray-shaded nucleotides), two G residues (GG-5′) were added to the 5′ end of constructs to improve yields from in vitro transcription. In-line probing markings reflect data generated (not shown) using constructs Thi1+T1 (riboswitch 1) and Thi2+T2 (riboswitch 2).
High-affinity binding of TPP by both aptamers from the B. anthracis tenA RNA
The tandem aptamer arrangements that have been studied previously exhibit either cooperative binding of two identical ligands (Mandal et al. 2004) or the independent binding of two different ligands (Sudarsan et al. 2006). The tenA RNA is distinct because it carries two complete riboswitches (aptamer and expression platform) that respond to the same ligand type. This arrangement must provide some selective advantage to the bacteria for it to be maintained. However, given the different architecture of the B. anthracis tenA 5′ UTR, we expected that the tenA RNA characteristics would be different from those of tandem aptamer precedents.
To establish that aptamer 1 and aptamer 2 (Fig. 3) both bind TPP, several constructs were generated that encompass only aptamer 1 (constructs Thi1 and Thi1+T1) or only aptamer 2 (constructs Thi2 and Thi2+T2), either without or with the downstream terminator structures (T1 and T2). The ligand-binding characteristics of these RNAs were established by using in-line probing (Soukup and Breaker 1999; Soukup et al. 2001; Winkler et al. 2002b), which reveals changes in the patterns of spontaneous RNA cleavage that occur upon ligand association (see also Materials and Methods). For example, construct Thi1 exhibits substantial changes in spontaneous cleavage at several locations in response to increasing TPP concentrations (Fig. 4A). TPP-induced structural modulation of aptamer 1 (Fig. 4A) and aptamer 2 (construct Thi2; data not shown) occurs at locations similar to those observed previously for members of this riboswitch class (Winkler et al. 2002b; Sudarsan et al. 2003a, 2005).
FIGURE 4.
Structural probing and TPP binding affinity for various B. anthracis tenA RNA constructs. (A) Representative in-line probing data for RNA construct Thi1 (Fig. 3, nt 14–109) encompassing only aptamer 1. Full-length (FL) 5′-32P-labeled RNAs were examined after no reaction (NR), after partial digestion with RNase T1 (T1) or alkali (−OH), or after incubation with concentrations of TPP ranging from 100 fM to 10 μM (see Materials and Methods for additional details). Selected products of RNase T1 partial digestion (cleavage after G residues; closed arrowheads), sites of TPP-dependent changes in spontaneous RNA cleavage (open arrowheads). Sites labeled 1 through 3 were quantitated to estimate the K D for the RNA-ligand complex. (B) Plot of the normalized fraction of full-length Thi1 RNA cleaved at sites 1–3 versus the logarithm of the molar concentration (c) of TPP. (C) Summary of the K D values for various B. anthracis tenA RNA constructs determined by using in-line probing.
The levels of spontaneous cleavage at three positions (site 1, A44; site 2, U91; site 3, A104) were quantitated and used to estimate the apparent dissociation constant (K D) for ligand binding (Fig. 4B). The K D value determined for aptamer 1 alone (210 pM) is >10-fold better than the values measured for other tight-binding riboswitch aptamers (e.g., Winkler et al. 2002a, 2003; Mandal et al. 2003). Similarly, the K D value for TPP binding by aptamer 2 (850 pM; Fig. 4C) also represents one of the tightest aptamer–metabolite interactions measured to date. These findings support our prediction that both B. anthracis tenA aptamers sense TPP.
Ligand binding by tandem TPP aptamers is not cooperative
Previous efforts to establish K D values for riboswitch aptamers have revealed that nucleotides downstream of the conserved core of the aptamer can influence ligand affinity (Winkler et al. 2002b). These added nucleotides commonly interfere with proper folding of the aptamer and thereby cause the construct to bind more weakly. This effect can be pronounced with some riboswitches wherein the expression platform functions by forming alternatively paired structures that preclude proper aptamer folding (Wickiser et al. 2005a,b; Lemay et al. 2006).
We observed that constructs including either terminator 1 (Thi1+T1) or terminator 2 (Thi2+T2) with their corresponding aptamers (Fig. 3) exhibit only slightly poorer affinities for TPP (Fig. 4C). These flanking nucleotides have a small effect on K D of the aptamer, most likely because terminator stem formation dominates over anti-terminator stem formation even in the absence of ligand during in-line probing. Therefore, structural heterogeneity does not substantially erode the affinity measured for the population of folded RNAs.
