Abstract
We previously found that the SINE-encoded mouse B2 RNA binds RNA polymerase II and represses mRNA transcription during the cellular heat-shock response. In vitro B2 RNA assembles into preinitiation complexes on promoter DNA via its interaction with the polymerase, thus rendering the complexes inactive. With the goal of understanding which regions of B2 RNA are important for high-affinity binding to RNA polymerase II and repression of transcription, we performed a structural and deletion analysis of a 178 nucleotide (nt) B2 RNA. We describe an experimentally derived secondary structure model for B2 RNA, and show that RNA polymerase II protects a specific region from RNase digestion. Deletion studies revealed that a 51-nt region of B2 RNA is sufficient for high-affinity binding to RNA polymerase II, association with preinitiation complexes, and repression of transcription in vitro, the latter of which involves a large predominately single-stranded region within the RNA. In addition, this piece of B2 RNA blocked the polymerase from properly associating with template DNA during the assembly of elongation complexes. Further deletion revealed that a 33-nt piece of B2 RNA binds RNA polymerase II, associates with preinitiation complexes, but cannot repress transcription. These results support a model in which RNA polymerase II contains a high-affinity ncRNA docking site to which a distinct region of B2 RNA binds, thereby allowing another region of the RNA to repress transcription. Moreover, the mechanism of transcriptional repression by B2 RNA likely involves disrupting critical contacts between RNA polymerase II and the promoter DNA.
Keywords: ncRNA, short interspersed elements, RNA polymerase II, transcription, repression, B2 RNA
INTRODUCTION
RNA polymerase II transcribes eukaryotic protein encoding genes into mRNA in collaboration with a series of general transcription factors (Orphanides et al. 1996). Gene-specific regulation of transcription occurs through the concerted action of a multitude of regulatory factors including promoter-specific activators and repressors, coregulatory complexes, and histone remodeling and modifying factors (Malik and Roeder 2000; Naar et al. 2001; Narlikar et al. 2002). These factors are primarily proteins, although recently noncoding RNA (ncRNA) regulators of transcription have been discovered (Storz et al. 2005; Goodrich and Kugel 2006). The total number of such ncRNAs is small, but their mechanisms of action are widely diverse, suggesting we have only sampled the breadth of ncRNA transcriptional regulators that exist in eukaryotic cells.
We previously discovered that mouse B2 RNA is a general repressor of RNA polymerase II transcription after heat shock in mouse cells (Allen et al. 2004; Espinoza et al. 2004). Upon heat shock many RNA polymerase II genes are repressed (e.g., actin and hexokinase II), while other heat-shock responsive genes are highly activated (e.g., hsp 70) (Findly and Pederson 1981; Gilmour and Lis 1985; O'Brien and Lis 1993; Sonna et al. 2002). Coincident with these changes in mRNA transcription, RNA polymerase III transcription of B2 RNA is up-regulated in response to heat shock (Fornace and Mitchell 1986; Liu et al. 1995; Li et al. 1999). We found that after heat shock in mouse cells, B2 RNA coimmunoprecipitated with RNA polymerase II (Allen et al. 2004). Moreover, an antisense oligonucleotide against B2 RNA attenuated the transcriptional repression of the actin and hexokinase II genes after heat shock, showing that B2 RNA is critical for the repression of mRNA genes during the mouse heat-shock response (Allen et al. 2004). In vitro experiments provided insight into the mechanism by which B2 RNA represses transcription; it binds directly to core RNA polymerase II with high affinity, specificity, and kinetic stability (Espinoza et al. 2004). Via this interaction, B2 RNA assembles into preinitiation complexes at promoter DNA along with RNA polymerase II and the general transcriptional factors and blocks all detectable RNA synthesis (Espinoza et al. 2004). Although we have begun to understand the general mechanism by which B2 RNA blocks transcription in vitro, the regions and structural elements in B2 RNA responsible for its function are unknown.
Mouse B2 RNA is encoded by short interspersed elements (SINEs), which are retrotransposons dispersed throughout the mouse genome, with ∼350,000 copies per cell (Kramerov and Vassetzky 2005). It is not yet known how many of the B2 SINE sequences encode functional transcriptional repressors. B2 SINEs are transcribed by RNA polymerase III to produce B2 RNAs that are ∼180 nucleotides (nt) in length (Kramerov and Vassetzky 2005). The promoter elements (e.g., the A box and B box) that drive transcription are downstream from the transcription start site and therefore contained within the early transcribed region of B2 SINEs. The 70 nt at the 5′ end of B2 RNAs are evolutionary related to tRNAs, and the very 3′ end of B2 RNAs contain an A-rich sequence conserved among all SINEs (Kramerov and Vassetzky 2005). In addition to heat shock, the level of B2 RNA in cells is up-regulated during development (Bachvarova 1988), in cancer cells (White et al. 1989; Kramerov et al. 1990), and in response to UV irradiation (Rudin and Thompson 2001), viral infection (Singh et al. 1985), and cycloheximide treatment (Liu et al. 1995). It remains unknown whether B2 RNA functions as a transcriptional repressor in response to these events.
