Abstract
Processing bodies (P-bodies) are subcellular ribonucleoprotein (RNP) granules that have been hypothesized to be sites of mRNA degradation, mRNA translational control, and/or mRNA storage. Importantly, P-bodies are conserved from yeast to mammals and contain a common set of evolutionarily conserved protein constituents. P-bodies are dynamic structures and their formation appears to fluctuate in correlation with alterations in mRNA metabolism. Despite these observations, little is understood about how P-body structures are formed within the cell. In this study, we demonstrate a relationship between P-bodies and microtubules in the budding yeast, Saccharomyces cerevisiae. First, we demonstrate that disruption of microtubules by treatment with the drug benomyl leads to aggregation of P-body components. Consistent with this finding, we also demonstrate that disruption of microtubules by a temperature-sensitive allele of the major α tubulin, TUB1 (tub1–724) stimulates P-body formation. Second, we find that the α-tubulin protein Tub1 colocalizes with P-bodies upon microtubule destabilization. Third, we determine that a putative tubulin tyrosine ligase, encoded by YBR094W, is a protein component of P-bodies, providing additional evidence for a physical connection between P-bodies and microtubules. Finally, we establish that P-bodies formed by microtubule destabilization fail to correlate with global changes in the stability of mRNA or in general mRNA translation. These findings demonstrate that the aggregation of P-body components is linked to the intracellular microtubule network, and, further, that P-bodies formed by disruption of microtubules aggregate independent of broad alterations in either mRNA decay or mRNA translation.
Keywords: P-bodies, microtubules, mRNA decay, mRNA translational control, mRNA stability
INTRODUCTION
P-bodies represent aggregations of RNA and proteins that are suggested to be involved in the processes of mRNA degradation, mRNA translational repression, and mRNA storage (Bashkirov et al. 1997; Ingelfinger et al. 2002; Lykke-Andersen 2002; van Dijk et al. 2002; Sheth and Parker 2003; Cougot et al. 2004; Brengues et al. 2005; Teixeira et al. 2005). In yeast, P-body constituents include the translational repressors, Dhh1p and Pat1p (Sheth and Parker 2003; Coller and Parker 2005); the activators of mRNA decapping, Lsm1p and Edc3p (Sheth and Parker 2003; Kshirsagar and Parker 2004), the decapping enzyme, Dcp1p/Dcp2p (Sheth and Parker 2003); the major 5′–3′ exoribonuclease activity, Xrn1p (Sheth and Parker 2003); and the machinery for nonsense-mediated decay, the Upf proteins (Sheth and Parker 2006). The observation that P-body aggregates contain intermediates of mRNA decay has led to the hypothesis that these structures serve as the location of 5′–3′ mRNA decay within the cell (Sheth and Parker 2003). In yeast it has also been proposed that P-bodies may be sites of mRNA storage (Brengues et al. 2005; Hilgers et al. 2006), a function that is reminiscent of mRNP granules that harbor maternal mRNAs as translationally quiescent species during early metazoan development (Coller et al. 2001; Coller and Parker 2004). In metazoans, P-bodies have been implicated in post-transcriptional gene silencing by RNA interference (RNAi), as they contain both the machinery for RNAi (i.e., RISC complex) and microRNAs (miRNAs) (Liu et al. 2005; Pillai et al. 2005; Sen and Blau 2005; Bhattacharyya et al. 2006). Moreover, proteins associated with P-bodies such as the Dhh1p homolog, RCK/p54, and the decapping proteins Dcp1 and Dcp2 have been implicated in miRNA-mediated gene silencing (Rehwinkel et al. 2005; Behm-Ansmant et al. 2006; Chu and Rana 2006). The conserved nature of cytoplasmic P-bodies in eukaryotic cells and their proposed role in mRNA metabolism have prompted a need to better understand the function of these aggregates and the cellular events that dictate their assembly.
Several observations suggest that P-bodies are dynamic mRNP-based structures whose size and abundance correlate with alterations in mRNA metabolism (Sheth and Parker 2003; Coller and Parker 2005; Teixeira et al. 2005). For instance, conditions of cellular stress such as glucose deprivation or osmotic shock result in a dramatic decrease in mRNA translation and inhibition of mRNA decay (Ashe et al. 2000; Hilgers et al. 2006). Both glucose deprivation and osmotic stress also result in a concomitant increase in P-body abundance (Teixeira et al. 2005). Moreover, mutations that reduce the rate of either decapping or exonucleolytic digestion of mRNA result in an increase in P-body number and size (Sheth and Parker 2003). Finally, yeast cells harboring mutations that inhibit translational repression exhibit a dramatic decrease in P-body abundance (Coller and Parker 2005). Based on these observations, it has been proposed that P-body formation is dependent on the availability of mRNA substrates upon which P-body proteins can assemble and, therefore, the size and abundance of P-bodies is directly proportional to alterations in mRNA metabolism (Coller and Parker 2004).
Despite a growing interest in the function of P-bodies, little is still known about the exact role of these cytoplasmic structures in mRNA metabolism and the suite of biological consequences that lead to their formation. Here we report a relationship between the cytoskeleton and P-body formation. Specifically, we demonstrate that conditions that result in microtubule destabilization lead to a redistribution of Dcp2 protein, a known P-body component, into foci within the cell. Moreover, the aggregations we observe under microtubule-destabilizing conditions are bona fide P-bodies, as they contain additional characterized protein constituents as well as an intermediate of mRNA decay. Furthermore, under microtubule-destabilizing conditions, we also find that the α-tubulin protein, Tub1p, colocalizes with P-bodies. We also report here the identification of a novel P-body component, Ybr094w protein. Interestingly, Ybr094wp is the yeast homolog of several characterized tubulin tyrosine ligases, which act to modify tubulin in vivo. Finally, we establish that although microtubule destabilization results in dramatic P-body formation, no appreciable change in either mRNA decay or global mRNA translation is observed. This latter observation indicates that under certain conditions, P-body formation can be uncoupled from global alterations in mRNA metabolism. Together, these data suggest that the aggregation of mRNPs into P-bodies can be influenced by microtubule integrity, possibly implicating higher-order cellular organization in controlling the dynamics associated with these structures, and that a mechanism for P-body formation exists that is independent of dramatic alterations in mRNA metabolism.