Most glycine riboswitches are atypical because they require nucleotides outside the conserved core of each aptamer to enhance binding affinity (Mandal et al. 2004). Glycine aptamers arranged in tandem require glycine to be docked in the adjacent aptamer for the adjoining aptamer to exhibit maximal affinity. To rule out the possibility that the tandem TPP aptamers are configured to bind two TPP molecules cooperatively, we determined the K D values for both aptamer 1 and aptamer 2 when co-residing in a single RNA construct (Complete; Fig. 4C). Both aptamers bind TPP ∼10-fold better when tested alone, compared with the K D values for these aptamers when positioned in tandem. This finding suggests that the two aptamers from B. anthracis tenA have not evolved to function cooperatively.
Tandem TPP riboswitches control transcription termination
Some previous studies of riboswitches that control transcription termination have employed in vitro transcription assays to monitor riboswitch function (e.g., Mironov et al. 2002; Winkler et al. 2002a, 2003; Epshtein et al. 2003; Grundy et al. 2003; McDaniel et al. 2003; Sudarsan et al. 2003b; Mandal et al. 2004; Wickiser et al. 2005a,b). These assays provide evidence that transcription termination is modulated by selective binding of metabolite to the aptamer. However, the extent of modulation of transcription termination in vitro is rarely maximal, which might be due to a poor correspondence between the conditions used for the assays and those experienced by riboswitches in vivo.
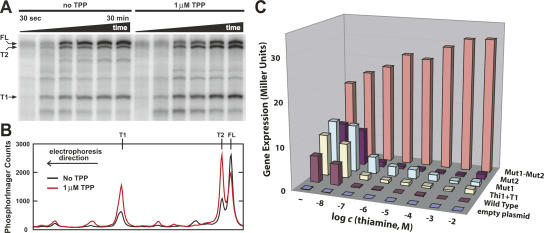
We used single-round transcription assays (Sudarsan et al. 2003b) to examine the products made by Escherichia coli RNA polymerase as it transcribes a DNA template corresponding to the 5′ UTR of the B. anthracis tenA RNA. Aliquots from in vitro transcription assays (see Materials and Methods) were removed at various time points to assess the RNA products. Although a number of premature termination and pause sites are evident as transcription progresses (Fig. 5A), the three most prominent RNA products correspond in size to those expected for termination in expression platform 1 (T1), termination in expression platform 2 (T2), and for full-length (FL) RNA.
FIGURE 5.
Transcription termination and gene control by the tandem TPP riboswitch from the tenA mRNA of B. anthracis. (A) In vitro transcription of the 5′ UTR of the B. anthracis tenA mRNA. Bands labeled T1, T2, and FL correspond to RNA products terminated at the first expression platform, the second expression platform, and full-length transcription, respectively. (B) Line plots comparing the amounts of RNA transcripts produced during in vitro transcription after 30 min incubation with or without added TPP, as indicated. Data were derived from the PAGE image in A, where the top of the gel corresponds to the right of the line plot. (C) Plot of the amount of β-galactosidase reporter gene expression resulting from fusions to several versions of the B. anthracis tenA mRNA 5′ UTR. The Thi1+T1 construct carries only the first riboswitch. Mut1 (G41C, A42U) and Mut2 (G210C, A211U) constructs carry mutations in the junction between P3 and P4 in the first or second aptamer (Fig. 3) that disrupt TPP binding (data not shown; Winkler et al. 2002b). Mut1-Mut2 construct carries both sets of disruptive mutations. Constructs were tested in transformed B. subtilis cells in minimal medium supplemented with various concentrations of thiamine as indicated.
In addition, the relative levels of these three major transcripts change in response to the addition of 1 μM TPP (Fig. 5B). The TPP-dependent decrease in the amount of full-length transcript and the corresponding increases in the amounts of both truncated products are expected if both riboswitches sense TPP and cause transcription termination.
Tandem TPP riboswitches control gene expression independently
To confirm our hypothesis that the two riboswitches function independently and respond to the same ligand, we tested a series of reporter gene constructs that carry variations of the wild-type tenA mRNA 5′ UTR. These constructs were integrated into thiamine auxotroph Bacillus subtilis, and levels of β-galactosidase reporter gene expression were measured after cell growth in a minimal medium supplemented with various concentrations of thiamine. Thiamine supplementation is expected to permit these cells to take up the vitamin, generate the phosphorylated derivative TPP, and down-regulate the TPP riboswitch-controlled reporter gene as was observed previously using E. coli (Winkler et al. 2002b).
As expected, cells carrying a reporter gene fused with the wild-type construct exhibit strong repression of the reporter gene when the medium is supplemented with increasing concentrations of thiamine (Fig. 5C, “Wild Type”). Also, the construct wherein both riboswitch aptamers carry deleterious mutations (Mut1-Mut2) yields robust reporter gene expression regardless of the amount of thiamine supplementation. It is notable that the levels of expression between these two constructs differ somewhat when cells are grown without thiamine supplementation. The cells were initially grown in a rich medium that contains thiamine prior to pelleting and resuspension in a minimal medium that lacks thiamine unless supplemented. Residual TPP in each cell should cause some reporter gene suppression of the wild-type fusion. Overall, these results indicate that the TPP riboswitches most likely are entirely responsible for the control of gene repression in response to changing TPP concentrations.