One other natural ncRNA, bacterial 6S RNA, has been shown to bind to an RNA polymerase in trans and regulate its activity (Wassarman and Storz 2000). 6S RNA was found to form a stable complex with the σ70 containing RNA polymerase holoenzyme and inhibit σ70-mediated transcription in the late stationary phase (Wassarman and Storz 2000). In addition, a synthetic RNA aptamer, FC, has been previously shown to bind Saccharomyces cerevisiae RNA polymerase II and repress transcription; however, it does not inhibit transcription by Schizosaccharomyces pombe or wheat germ RNA polymerase II (Thomas et al. 1997). Both 6S RNA (Barrick et al. 2005; Trotochaud and Wassarman 2005) and FC RNA (Kettenberger et al. 2006) have been proposed to repress transcription by disrupting contacts between the promoter DNA and the polymerase. 6S RNA was recently found to stably associate with Escherichia coli RNA polymerase, and serve as a template for synthesis of short RNA products (Wassarman and Saecker 2006).
With the goal of further understanding the mechanism by which B2 RNA binds RNA polymerase II and represses transcription, we performed a structural and deletion analysis of B2 RNA. We experimentally arrived at a model for the secondary structure of B2 RNA. RNase footprinting indicated that RNA polymerase II binds to a 3′ region of B2 RNA. Systematic deletions resulted in the identification of a 51 nt piece of B2 RNA that is sufficient to bind RNA polymerase II with high affinity and potently repress transcription in vitro using a predominately single-stranded region of the molecule. Deletion of a stem–loop from the 51 nt piece resulted in a 33-nt RNA that bound RNA polymerase II but was unable to repress transcription. This suggests that distinct regions of B2 RNA function in binding RNA polymerase II and repressing transcription. Finally, we found that the 51-nt piece of B2 RNA blocked the assembly of elongation complexes. Together our results support a model in which a region of B2 RNA binds a high-affinity ncRNA docking site on RNA polymerase II, and transcriptional repression occurs through a distinct region of the B2 RNA. Moreover, the mechanism of transcriptional repression likely involves B2 RNA disrupting important contacts between RNA polymerase II and the promoter DNA.
RESULTS
The elements sufficient for binding to RNA polymerase II and repressing transcription are contained in the 3′ region of B2 RNA
To gain further insight into the mechanism by which B2 RNA functions, we wanted to identify the regions of the RNA that are responsible for binding RNA polymerase II and repressing transcription. Previous work showed that the region from 3 to 149 of the 178 nt B2 RNA was fully functional for binding RNA polymerase II and repressing transcription, while the region from 3 to 74 neither bound the polymerase nor repressed transcription (Espinoza et al. 2004). This led us to predict that in isolation the region from 75 to 149 would be sufficient to function as a transcriptional repressor.
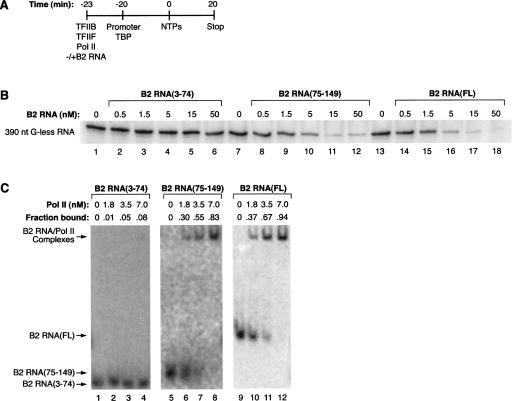
To test this we constructed B2 RNA(75–149) and tested it for the ability to repress transcription in vitro. We used a highly purified transcription system composed of a minimum set of factors that allow promoter-specific transcription: TBP, TFIIB, TFIIF, RNA polymerase II, and the adenovirus major late promoter (AdMLP) contained on a negatively supercoiled plasmid (Kugel and Goodrich 2003). As diagrammed in Figure 1A, different B2 RNA deletions were incubated with protein factors, then added to promoter-bound TBP. After allowing preinitiation complexes to form on the promoter for 20 min, nucleotides were added to initiate transcription, and a 390-nt G-less RNA transcript was monitored. As shown in Figure 1B, B2 RNA(75–149) repressed transcription with a potency similar to that of full-length B2 RNA. As expected, B2 RNA(3–74) did not repress transcription. We also tested these RNAs for the ability to bind RNA polymerase II using electrophoretic mobility shift assays (EMSAs). As shown in Figure 1C, B2 RNA(75–149) bound the polymerase with an affinity similar to that of full-length B2 RNA. As a negative control, B2 RNA(3–74) did not appreciably bind to RNA polymerase II. Therefore, region 75–149 contains the primary determinants for RNA polymerase II binding and transcriptional repression.