RESULTS AND DISCUSSION
Microtubule disruption stimulates formation of Dcp2p-containing cytoplasmic foci
In all eukaryotes, microtubule networks are important structural components of cells (Winsor and Schiebel 1997). In yeast, an α-tubulin protein, either Tub1p or Tub3p, forms a heterodimer with the β-tubulin protein, Tub2p, and heterodimers are compiled into very large, cylindrical microtubules. These highly ordered protein structures play several vital roles in the cell, most notably in the maintenance of cell shape and in proper chromosome segregation during mitosis. Microtubules also serve as tracks within the cell cytoplasm upon which vesicles and proteins are trafficked via motor-driven transport. Microtubules are dynamic structures, capable of being rapidly polymerized or depolymerized depending, for example, upon the stage of the cell cycle or the cellular environment.
Several findings from previous studies suggest that in yeast a relationship exists between P-bodies and microtubules. For example, genetic disruption of the gene encoding the P-body component Pat1p confers synthetic lethality to cells harboring deletions of either BIK1 or NUM1, two genes implicated in microtubule biogenesis (Tong et al. 2004). Moreover, the P-body protein, Edc3p, has been shown to physically interact with the tubulin proteins Tub2p and Tub3p, while a component of the mRNA decapping enzyme, Dcp1p, also a P-body constituent, interacts with Tub2p (Gavin et al. 2006). Further, Edc3p and Dcp1p physically interact with the polypeptide encoded by YBR094W (Krogan et al. 2006), a homolog of mammalian tubulin tyrosine ligase, a protein implicated in microtubule modification (Raybin and Flavin 1977). Finally, Xrn1p, the exoribonuclease activity catalyzing 5′–3′ mRNA degradation, has been localized to P-bodies (Sheth and Parker 2003) and also represents a microtubule-associated protein that stimulates microtubule growth in vitro (Interthal et al. 1995). These genetic and physical interactions between P-body components and microtubules imply that an important and functional relationship may exist between these cytoplasmic structures that warrants investigation.
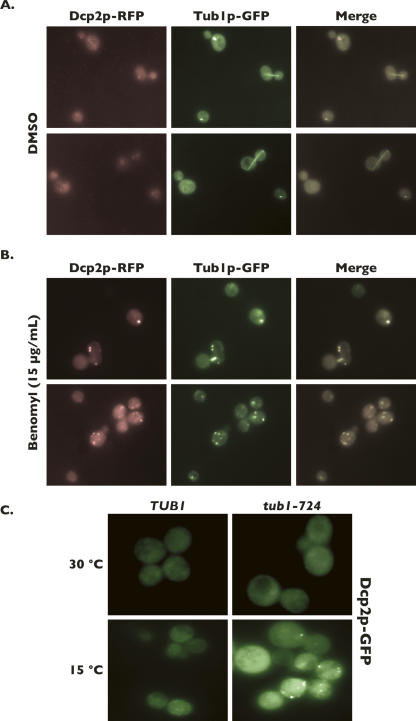
In order to determine if microtubule structure can influence P-body aggregation, we treated yeast cells with the microtubule-destabilizing drug benomyl. Benomyl binds to β-tubulin in vivo and alters the conformation of the α/β-tubulin heterodimer such that microtubules are destabilized (Kilmartin 1981). Consistent with this, benomyl has also been shown to inhibit microtubule polymerization in vitro (Gupta et al. 2004). In our studies, microtubules were detected microscopically in logarithmically growing yeast cells by visualizing a chimeric form of Tub1p, in which the green fluorescent protein (GFP) is fused to the C terminus (Tub1p-GFP). Treatment of cells with 15 μg/mL benomyl led to a dramatic destabilization of microtubules and resulted in the redistribution of Tub1p into foci (Fig. 1B, middle panels). The formation of Tub1p-GFP foci is in contrast to the linear, elongated microtubule filaments that are commonly present in untreated cells and that are observed in cells treated with DMSO alone (Fig. 1A, middle panel).
FIGURE 1.
Both benomyl and tub1-724-mediated microtubule disruption leads to the formation of Dcp2p-containing foci. Cultured cells encoding a chromosomal copy of TUB1-GFP transformed with DCP2-RFP encoded on a plasmid were treated with DMSO alone (A) or benomyl (B), followed by imaging of the cells by fluorescence microscopy. The two sets of panels per treatment represent different images from the same preparation. (C) TUB1 and tub1-724 cell cultures were transformed with DCP2-GFP encoded on a plasmid, and were grown at either the permissive temperature (30°C) or the restrictive temperature (15°C), followed immediately by imaging of the cells by fluorescence microscopy.
A plasmid expressing a fusion between the major mRNA decapping enzyme, Dcp2p, and the red fluorescent protein (Dcp2p-RFP) was introduced into Tub1p-GFP-expressing cells. Dcp2p is a characterized constituent of P-bodies and serves as a molecular marker for these structures (Sheth and Parker 2003; Coller and Parker 2005; Teixeira et al. 2005). Log-phase cells expressing both Tub1p-GFP and Dcp2p-RFP were treated with DMSO (control) or with 15 μg/mL benomyl prior to visualization using epifluorescent phase-contrast microscopy. Mock-treated cells show very faint Dcp2p-RFP foci (Fig. 1A, left panels) consistent with previous observations (Teixeira et al. 2005). In contrast, treatment with benomyl led to the dramatic accumulation of large Dcp2p-RFP foci in cells (Fig. 1B, left panels). Inspection of the cytoplasmic foci determined that Tub1p-GFP colocalized regularly with Dcp2p-RFP foci in cells treated with benomyl but not with DMSO alone (Fig. 1B, right panels). These results indicate that upon microtubule destabilization by treatment of cells with benomyl, the tubulin protein, Tub1p, can associate with cytoplasmic Dcp2p foci. This finding is consistent with previous findings that tubulin demonstrates physical associations with several P-body components.
Our observation that microtubule destabilization induced by the drug benomyl led to the aggregation of Dcp2p-containing foci did not identify whether the result was directly due to microtubule depolymerization or a secondary consequence due to a potential pleiotropic effect of benomyl treatment on cells. Indeed, it has been previously reported that multiple stress conditions facilitate P-body aggregation (Teixeira et al. 2005). As an additional means to determine if microtubule destabilization impacts Dcp2p aggregation, we used a strain containing a cold-sensitive allele of TUB1, tub1-724, which results in depletion of polymerized microtubules when grown at the restrictive temperature of 15°C (Vega et al. 1998). Wild-type (TUB1) and tub1-724 strains were transformed with plasmid DNA encoding Dcp2p-GFP, and cells were grown at 15°C for the required time prior to monitoring Dcp2p-GFP localization by microscopy (Vega et al. 1998). Growth of cells at the restrictive temperature led to the formation of Dcp2p-containing foci in the tub1-724 mutant, while in contrast, Dcp2p foci did not form in wild-type cells (Fig. 1C, bottom panels). Importantly, Dcp2p-GFP foci were not formed in either TUB1 or tub1-724 cells when grown at the permissive temperature of 30°C (Fig. 1C, top panels). These results demonstrate that disruption of microtubules by a genetic lesion also facilitates the aggregation of cytoplasmic foci containing Dcp2p.