Importantly, variants of the tenA 5′ UTR that carry only the first riboswitch (Thi1+T1), or that carry either a defective first (Mut1) or second (Mut2) aptamer, also exhibit gene control characteristics that are nearly identical with wild type. Although the action of a second riboswitch might further reduce the number of mRNAs that reach full length in the population of bacteria, the observed effect of the loss of a second riboswitch is almost negligible, suggesting that a single riboswitch is sufficient to yield nearly total repression of reporter gene expression.
Moreover, the majority of repression occurs when the medium is supplemented with 100 nM thiamine, regardless of which reporter gene construct was examined. These results indicate that either the first or the second TPP riboswitch can function independently to achieve a near total inactivation of reporter gene expression in response to similar levels of thiamine supplementation. Therefore, it is unlikely that the tandem riboswitch arrangement is present to increase the dynamic range for gene control, or to yield a composite genetic switch that responds to a wider range of ligand concentrations.
DISCUSSION
Tandem riboswitch functions
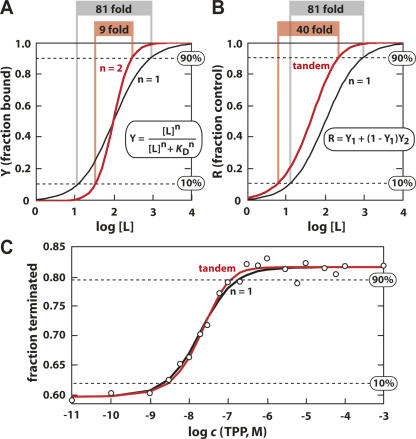
The utility of some tandem riboswitch systems is unambiguous and well established (Mandal et al. 2004; Sudarsan et al. 2006). The close adjoining of two aptamers that function cooperatively to bind two identical ligands yields a gene control element that is more responsive to smaller changes in ligand concentration (Mandal et al. 2004). If the two aptamers are perfectly cooperative, they would reduce from 81-fold to ninefold the change in ligand concentration needed to progress from 10% to 90% modulation of gene expression (Fig. 6A).
FIGURE 6.
Theoretical ligand binding and gene control characteristics of two types of tandem riboswitch architectures. (A) Plot of the fractional aptamer occupancy versus the logarithm of the concentration of ligand for a single aptamer (n = 1) and for two aptamers that are perfectly cooperative (n = 2). The values at the top of the plot report the fold change in ligand concentration needed for the receptor to progress from 10% to 90% receptor occupancy (or gene modulation) as indicated (dashed lines). Curves and values were obtained using the equation provided in the inset (see also Materials and Methods). (B) Plot of the fractional gene control for a single riboswitch (n = 1) and for tandem riboswitches that function independently and exhibit identical K D values (Tandem). Other details are as described for A. (C) Dependence of in vitro transcription termination of B. anthracis tenA mRNA on the concentration of TPP. Theoretical curves for single and tandem riboswitch-mediated transcription termination (as depicted in B) are overlaid to best fit the actual transcription data for comparison. Other details are as described for A.
Although not perfectly cooperative, the tandem glycine aptamers found in Vibrio cholerae (Mandal et al. 2004) exhibit a level of cooperativity (n = 1.64) that requires a ligand concentration change of only 14-fold to progress from 10% to 90% modulation. This increased sensitivity to changes in glycine concentration most likely allows bacteria to efficiently use this amino acid as an energy source without depleting it to the point where protein synthesis would be compromised.
In contrast, noncooperative tandem riboswitches that sense two different ligands would allow cells to control genes in response to changing concentrations of two metabolite types. This strategy is used by strains of B. clausii to sense SAM and AdoCbl to modulate the production of enzymes to optimally biosynthesize methionine (Sudarsan et al. 2006). However, the common arrangement represented by the TPP riboswitches in the tenA RNA from B. anthracis senses the same ligand type in both aptamers, so the biological utility of this arrangement must be different.
Possible functions of tandem riboswitches that sense the same ligand type
There are three possible characteristics that might be advantageous for gene control by independent tandem riboswitches that sense the same ligand. First, the two riboswitches might increase the dynamic range of gene expression. For example, if each riboswitch has a 0.9 probability of terminating transcription when bound to ligand, then the presence of only one riboswitch in an mRNA would allow ∼10-fold change in gene expression. But the combined action of both riboswitches in a single mRNA would allow ∼100-fold change in gene expression. This possible utility for the B. anthracis tenA riboswitches seems unlikely, given that even a single functional TPP riboswitch gives essentially full gene repression when fused to a reporter gene in B. subtilis (Fig. 5C).
Second, it has been pointed out previously (Rodionov et al. 2004) that the tandem arrangement of two independent riboswitches will produce a genetic switch that requires a lower ligand concentration to trigger gene control. The authors also pointed out that it would be unusual for tandem riboswitches to be used to achieve this goal instead of acquiring mutations that would improve ligand affinity. We have calculated the effect that two independent riboswitches would have on gene control (Fig. 6B), and our results support the earlier speculation that an increase in ligand sensitivity indeed will occur. However, this increase corresponds to only a 1.6-fold improvement in the concentration of ligand needed to half-maximally trigger gene control.
To further stress the unlikelihood that this characteristic provides the selective advantage that cells acquire from this type of tandem riboswitch arrangement, we point out that very small changes in either the rate of ligand association or the speed of RNA transcription could easily permit a single riboswitch to respond more sensitively to ligands (Wickiser et al. 2005a,b). Therefore, it seems very unlikely that the most common architecture for tandem riboswitches is used to modestly improve ligand sensitivity when other strategies that are far more productive and far less information intensive are possible.
By using in-line probing, we have established that the K D values for the tandem TPP aptamers from B. anthracis are 210 pM and 850 pM. These values are among the tightest known metabolite interactions with natural aptamers, and are well below the concentration of thiamine in culture medium that triggers reporter gene repression (Fig. 5C). Although the concentration of TPP inside the cells during growth of reporter gene transformants is unknown, it seems unlikely that these aptamers with K D values in the mid- to high-picomolar range would require a 1.6-fold improvement in ligand sensitivity to sense biologically relevant concentrations of this metabolite.
A third characteristic that is inherent to independently functioning tandem riboswitches with similar K D values is an increase in the digital character of the genetic switch (Fig. 6B). The overall level of gene modulation is a function of the probabilities of the two riboswitches acting as concentrations of metabolite increase (Fig. 6B, inset). Their combined action reduces from 81-fold to ∼40-fold the change in metabolite concentration needed to progress from 10% to 90% modulation, assuming the K D values (or perhaps more appropriately the T50 values; see below) for the two riboswitches are equal. If the K D or T50 values are different, this more digital character is diminished (see Materials and Methods).
In an effort to determine whether transcription termination in the tandem riboswitch system is more responsive to TPP concentration changes, we have conducted single-round transcription termination assays at various concentrations of TPP (Fig. 6C). The construct used provides a modest level of transcription control from ∼60% termination to ∼80% termination in the absence and presence of TPP, respectively. The concentration of TPP needed to half-maximally modulate transcription termination (T50) (Wickiser et al. 2005b) is ∼20 nM. Unfortunately, the predicted differences in the profiles for TPP-mediated transcription termination between the single- and tandem-riboswitch cases are not sufficiently large to determine which model best matches our transcription termination data (Fig. 6C). Similarly, the in vivo reporter gene data (Fig. 5C) cannot be used to support either model, in part due to the fact that the concentrations of TPP inside cells under various growth conditions are unknown.
Conclusions
The predicted reduction in the dynamic range of ligand concentration is not as substantial as a perfectly cooperative tandem system (Fig. 6A) and is less than that observed with cooperative glycine aptamers (Mandal et al. 2004). However, the arrangement of identically functioning riboswitches as observed with the B. anthracis tenA mRNA provides a simple mechanism for compressing the dynamic range of ligand concentration and producing more digital gene control responses. Interestingly, the tandem arrangement of riboswitches with identical ligand specificities appears to be more common for tRNA-sensing T-boxes (Gutierrez-Preciado et al. 2005) than for metabolite-sensing riboswitches. It seems likely that cells will tolerate ∼100-fold changes in metabolite concentrations better than similar changes in the concentrations of uncharged tRNAs. If true, then we would expect to find more tandem T-box RNAs than metabolite-sensing riboswitches.
In summary, the assembly of independently functioning riboswitches appears to be a relatively simple way that organisms can construct genetic switches made of RNA that are more responsive to smaller changes in metabolite concentrations. Although greater dynamic range for gene control and greater sensitivity to ligand concentrations also emerge on assembling riboswitches with identical ligand specificities in tandem, these properties could easily be manifested by lone riboswitches. Therefore, the ligand-binding characteristics, gene control functions, and genetic distribution of tandem TPP riboswitches are consistent with our hypothesis that these systems exist to provide a small increase in digital gene control character.