Microtubule destabilization enhances P-body formation
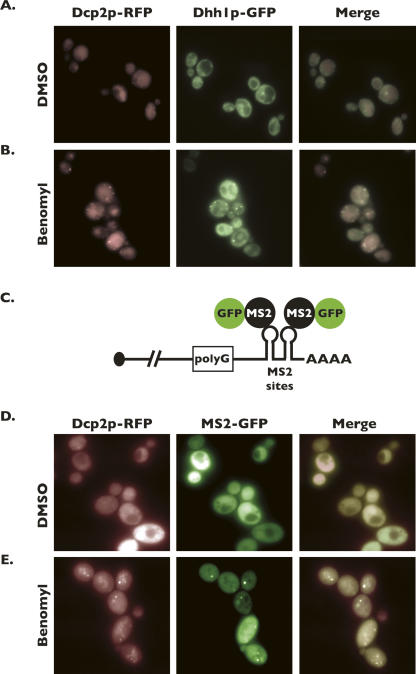
We observed that disruption of microtubules by treatment of cells with benomyl led to the aggregation of Dcp2p-containing foci and that Tub1p also colocalized with these foci. Dcp2p represents one of numerous proteins that have been characterized as constituents of P-bodies, and although Dcp2p aggregation into foci has never been shown to occur independent of these other proteins, we sought to confirm that microtubule destabilization leads to the accumulation of bona fide P-bodies. Dcp2p-RFP-expressing yeast cells were transformed with a plasmid expressing the translational repressor Dhh1p fused with GFP (Dhh1p-GFP), a second marker for P-bodies (Sheth and Parker 2003; Coller and Parker 2005). In cells treated with benomyl, Dhh1p-GFP forms cytoplasmic foci similar to those seen for Dcp2p-RFP (Fig. 2B). In contrast, treatment of cells with DMSO alone fails to result in significant formation of Dhh1p-GFP foci (Fig. 2A). Importantly, the majority of Dhh1p-GFP foci formed upon treatment with benomyl colocalize with Dcp2p-RFP, as indicated by the merged images (Fig. 2B, right panel).
FIGURE 2.
Foci formed within cells as a consequence of microtubule-destabilizing conditions are P-bodies. Wild-type cells were transformed with plasmids encoding DHH1-GFP and DCP2-RFP, and cultures of these cells were treated with DMSO alone (A) or benomyl (B), and then cells were imaged by fluorescence microscopy. The MFA2-pG-MS2 reporter used to detect the intermediate of mRNA decay is diagrammed in C. The poly(G) tract provides an efficient block to Xrn1p-catalyzed 5′–3′ exonucleolytic digestion of the mRNA. The MS2-binding sites are bound by MS2-GFP, thus allowing visualization of the reporter. Wild-type cells were transformed with MFA2-pG-MS2, MS2-GFP, and DCP2-RFP (all encoded on plasmids), and cultures of these cells were treated with DMSO alone (D) or benomyl (E), and then the cells were imaged by fluorescence microscopy.
Previous studies determined that an intermediate of mRNA decay colocalizes with cytoplasmic P-bodies, thereby providing an additional marker for the accurate identification of these aggregates (Sheth and Parker 2003). The presence of the intermediate of mRNA decay is monitored microscopically using an MFA2 mRNA reporter harboring two tandem binding sites for the bacteriophage MS2 coat protein in its 3′-untranslated region (Fig. 2C). This reporter also harbors a polyguanosine (poly[G]) tract of 18 nucleotides (nt) embedded upstream of the two MS2-binding sites, which blocks progression of the 5′–3′ exonuclease, Xrn1p, facilitating the accumulation of a decay intermediate representing the extreme 3′ end of the MFA2 mRNA (Muhlrad et al. 1994). The localization of the decay intermediate is detected in cells that also express the MS2 coat protein fused to GFP (MS2-GFP).
To determine if the foci we observe to aggregate upon treatment of cells with benomyl also contain mRNA decay intermediates, yeast cells were transformed with plasmids harboring the DCP2-RFP, MFA2pG-MS2, and MS2-GFP gene constructs, and cells were either treated with DMSO or with 15 μg/mL benomyl for 1 h. As seen in Figure 2D, treatment of cells with DMSO alone did not lead to the formation of either Dcp2p-RFP or MS2-GFP foci. In contrast, treatment of cells with benomyl facilitates the formation of both Dcp2p-RFP and MS2-GFP foci, and these foci were found to colocalize within the cell (Fig. 2E). Importantly, the MS2-GFP foci represent the decay intermediate, since no foci were observed to form in the absence of the reporter mRNA harboring the MS2-binding sites (data not shown). The foci we observe forming as a result of microtubule destabilization contain a component of the decapping complex, Dcp2p; the translational repressor, Dhh1p; and mRNA decay intermediates (Sheth and Parker 2003); therefore, we can conclude that these foci are P-bodies.
The yeast homolog of a tubulin tyrosine ligase aggregates in P-bodies
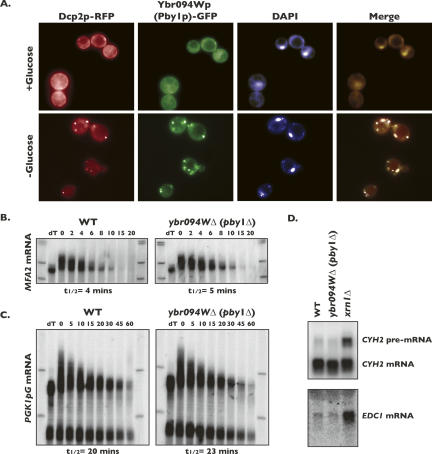
It was determined previously that the yeast homolog of a characterized tubulin tyrosine ligase from mammals, encoded by the uncharacterized open reading frame, YBR094W, physically associates with two protein components of cytoplasmic P-bodies, Edc3p and Dcp1p (Gavin et al. 2006). Mammalian tubulin tyrosine ligases have been shown to add tyrosine to the C terminus of α-tubulin (Raybin and Flavin 1977). While the exact function of this modification is unclear, findings suggest it may be important for controlling microtubule dynamics (Raybin and Flavin 1977). Consistent with the hypothesis that Ybr094wp is associated with microtubules, it has previously been demonstrated that ybr094wΔ mutants exhibit hypersensitive growth inhibition in the presence of benomyl (Parsons et al. 2004), a phenotype exhibited by mutations in many microtubule-associated proteins. Based on these observations, we determined if Ybr094wp was a constituent of cytoplasmic P-bodies. Cells expressing a chromosomal copy of YBR094W engineered to express a C-terminally tagged Ybr094wp-GFP fusion protein were transformed with a plasmid expressing Dcp2p-RFP, and P-bodies were monitored under glucose starvation, a condition that significantly enhances P-body aggregation (Teixeira et al. 2005). Log-phase cells were deprived of glucose for 10 min and the localization of Ybr094wp-GFP was monitored by fluorescence microscopy. As previously observed, P-body aggregation was dramatically stimulated in glucose-deprived cells, as monitored by the accumulation of Dcp2p-RFP in foci (Fig. 3A, first panel). Inspection of the localization of Ybr094wp-GFP determined that glucose deprivation also led to the aggregation of this protein into foci (Fig. 3A, second panel). Importantly, the foci formed by Ybr094wp-GFP exhibited strong colocalization with Dcp2p-RFP, indicating that Ybr094wp is a component of P-bodies (Fig. 3A, fourth panel). The localization patterns of both Ybr094wp-GFP and Dcp2p-RFP were also monitored in the presence of glucose; neither Ybr094wp-GFP nor Dcp2p-RFP was found to significantly aggregate in these cells (Fig. 3A, upper panels). Given the association of Ybr094wp with cytoplasmic P-bodies upon glucose deprivation, we have named this previously uncharacterized ORF PBY1, for P-body protein 1.
FIGURE 3.
Ybr094wp (Pby1p) localizes in P-bodies and plays no obvious role in 5′–3′ mRNA decay, NMD, or decay of EDC1 mRNA. (A) Cells encoding a chromosomal copy of YBR094W-GFP were transformed with DCP2-RFP encoded on a plasmid, and cultures were treated with medium either containing or lacking glucose for 10 min, at which time DAPI was added directly to the culture medium. Cells were then imaged by fluorescence microscopy. Half-lives of MFA2pG mRNA (B) and PGK1pG mRNA (C) were determined in cultures of wild-type or pby1Δ cells by transcriptional shut-off by glucose repression. RNA was isolated at various time points after repression (indicated in the figure) and analyzed by Northern blot. Steady-state levels of CYH2 pre-mRNA and EDC1 mRNA were determined in cultures of wild-type, pby1Δ, and xrn1Δ cells by RNA isolation followed by Northern blot analysis (D).
Many of the characterized constituents of yeast cytoplasmic P-bodies play a role in mRNA decay; therefore, we reasoned that Pby1p might be involved in some aspect of mRNA metabolism. To test this, we constructed a yeast strain deleted for PBY1 and performed analysis of mRNA decay through transcription shut-off experiments utilizing plasmid-encoded reporter mRNAs under the transcriptional control of the galactose-inducible promoter (Muhlrad et al. 1994). The intrinsic instability of MFA2 mRNA allows for monitoring of any inhibition of mRNA decay, while the stability of PGK1 mRNA allows detection of acceleration in the rate of mRNA decay. Analysis of the half-lives for either MFA2 or PGK1 reporter mRNA determined that they were not dramatically altered in pby1Δ cells compared to wild type (Fig. 3B,C). mRNA decay rates were also assayed in cells in which the pby1Δ allele was introduced into a hypersensitized genetic background that allows subtle aberrations in mRNA decay to be observed (i.e., combining pby1Δ with the dcp1-2 allele) (Schwartz et al. 2003; Kshirsagar and Parker 2004). In such cells, mRNA decay rates also did not differ compared to the rate of decay of mRNA from cells harboring the dcp1-2 allele alone (data not shown).
We also monitored the effect of the pby1Δ mutation on the steady-state RNA levels of a substrate of the nonsense-mediated mRNA decay pathway, CYH2 pre-mRNA (He et al. 1993), and EDC1 mRNA, an mRNA whose levels are very sensitive to changes in mRNA decapping and 5′–3′ mRNA decay (Muhlrad and Parker 2005). Levels of either CYH2 pre-mRNA or EDC1 mRNA were unchanged in pby1Δ cells in comparison to levels in wild-type cells; in contrast, elevated levels of both RNAs were observed in cells harboring a deletion in the 5′–3′ exonuclease, XRN1 (Fig. 3D). Taken together, our findings suggest that Pby1p does not play a discernible role in the degradation of various classes of mRNAs within the cell. Notwithstanding, the association of Pby1p, a putative tubulin tyrosine ligase, with cytoplasmic P-bodies provides additional evidence for a link between the mRNP aggregates and microtubules within the cell.
P-body aggregation as a consequence of microtubule destabilization occurs independent of changes in mRNA metabolism
Numerous studies have established that changes in P-body aggregation are intimately linked to large-scale alterations in mRNA decay and translation (Sheth and Parker 2003; Brengues et al. 2005; Coller and Parker 2005; Teixeira et al. 2005). Specifically, conditions that augment P-body formation, such as glucose deprivation or osmotic stress, are known to be hallmarked by global translational repression of mRNA and inhibition of mRNA decay (Ashe et al. 2000; Hilgers et al. 2006). Moreover, cells incapable of inducing global translational repression during stress demonstrate a general decrease in P-bodies (Coller and Parker, 2005). Finally, mutations that inhibit mRNA decay also increase the size and abundance of P-bodies within the cell (Sheth and Parker 2003).