MATERIALS AND METHODS
Chemicals and oligonucleotides
Thiamine and TPP were purchased from Sigma, and the dinucleotide ApA was purchased from BIOLOG Life Science Institute. The intergenic region upstream of the tenA thiamine biosynthesis operon of B. anthracis (Sterne strain) was PCR amplified (by Dr. Sean Rollins, Department of Infectious Diseases, Massachusetts General Hospital) using the DNA primers 5′-TAATACGACTCACTATAGGGATAATAGAGACCTCCTCTAGTTGTATAG (T7 RNA polymerase promoter and three additional G residues are underlined) and 5′-TTCTCTATCCCCTTCCGGTATGTG. The resulting DNA construct was cloned into a TOPO-TA vector (Invitrogen), transformed into Top10 E. coli cells (Invitrogen), and the resulting plasmid was isolated from cells using the Qiaprep Spin Miniprep Kit (QIAGEN) following protocols supplied by the manufacturers. DNA sequencing (The Keck Foundation Biotechnology Resource Center, Yale University) confirmed successful cloning, and all constructs used in this work were generated from this plasmid by PCR with appropriate synthetic DNA primers using methods similar to those described elsewhere (Puerta-Fernandez et al. 2006).
In-line probing
RNA constructs used for in-line probing were transcribed in vitro from PCR-amplified DNA, dephosphorylated, and 5′-32P-labeled using protocols similar to those published previously (Seetharaman et al. 2001). To improve transcription efficiency with T7 RNA polymerase, additional G residues were introduced into the DNA template as noted. In-line probing was performed as described previously (Winkler et al. 2002b). Briefly, 5′-32P-labeled RNA (<1 nM) was incubated for 40 h at 25°C in 10 μL of in-line probing buffer (50 mM Tris-HCl [pH 8.3 at 25°C], 20 mM MgCl2, and 100 mM KCl) containing concentrations of TPP as defined for each reaction. After incubation, 10 μL of 10 M urea was added and the products were separated using denaturating 10% polyacrylamide gel electrophoresis (PAGE). Gels were dried and analyzed using a Storm PhosphorImager (GE Healthcare). To obtain K D values in the subnanomolar range, the in-line assay was conducted with RNA concentrations <50 pM. Therefore, the concentration of ligand needed to cause half-maximal modulation of spontaneous cleavage yield reflects the K D for the RNA–ligand interaction.
Synchronized in vitro transcriptions
Synchronized transcription reactions were performed using a protocol similar to that published previously (Sudarsan et al. 2003b). Initial transcription experiments with PCR-generated template DNAs comprising the complete intergenic region upstream of the B. anthracis tenA gene and E. coli RNA polymerase were used to reveal the putative native promoter and transcription start site. For efficient transcription initiation, an additional A residue was inserted upstream of A1 by PCR to permit initiation by the dinucleotide ApA. The initiation reaction mixture contained 100 nM double-stranded DNA template, NTPs (1 μM ATP and GTP, 0.4 μM UTP), 200 μM ApA, 10 μg/mL BSA, and 33 nM α-32P-radiolabeled UTP in transcription buffer (50 mM Tris HCl [pH 7.5 at 25°C], 10 mM MgCl2, 50 mM KCl). Transcription was initiated by the addition of E. coli RNA polymerase holoenzyme (EpiCentre) at 88 nM final concentration. The reaction was incubated for 10 min at 37°C, and subsequently transcription elongation was started by transferring 10 μL aliquots to a microfuge tube containing 2.5 μL of prewarmed elongation mix (transcription buffer, 1 mg/mL heparin, 50 μM each NTP, and a TPP concentration as noted for each experiment). Reactions were terminated after 20 min at 37°C by adding 12.5 μL of 7 M urea and storage on ice. Product analysis was performed as described for in-line probing reactions.
In vivo reporter assays
DNA constructs containing one or both B. anthracis tenA TPP riboswitches were fused to a lacZ reporter and transformed into B. subtilis cells using protocols similar to those published previously (Sudarsan et al. 2003b). Briefly, DNA constructs encompassing the putative native promoter and either the first or both riboswitches (nucleotides −430 to −183 or nucleotides −430 to −10 relative to the translation start site) were PCR-amplified using DNA primers 5′-CAGGAATTCGACAATTGAGAAAATTAAAACAGTTGATGATGTC and 5′-CATGGATCCAATATGAAATTTTCAAATAAAAAAGCCCCGTTTC that introduce EcoRI and BamHI restriction sites, respectively. Mutations were introduced by two-step PCR using appropriate primers.