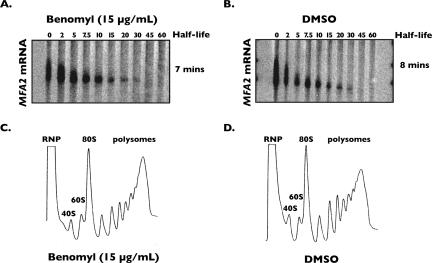
Since we observed a dramatic change in P-body abundance in response to microtubule destabilization, we evaluated if the increase in P-body aggregation corresponded to either an inhibition of mRNA decay or global repression of translation upon treatment of cells with benomyl. In yeast, decay of most mRNA is initiated by removal of the mRNA 3′ poly(A) tail, followed by decapping, and 5′–3′ exonucleolytic decay (for review, see Coller and Parker 2004). To determine the rate of mRNA decay in cells treated with benomyl, a transcriptional shut-off assay was performed in a strain harboring a temperature-sensitive allele of RNA polymerase II (Nonet et al. 1987). Shifting these cells to the restrictive growth temperature results in rapid and dramatic repression of global transcription catalyzed by RNA polymerase II, thereby allowing the deadenylation and decay rate of individual mRNAs to be monitored (Muhlrad et al. 1994). We determined if microtubule destabilization altered mRNA decay rates by treating cells with either benomyl or DMSO, and performing shut-off analysis. For each experiment, P-body formation as a consequence of benomyl treatment was confirmed by monitoring the aggregation of Dcp2p-GFP in these strains (data not shown). As seen in Figure 4B, endogenous MFA2 mRNA deadenylates rapidly and decays with a half-life of 8 min in DMSO-treated cells, consistent with previous findings (Decker and Parker 1993). In cells treated with benomyl, the rate of deadenylation and decay of MFA2 mRNA was nearly identical to DMSO-treated cells (t 1/2 = 7 min) (Fig. 4A). Similar results were observed for additional endogenously encoded mRNAs including CYH2 and RPL41a (data not shown). These results demonstrate that despite a dramatic increase in P-body abundance that corresponds with microtubule destabilization upon treatment of cells with benomyl, the rate of decay of several cellular mRNAs was unaltered.
FIGURE 4.
Benomyl treatment of cells does not alter mRNA decay or global translation. rpb1-1 cells were treated with either benomyl (A) or DMSO alone (B), and the half-life of MFA2 mRNA was determined by transcriptional shut-off by temperature shift. RNA was isolated at various time points after temperature shift (indicated in the figure) and analyzed by Northern blot. Cells containing a chromosomal copy of TUB1-GFP were treated with either benomyl (C) or DMSO alone (D), and sucrose gradient fractionation followed by polysome analysis was carried out as described in the Materials and Methods. Curves represent a continuous plot of absorbance at 254 nm (y-axis) versus position in the sucrose gradient (x-axis). 40S and 60S ribosomal subunit peaks as well as 80S monosome peaks, polysome peaks, and RNP peaks are labeled on the curves.
We next assayed the effect of microtubule destabilization on general mRNA translation using sucrose gradient density fractionation to monitor the size and distribution of polyribosomes within the cell. Cells were treated with either DMSO alone or benomyl for 1 h and polysomes were extracted and applied to 7%–47% sucrose gradients as previously described (Coller and Parker 2005). The effect of both DMSO and benomyl treatment on microtubule destabilization was monitored prior to polyribosome analysis to ensure that the results were similar to those presented in Figure 1. Following ultracentrifugation, the presence of polyribosomes was monitored by continual UV absorbance of rRNA during gradient fractionation. As shown in Figure 4C, cells in which microtubules have been disrupted by treatment with benomyl demonstrate a large abundance of polyribosomes that do not differ from previous findings for wild-type cells (Coller and Parker 2005) or those observed for DMSO-treated cells (Fig. 4D). These observations demonstrate that global change in mRNA translation does not occur upon microtubule destabilization, despite the enhanced aggregation of P-bodies. In summary, our results suggest that the dramatic aggregation of P-body components that occurs as a consequence of microtubule destabilization is uncoupled from significant alterations in either mRNA decay or global mRNA translation. These findings further suggest that an alternative route exists for P-body formation that is independent of perturbations in mRNA metabolism.
To further evaluate our observations that microtubule disruption represents a unique cellular condition leading to enhanced P-body formation, we analyzed microtubule polymerization under glucose deprivation, a condition previously established to enhance P-body aggregation (Teixeira et al. 2005). To assess the status of both microtubules and P-body aggregation in the same cell, cells harboring a chromosomal copy of TUB1-GFP were transformed with plasmid DNA encoding DCP2-RFP. Depriving cells of glucose for 10 min led to the formation of large P-bodies as monitored by Dcp2p-RFP foci compared to cells grown in glucose-containing medium (Supplemental Fig. 1, first column). In both glucose-fed and glucose-deprived cells, microtubules were long and filamentous in nature as established by localization of Tub1p-GFP (Supplemental Fig. 1, middle column). These results establish that glucose deprivation does not significantly impact the status of microtubules in the cell, indicating that the two conditions influence P-body aggregation by distinct mechanisms.
Conclusions
In this report we present evidence that microtubule destabilization stimulates the formation of cytoplasmic P-bodies independent of global changes in mRNA metabolism. First, we demonstrate that treatment of cells with the drug benomyl stimulates the formation of foci that contain Dcp2p, Dhh1p, and an intermediate of mRNA decay, three constituents of bona fide P-bodies (Sheth and Parker 2003). Second, a genetic lesion in the TUB1 gene (tub1-724) that has been demonstrated to disrupt microtubules under nonpermissive growth conditions (Vega et al. 1998) also manifests enhanced formation of P-bodies at the restrictive growth temperature. Finally, we find that disruption of microtubules does not lead to detectable alterations in either mRNA decay or global mRNA translation.
One possible model explaining our observations that P-body aggregation is stimulated by depolymerization of microtubules is that microtubules act, in part, to physically restrict the formation of microscopically detectable P-bodies by either direct interaction with one or more of the constituents of P-bodies or through an unidentified adaptor protein. Interestingly, two P-body proteins, Dcp1p and Edc3p, interact with Tub2p and Tub3p (Gavin et al. 2006). In this model, destabilization of microtubules might relieve a physical impediment preventing the spontaneous aggregation of P-body components, or may prevent P-body components from trafficking away from P-bodies along microtubules. As evidence for a direct link between microtubules and P-body dynamics, we determined that the α-tubulin, Tub1p, localizes to P-bodies upon destabilization of microtubules (Fig. 1B). Additionally, we show that the yeast homolog of tubulin tyrosine ligase, Ybr094wp (Pby1p), is a component of cytoplasmic P-bodies (Fig. 3A). While our data argue that a direct relationship exists between microtubules and P-body dynamics, P-body formation is known to be stimulated by cellular stresses such as glucose deprivation or osmotic stress (Teixeira et al. 2005). Therefore, the possibility exists that disruption of microtubules by either treatment of cells with benomyl or growth of tub1-724 cells at the restrictive temperature leads to activation of a cellular stress pathway that, in turn, induces P-body aggregation. Induction of a stress pathway seems unlikely, however, as the characterized stress conditions that enhance P-body formation result in a general decrease in global mRNA translation and/or an inhibition of mRNA decay (Ashe et al. 2000; Hilgers et al. 2006). Microtubule destabilization does not appear to alter general mRNA metabolism; therefore, the formation of P-bodies under this condition does not mimic what occurs during other types of stress.
The intimate relationship that we observe between microtubules and P-body dynamics is supported by additional observations in the literature. First, several components of the Saccharomyces pombe RNAi machinery localize into P-body-like structures, whose size and abundance increase in response to thiabendazole, a microtubule poison (Carmichael et al. 2006). Second, the Argonaute I homolog from sea urchins, Seawi, has been identified as a microtubule-associated protein and localizes in cytoplasmic puncta (Rodriguez et al. 2005). Third, the oskar mRNP in Drosophilia oocytes, which contains several P-body proteins, mislocalizes in mutants in which microtubule organization is abnormal (Lin et al. 2006). While a precise function for P-bodies in mRNA metabolism remains ambiguous, taken together, these results suggest that P-bodies and similar mRNP granular structures are influenced by the microtubule network in the cell, and P-body assembly is not obligatorily dependent on global alterations in mRNA metabolism. The significance of this relationship remains enigmatic, but suggests that P-body formation is much more complicated than previously believed.
MATERIALS AND METHODS
Yeast strains, plasmids, and growth conditions
Strains used are as follows: yJC245, MAT a ura3 leu2 his3 met15 TUB1-GFP::HIS3; yJC233, MAT a ura3 leu2 his3 met15 YBR094W-GFP::HIS3; yJC151, MAT a ura3 leu2 his3 met15; yJC272 (FSY157), MAT a his3Δ200 leu2-3,112 lys2-801 ura3-52 trp1 Δtub1::HIS3, Δtub3::TRP1 [tub1-724, LEU2, CEN4, ARS1]; yJC271 (FSY182), MAT a his3Δ200 leu2-3,112 lys2-801 ura3-52 trp1 Δtub1::HIS3, Δtub3::TRP1 [TUB1, LEU2, CEN4, ARS1]; yRP1724, MAT a leu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG DHH1-GFP::NEO; yJC244 (N218), MAT a ura3-52 his3-200 leu2-3,112 rpb1-1; yJC240, MAT a ura3 leu2 his3 met15 ybr094w::NEO; yJC182, MAT a ura3 leu2 his3 met15 xrn1::NEO; yJC205, MAT a dcp1-2::TRP1 his3 leu2 lys2 trp1 ura3; yJC259, MAT a dcp1-2::TRP1 ybr094w::NEO his3 leu2 lys2 trp1 ura3. yJC245 and yJC233 were purchased from Invitrogen. yJC240 was prepared by standard methods (Longtine et al. 1998).
Plasmids used are as follows: pJC234 (pRP1167) [DCP2-RFP, LEU2]; pJC147 (pRP1175) [DCP2-GFP, URA3]; pJC139 [DHH1-GFP, URA3]; pJC243 (pRP1081) [MFA2-pG-MS2, URA3]; pJC245 (pCP-GFP) [MS2-GFP, HIS3]; pRP485 [Pgal-MFA2pG, URA3]; pRP469 [Pgal-PGK1pG, URA3]. pRP1175 (Coller and Parker 2005), pRP1081 and pCP-GFP (Sheth and Parker 2003), and pRP485 and pRP469 (Decker and Parker 1993) have all been previously described. pJC139 was prepared by PCR amplification of DHH1-GFP from genomic DNA isolated from strain yRP1724 using oligonucleotides 5′-CCTGTAGGTCTTCTAGACCATCAGATGG-3′ and 5′-CTAGCGGATCTGCCTCTAGAGGTGTGGT-3′ (XbaI sites used for cloning in italics), and ligated into XbaI-linearized vector YCpLac33 (Gietz and Sugino 1988). Yeast transformations were performed using the lithium acetate/single-stranded carrier DNA/polyethylene glycol method (Gietz and Woods 2002).
Strains were cultured in either yeast peptone dextrose (YPD) or synthetic complete (SC) medium lacking the appropriate amino acids. All strains were cultured overnight, then diluted to an OD600=0.05, followed by growth to the desired OD600. A benomyl stock solution (10 mg/mL) was prepared in dimethylsulfoxide (DMSO) and stored at −20°C. Benomyl was added to cultures grown in YPD to a final concentration of 15 μg/mL. DMSO was added to cultures to a final concentration of 0.15%. Cultures treated with benomyl required resuspension in YPD due to the insolubility of benomyl in SC-based medium after several minutes of shaking.
Microtubule destabilization by benomyl treatment of cells
Cell cultures were grown in SC medium at 30°C until an OD600=0.4 was reached. Cells were harvested by centrifugation at 4000 rpm for 3 min, and then resuspended in YPD medium, at which time benomyl or DMSO were added to the cultures. Cultures were incubated at 30°C for 1 h with shaking, after which time a 1 mL aliquot of cell culture was removed, centrifuged at 13,300 rpm for 30 sec, and resuspended in 1 mL of SC medium containing either benomyl or DMSO. Cells were concentrated into 50 μL of SC medium containing either benomyl or DMSO, vortexed, and 3 μL of suspension placed on a glass slide was immediately subjected to microscopy.
Microtubule destabilization by tub1-724
TUB1 or tub1-724 cells transformed with a plasmid encoding DCP2-GFP were grown in SC medium at 30°C to an OD600=0.4, and then either shifted to 15°C or kept at 30°C. Cells were allowed to grow for 24 h, and then were diluted back to an OD600=0.3. Cells were also diluted to an OD600=0.3 at 48 and 72 h. After 96 h at 15°C, tub1-724 cells maintained the same OD600=0.3 after being diluted back to an OD600=0.3 at 72 h. This was taken as the point at which microtubules must be altered, as cells could no longer grow. TUB1 cultures growing at 15°C and 30°C, and tub1-724 cultures growing at 30°C were all diluted to an OD600=0.05 after 92 h, and were grown to an OD600=0.3 (roughly 96 total hours of growth). After each strain grew for 96 total hours, 1 mL of cell culture was centrifuged at 13,300 rpm for 30 sec, and then all medium except for 50 μL was removed. Cells were briefly vortexed, and 3 μL of suspension placed on a glass slide was immediately subjected to microscopy.
Detection of Ybr094wp (Pby1p) localization
Cells encoding a chromosomal copy of YBR094w-GFP transformed with DCP2-RFP encoded on a plasmid were grown in SC medium at 30°C to an OD600=0.4. Cells were then shifted to SC medium either containing or lacking glucose, and cells were incubated for 10 min at room temperature. DAPI was added to cultures to a final concentration of 0.1 μg/mL. A 1 mL aliquot of cells was centrifuged at 13,300 rpm for 30 sec, and all but 50 μL of medium were removed. Cells were briefly vortexed, and 3 μL of suspension placed on a glass slide were immediately subjected to microscopy.