Constructs were cloned into plasmid pDG1661 (Bacillus Genetic Stock Center, The Ohio State University) upstream of the lacZ reporter gene. Subsequent transformation into the amyE locus of a thiamine auxotroph strain of B. subtilis (1A251; Bacillus Genetic Stock Center, The Ohio State University) was confirmed by sequencing. Fresh overnight cultures in 2XYT medium (16 g/L tryptone, 10 g/L yeast extract, 5 g/L NaCl) were diluted to 0.05 OD600 mL−1 with 2XYT medium and grown for an additional 2 h by shaking at 37°C. Cultures were pelleted (10 min, 2500 rpm) and suspended in minimal medium (100 mM K2HPO4, 44 mM KH2PO4, 4 mM trisodium citrate, 0.8 mM MgSO4, 14 mM Na2SO4, 50 μM FeCl3, 0.4% glucose, 0.2% glutamate, 0.1% DIFCO vitamin assay casamino acids) supplemented with 500 mg/L adenine, and grown to 0.3 OD600 mL−1. After adding 50 μL of cells to 100 μL of minimal medium supplemented with adenine and containing different thiamine concentrations, cultures were shaken for 4 h at 37°C. β-galactosidase assays were performed in 96-well microplates following a procedure published elsewhere (Blount et al. 2007).
Calculations of tandem riboswitch effects
Described below is an example calculation of the dependence of the change in ligand concentration needed to shift between 10% and 90% gene repression (example given for gene repression by transcription termination) for a K D ratio of x between two independently functioning tandem riboswitches. The lowest ratio (the most “digital” response curve) is found when x = 1, or K D1 = K D2.
The fraction of transcripts undergoing termination (R) is given by the following equation:
where Y 1 equals fractional saturation of aptamer 1 with ligand and Y 2 equals fractional saturation of aptamer 2 with ligand. The fractional saturation of a single receptor equals
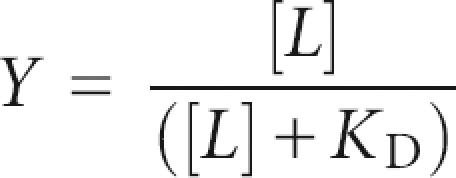 |
for a simple receptor–ligand interaction where [L] equals the ligand concentration.
Then:
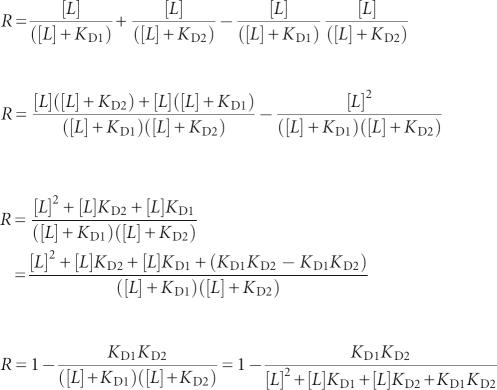 |
For x = K D2/K D1:
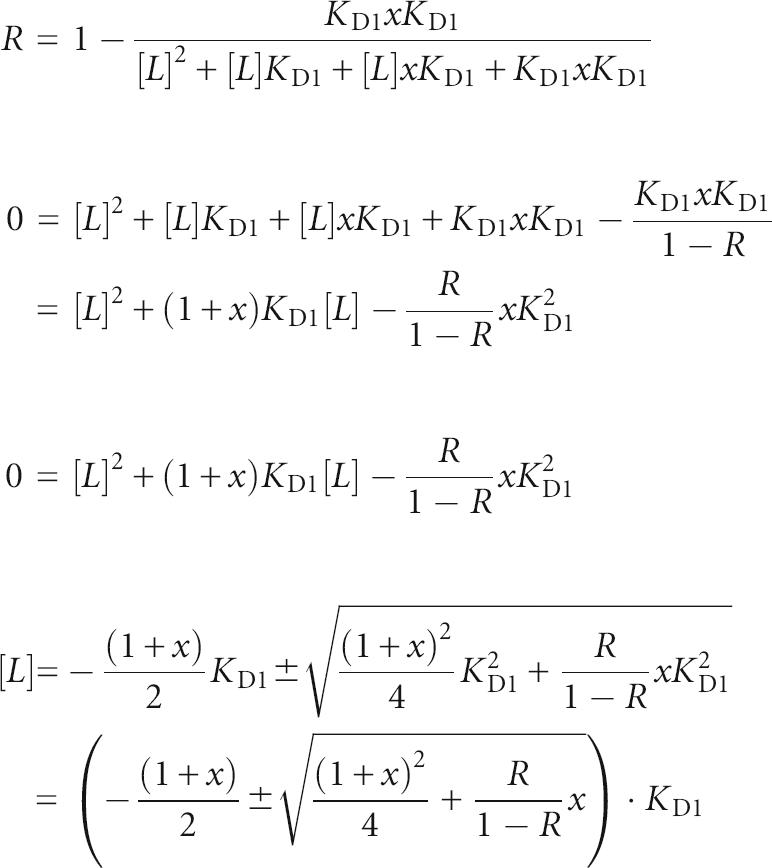 |
Since [L] > 0, x ≥ 0, KD > 0 and 0 ≤ r ≤ 1;
 |
Examples of the dynamic ranges of ligand concentration needed to shift from 10% to 90% gene modulation:
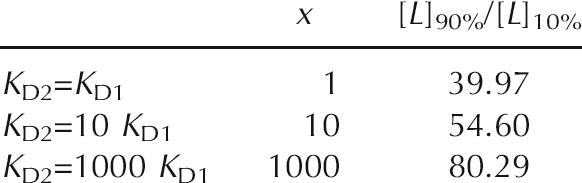
ACKNOWLEDGMENTS
We thank members of the Breaker laboratory and Dr. Donald Crothers for helpful discussions, and Drs. Sean Rollins and Edward Ryan for preparing and supplying the B. anthracis tenA PCR DNA. This work was supported by funding to R.R.B. from the NIH (GM068819), NSF (EIA 0323510), and the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.407707.