Microscopy
Imaging was performed with a Leica DM 2500 epifluorescent phase contrast microscope attached to a QImaging Retiga Exi Fast 1394 camera. Colocalization was performed using the GFP filter first, followed immediately by using the RFP filter. Color addition and image merging for colocalization were performed using Adobe Photoshop.
mRNA decay analysis by transcriptional shut-off
Wild-type (yJC151), pby1Δ (yJC240), dcp1-2 (yJC205), or dcp1-2 pby1Δ (yJC259) cells were transformed with either pRP485 or pRP469. These plasmids encode MFA2pG and PGK1pG, both under control of the galactose-inducible promoter. Transcriptional shut-off by glucose repression was performed as previously described (Decker and Parker 1993), and aliquots of cell culture were taken at the time points indicated in Figure 3, then cell pellets were flash-frozen in liquid nitrogen. RNA was isolated as previously described (Caponigro et al. 1993). RNA isolated from cells expressing PGK1pG was first digested with RNase H and oJC304 (5′-CGGATAAGAAAGCAACACCTGG-3′) to determine poly(A) status of the PGK1pG mRNA. A total of 20 μg of RNA from each time point was separated by 6% PAGE, and Northern analysis was carried out using a 32P 5′-end-labeled oligonucleotide (5′-AATTCCCCCCCCCCCCCCCCCCA-3′) as the probe to detect reporter mRNA containing the poly(G) tract.
yJC244 (rpb1-1) cells were grown in YPD at 24°C to an OD600=0.4, at which time either benomyl or DMSO was added to the cell cultures, and the cultures were incubated for 1 h at 24°C. Transcriptional shut-off by temperature shift was performed as previously described (Decker and Parker 1993) and aliquots of cell culture were removed at time points following the temperature shift indicated in Figure 4, then cell pellets were flash-frozen in liquid nitrogen. RNA was isolated as above, 20 μg of RNA from each time point were separated by 6% PAGE, and Northern analysis was performed using an MFA2 riboprobe generated by in vitro transcription to prepare anti-sense MFA2 RNA (Muhlrad et al. 1994). Blots were stripped and subsequently reprobed using the 32P 5′-end-labeled oligonucleotides 5′-CCATACCTCTACCACCGGGGTGCTTTCTGTGCTTACCG-3′ and 5′-TTAGAGTTATTTACTCATAATCCGC-3′ to detect CYH2 mRNA and RPL41A mRNA, respectively.
Steady-state mRNA analysis
To assay the effect of pby1Δ on nonsense-mediated decay and EDC1 mRNA decay, wild-type (yJC151), pby1Δ (yJC240), and xrn1Δ cells were grown in YPD at 30°C to an OD600=0.4. Cells were collected by centrifugation, then RNA was isolated as above, and 40 μg of each RNA sample were loaded onto agarose-formaldehyde gels. RNA was transferred to nylon membranes, and CYH2 mRNA was probed for using the oligonucleotide described above. Blots were stripped and were subsequently probed using a 32P 5′-end-labeled oligonucleotide (5′-AATTGCTTTGGATGACCAGATCC-3′) specific for EDC1 mRNA.
Polysome analysis
TUB1-GFP cells (yJC245) were grown to an OD600=0.4 at 30°C. Benomyl or DMSO was then added to the cultures and the cultures were incubated for 1 h at 30°C, at which point cycloheximide was added to each culture to a final concentration of 50 μg/mL. Cells were harvested by centrifugation at 4000 rpm for 2 min, followed by removal of the supernatant and freezing of the cells on dry ice. A small aliquot of each sample was subject to microscopy to confirm that Tub1p-GFP had been disrupted (data not shown). Cell pellets were stored at −80°C until used. Cells were lysed in breaking buffer (10 mM Tris-HCl at pH 7.4, 100 mM NaCl, 30 mM MgCl2, 0.5 mg/mL heparin, 200 μM DTT, 50 μg/mL cycloheximide) by vortexing with 1 vol of glass beads three times for 3 min each at 4°C, with 2 min on ice between each round of vortexing. For each sample, 20 OD260 U were loaded onto a 7%–47% sucrose gradient, and gradients were centrifuged at 37,000 rpm for 3 h at 4°C. Following ultracentrifugation, polysome profiles were generated by pumping the gradient at 1.5 mL/min through an optical cell that measures absorbance at 254 nm.
Supplemental Methods
Cells encoding a chromosomal copy of TUB1-GFP transformed with DCP2-RFP encoded on a plasmid were grown in SC medium at 30°C to an OD600=0.4. Cells were then shifted to SC medium either containing or lacking glucose, and cells were incubated for 10 min at room temperature. A 1 mL aliquot of cells was centrifuged at 13,300 rpm for 30 sec, and all but 50 μL of medium was removed. Cells were briefly vortexed, and 3 μL of suspension placed on a glass slide was immediately subjected to microscopy.
SUPPLEMENTAL DATA
Supplemental Figure 1 and information can be found online at: http://www.case.edu/med/coller/sweetetal07suppl.html.
ACKNOWLEDGMENTS
We thank Drs. Roy Parker (University of Arizona), Frank Solomon (MIT), and Richard Young (MIT) for their generous gift of reagents and helpful advice. We also thank members of the RNA Center for discussions. Funding was provided by the Cellular and Molecular Biology Training Grant for T.J.S., and a grant from the American Cancer Society for J.C.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.355807.