REFERENCES
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, K.F., Wang, J.X., Lim, J., Sudarsan, N., Breaker, R.R. Antibacterial lysine analogs that target lysine riboswitches. Nat. Chem. Biol. 2007;1:44–49. doi: 10.1038/nchembio842. [DOI] [PubMed] [Google Scholar]
- Corbino, K.A., Barrick, J.E., Lim, J., Welz, R., Tucker, B.J., Puskarz, I., Mandal, M., Rudnick, N.D., Breaker, R.R. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol. 2005;6:R70. doi: 10.1186/gb-2005-6-8-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, T.E., Ferré-D'Amaré, A.R. Crystal structures of the thi-box riboswitch bound to thiamine pyrophosphate analogs reveal adaptive RNA-small molecule recognition. Structure. 2006;14:1459–1468. doi: 10.1016/j.str.2006.07.008. [DOI] [PubMed] [Google Scholar]
- Epshtein, V., Mironov, A.S., Nudler, E. The riboswitch-mediated control of sulfur metabolism in bacteria. Proc. Natl. Acad. Sci. 2003;100:5052–5056. doi: 10.1073/pnas.0531307100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy, F.J., Lehman, S.C., Henkin, T.M. The L box regulon: Lysine sensing by leader RNAs of bacterial lysine biosynthesis genes. Proc. Natl. Acad. Sci. 2003;100:12057–12062. doi: 10.1073/pnas.2133705100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov, I., Nudler, E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- Gutierrez-Preciado, A., Jensen, R.A., Yanofsky, C., Merino, E. New insights into regulation of the tryptophan biosynthetic operon in Gram-positive bacteria. Trends Genet. 2005;21:432–436. doi: 10.1016/j.tig.2005.06.001. [DOI] [PubMed] [Google Scholar]
- Kubodera, T., Watanabe, M., Yoshiuchi, K., Yamashita, N., Nishimura, A., Nakai, S., Gomi, K., Hanamoto, H. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/s0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- Lemay, J.F., Penedo, J.C., Tremblay, R., Lilley, D.M.J., Lafontaine, D.A. Folding of the adenine riboswitch. Chem. Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Breaker, R.R. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., Breaker, R.R. Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell. 2003;113:577–586. doi: 10.1016/s0092-8674(03)00391-x. [DOI] [PubMed] [Google Scholar]
- Mandal, M., Lee, M., Barrick, J.E., Weinberg, Z., Emilsson, G.M., Ruzzo, W.L., Breaker, R.R. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- Marchler-Bauer, A., Anderson, J.B., Cherukuri, P.F., DeWeese-Scott, C., Geer, L.Y., Gwadz, M., He, S., Hurwitz, D.I., Jackson, J.D., Ke, Z., et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33:D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDaniel, B.A.M., Grundy, F.J., Artsimovitch, I., Henkin, T.M. Transcription termination control of the S box system: Direct measurement of S-adenosylmethionine by the leader RNA. Proc. Natl. Acad. Sci. 2003;100:3083–3088. doi: 10.1073/pnas.0630422100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Rios, J., Navarro, M., Soberón, M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. 2001;98:9736–9741. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, A.S., Gusarov, I., Rafikov, R., Lopez, L.E., Shatalin, K., Kreneva, R.A., Perumov, D.A., Nudler, E. Sensing small molecules by nascent RNA: A mechanism to control transcription in bacteria. Cell. 2002;111:747–756. doi: 10.1016/s0092-8674(02)01134-0. [DOI] [PubMed] [Google Scholar]
- Nahvi, A., Sudarsan, N., Ebert, M.S., Zou, X., Brown, K.L., Breaker, R.R. Genetic control by a metabolite binding RNA. Chem. Biol. 2002;9:1043–1049. doi: 10.1016/s1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Nahvi, A., Barrick, J.E., Breaker, R.R. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 2004;32:143–150. doi: 10.1093/nar/gkh167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puerta-Fernandez, E., Barrick, J.E., Roth, A., Breaker, R.R. Identification of a large noncoding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci. 2006;103:19490–19495. doi: 10.1073/pnas.0607493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov, D.A., Vitreschak, A.G., Mironov, A.A., Gelfand, M.S. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- Rodionov, D.A., Dubchak, I., Arkin, A., Alm, E., Gelfand, M.S. Reconstruction of regulatory and metabolic pathways in metal-reducing δ-proteobacteria. Genome Biol. 2004;5:R90. doi: 10.1186/gb-2004-5-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seetharaman, S., Zivarts, M., Sudarsan, N., Breaker, R.R. Immobilized RNA switches for the analysis of complex chemical and biological mixtures. Nat. Biotechnol. 2001;19:336–341. doi: 10.1038/86723. [DOI] [PubMed] [Google Scholar]