REFERENCES
- Ashe, M.P., De Long, S.K., Sachs, A.B. Glucose depletion rapidly inhibits translation initiation in yeast. Mol. Biol. Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashkirov, V.I., Scherthan, H., Solinger, J.A., Buerstedde, J.M., Heyer, W.D. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behm-Ansmant, I., Rehwinkel, J., Doerks, T., Stark, A., Bork, P., Izaurralde, E. mRNA degradation by miRNAs and GW182 requires both CCR4:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Dev. 2006;20:1885–1898. doi: 10.1101/gad.1424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya, S.N., Habermacher, R., Martine, U., Closs, E.I., Filipowicz, W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- Brengues, M., Teixeira, D., Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caponigro, G., Muhlrad, D., Parker, R. A small segment of the MAT α 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: A stimulatory role for rare codons. Mol. Cell. Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael, J.B., Stoica, C., Parker, H., McCaffery, J.M., Simmonds, A.J., Hobman, T.C. RNA interference effector proteins localize to mobile cytoplasmic puncta in Schizosaccharomyces pombe . Traffic. 2006;7:1032–1044. doi: 10.1111/j.1600-0854.2006.00441.x. [DOI] [PubMed] [Google Scholar]
- Chu, C.Y., Rana, T.M. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J., Parker, R. Eukaryotic mRNA decapping. Annu. Rev. Biochem. 2004;73:861–890. doi: 10.1146/annurev.biochem.73.011303.074032. [DOI] [PubMed] [Google Scholar]
- Coller, J., Parker, R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller, J.M., Tucker, M., Sheth, U., Valencia-Sanchez, M.A., Parker, R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot, N., Babajko, S., Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004;165:31–40. doi: 10.1083/jcb.200309008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker, C.J., Parker, R. A turnover pathway for both stable and unstable mRNAs in yeast: Evidence for a requirement for deadenylation. Genes & Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- Gavin, A.C., Aloy, P., Grandi, P., Krause, R., Boesche, M., Marzioch, M., Rau, C., Jensen, L.J., Bastuck, S., Dumpelfeld, B., et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Sugino, A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gietz, R.D., Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gupta, K., Bishop, J., Peck, A., Brown, J., Wilson, L., Panda, D. Antimitotic antifungal compound benomyl inhibits brain microtubule polymerization and dynamics and cancer cell proliferation at mitosis, by binding to a novel site in tubulin. Biochemistry. 2004;43:6645–6655. doi: 10.1021/bi036112v. [DOI] [PubMed] [Google Scholar]
- He, F., Peltz, S.W., Donahue, J.L., Rosbash, M., Jacobson, A. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1-mutant. Proc. Natl. Acad. Sci. 1993;90:7034–7038. doi: 10.1073/pnas.90.15.7034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilgers, V., Teixeira, D., Parker, R. Translation-independent inhibition of mRNA deadenylation during stress in Saccharomyces cerevisiae . RNA. 2006;12:1835–1845. doi: 10.1261/rna.241006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingelfinger, D., Arndt-Jovin, D.J., Luhrmann, R., Achsel, T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- Interthal, H., Bellocq, C., Bahler, J., Bashkirov, V.I., Edelstein, S., Heyer, W.D. A role of Sep1 (= Kem1, Xrn1) as a microtubule-associated protein in Saccharomyces cerevisiae . EMBO J. 1995;14:1057–1066. doi: 10.1002/j.1460-2075.1995.tb07088.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin, J.V. Purification of yeast tubulin by self-assembly in vitro. Biochemistry. 1981;20:3629–3633. doi: 10.1021/bi00515a050. [DOI] [PubMed] [Google Scholar]
- Krogan, N.J., Cagney, G., Yu, H., Zhong, G., Guo, X., Ignatchenko, A., Li, J., Pu, S., Datta, N., Tikuisis, A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae . Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kshirsagar, M., Parker, R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae . Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, M.D., Fan, S.J., Hsu, W.S., Chou, T.B. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev. Cell. 2006;10:601–613. doi: 10.1016/j.devcel.2006.02.021. [DOI] [PubMed] [Google Scholar]
- Liu, J., Valencia-Sanchez, M.A., Hannon, G.J., Parker, R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie A., 3rd, Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., Pringle, J.R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae . Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen, J. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 2002;22:8114–8121. doi: 10.1128/MCB.22.23.8114-8121.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Parker, R. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J. 2005;24:1033–1045. doi: 10.1038/sj.emboj.7600560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad, D., Decker, C.J., Parker, R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′→3′ digestion of the transcript. Genes & Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. [DOI] [PubMed] [Google Scholar]
- Nonet, M., Scafe, C., Sexton, J., Young, R. Eucaryotic RNA polymerase conditional mutant that rapidly ceases mRNA synthesis. Mol. Cell. Biol. 1987;7:1602–1611. doi: 10.1128/mcb.7.5.1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons, A.B., Brost, R.L., Ding, H., Li, Z., Zhang, C., Sheikh, B., Brown, G.W., Kane, P.M., Hughes, T.R., Boone, C. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 2004;22:62–69. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- Pillai, R.S., Bhattacharyya, S.N., Artus, C.G., Zoller, T., Cougot, N., Basyuk, E., Bertrand, E., Filipowicz, W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Raybin, D., Flavin, M. Enzyme which specifically adds tyrosine to the α chain of tubulin. Biochemistry. 1977;16:2189–2194. doi: 10.1021/bi00629a023. [DOI] [PubMed] [Google Scholar]
- Rehwinkel, J., Behm-Ansmant, I., Gatfield, D., Izaurralde, E. A crucial role for GW182 and the DCP1:DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640–1647. doi: 10.1261/rna.2191905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez, A.J., Seipel, S.A., Hamill, D.R., Romancino, D.P., Di Carlo, M., Suprenant, K.A., Bonder, E.M. Seawi–a sea urchin piwi/argonaute family member is a component of MT-RNP complexes. RNA. 2005;11:646–656. doi: 10.1261/rna.7198205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz, D., Decker, C.J., Parker, R. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA. 2003;9:239–251. doi: 10.1261/rna.2171203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, G.L., Blau, H.M. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Decapping and decay of messenger RNA occur in cytoplasmic processing bodies. Science. 2003;300:805–808. doi: 10.1126/science.1082320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheth, U., Parker, R. Targeting of aberrant mRNAs to cytoplasmic processing bodies. Cell. 2006;125:1095–1109. doi: 10.1016/j.cell.2006.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira, D., Sheth, U., Valencia-Sanchez, M.A., Brengues, M., Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA. 2005;11:371–382. doi: 10.1261/rna.7258505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong, A.H., Lesage, G., Bader, G.D., Ding, H., Xu, H., Xin, X., Young, J., Berriz, G.F., Brost, R.L., Chang, M., et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- van Dijk, E., Cougot, N., Meyer, S., Babajko, S., Wahle, E., Seraphin, B. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 2002;21:6915–6924. doi: 10.1093/emboj/cdf678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, L.R., Fleming, J., Solomon, F. An α-tubulin mutant destabilizes the heterodimer: Phenotypic consequences and interactions with tubulin-binding proteins. Mol. Biol. Cell. 1998;9:2349–2360. doi: 10.1091/mbc.9.9.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor, B., Schiebel, E. Review: An overview of the Saccharomyces cerevisiae microtubule and microfilament cytoskeleton. Yeast. 1997;13:399–434. doi: 10.1002/(SICI)1097-0061(199704)13:5<399::AID-YEA126>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]