- Serganov, A., Polonskaia, A., Phan, A.T., Breaker, R.R., Patel, D.J. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, G.A., Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, J.K., Soukup, G.A. Riboswitches exert genetic control through metabolite-induced conformational change. Curr. Opin. Struct. Biol. 2004;14:344–349. doi: 10.1016/j.sbi.2004.04.007. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., DeRose, E.C., Koizumi, M., Breaker, R.R. Generating new ligand-binding RNAs by affinity maturation and disintegration of allosteric ribozymes. RNA. 2001;7:524–536. doi: 10.1017/s1355838201002175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan, N., Barrick, J.E., Breaker, R.R. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA. 2003a;9:644–647. doi: 10.1261/rna.5090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan, N., Wickiser, J.K., Nakamura, S., Ebert, M.S., Breaker, R.R. An mRNA structure in bacteria that controls gene expression by binding lysine. Genes & Dev. 2003b;17:2688–2697. doi: 10.1101/gad.1140003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan, N., Cohen-Chalamish, S., Nakamura, S., Emilsson, G.M., Breaker, R.R. Thiamine pyrophosphate riboswitches are targets for the antimicrobial compound pyrithiamine. Chem. Biol. 2005;12:1325–1335. doi: 10.1016/j.chembiol.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Sudarsan, N., Hammond, M.C., Block, K.F., Welz, R., Barrick, J.E., Roth, A., Breaker, R.R. Tandem riboswitch architectures exhibit complex gene control functions. Science. 2006;314:300–304. doi: 10.1126/science.1130716. [DOI] [PubMed] [Google Scholar]
- Thore, S., Leibundgut, M., Nan, N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science. 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- Vitreschak, A.G., Rodionov, D.A., Mironov, A.A., Gelfand, M.S. Regulation of riboflavin biosynthesis and transport genes in bacteria by transcriptional and translational attenuation. Nucleic Acids Res. 2002;30:3141–3151. doi: 10.1093/nar/gkf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitreschak, A.G., Rodionov, D.A., Mironov, A.A., Gelfand, M.S. Regulation of the vitamin B12 metabolism and transport in bacteria by a conserved RNA structural element. RNA. 2003;9:1084–1097. doi: 10.1261/rna.5710303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg, Z., Ruzzo, W.L. Sequence-based heuristics for faster annotation of non-coding RNA families. Bioinformatics. 2006;22:35–39. doi: 10.1093/bioinformatics/bti743. [DOI] [PubMed] [Google Scholar]
- Wickiser, J.K., Cheah, M.T., Breaker, R.R., Crothers, D.M. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry. 2005a;44:13404–13414. doi: 10.1021/bi051008u. [DOI] [PubMed] [Google Scholar]
- Wickiser, J.K., Winkler, W.C., Breaker, R.R., Crothers, D.M. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol. Cell. 2005b;18:49–60. doi: 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Breaker, R.R. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Cohen-Chalamish, S., Breaker, R.R. An mRNA structure that controls gene expression by binding FMN. Proc. Natl. Acad. Sci. 2002a;99:15908–15913. doi: 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W., Nahvi, A., Breaker, R.R. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002b;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., Breaker, R.R. An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 2003;10:701–707. doi: 10.1038/nsb967. [DOI] [PubMed] [Google Scholar]
- Yamauchi, T., Miyoshi, D., Kubodera, T., Nishimura, A., Nakai, S., Sugimoto, N. Roles of Mg2+ in TPP-dependent riboswitch. FEBS Lett. 2005;579:2583–2588. doi: 10.1016/j.febslet.2005.03.074. [DOI] [PubMed] [Google Scholar]
- Yarnell, W.S., Roberts, J.W. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]