FIGURE 1.
The 3′ region of B2 RNA is sufficient for high-affinity binding to RNA polymerase II and potent transcriptional repression. (A) Schematic depicting how in vitro transcription reactions were performed. See text for complete explanation. (B) B2 RNA(75–149) potently represses transcription. B2 RNA(3–74), B2 RNA(75–149), and full-length B2 RNA were added to in vitro transcription reactions at the concentrations indicated. G-less transcript (390 nt) is shown. (C) B2 RNA(75–149) binds RNA polymerase II with high affinity. 32P-labeled B2 RNA(3–74), B2 RNA(75–149), and full-length B2 RNA (each at 0.25 nM) were incubated with RNA polymerase II at the concentrations indicated, and complexes were resolved by EMSA. The positions of bands containing unbound RNA and the complexes are labeled. The exposures for B2 RNA(3–74) and B2 RNA(75–149) are darkened compared to that of full-length B2 RNA to allow the labeled RNAs to be more readily seen. The fraction of the RNA bound is shown above each lane. The apparent K D values for the interaction of RNA polymerase II with B2 RNA(75–149) and full-length B2 RNA are estimated to be <3.5 nM.
A model for the secondary structure of B2 RNA
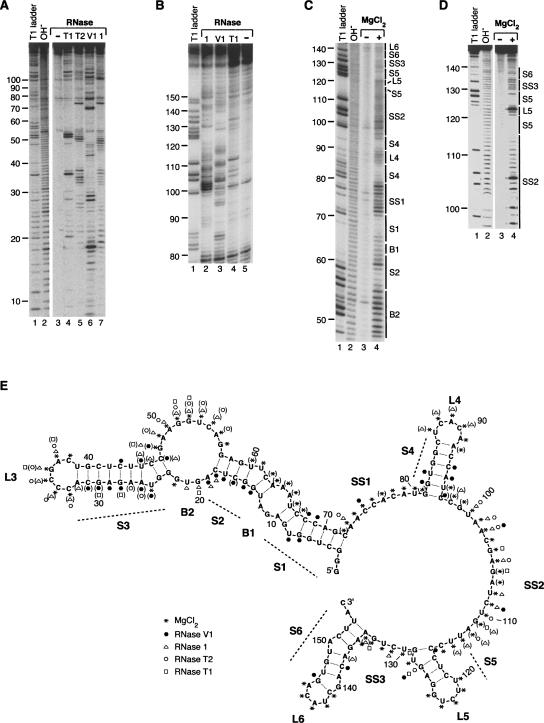
Before further delimiting the regions of B2 RNA involved in binding to RNA polymerase II and transcriptional repression, we determined a model for the secondary structure of the full-length B2 RNA using two techniques: RNase digestion and in-line probing. For the former technique, folded B2 RNA that was 32P labeled on the 5′ end was briefly treated with an RNase and the products were analyzed by denaturing PAGE. Figure 2, A and B, shows representative data for treatment with RNase T1, RNase T2, and RNase 1, which are all single-strand specific, as well as RNase V1, which is double-strand specific. Figure 2A provides resolution for the 5′ end of the RNA and Figure 2B provides resolution for the 3′ end. In-line probing was used to assay for regions of the secondary structure that are single stranded because the greater conformational flexibility of single-stranded RNA makes it more susceptible to cleavage via intramolecular transesterification (Soukup and Breaker 1999). Figure 2, C and D, shows representative in-line probing data; the data in the former panel were generated with full-length B2 RNA and the data in the latter panel with B2 RNA(75–149) to provide resolution of the 3′ end. The gaps in the cleavage patterns clearly show the formation of double-stranded regions.
FIGURE 2.
A model for the secondary structure of B2 RNA derived from RNase and in-line probing experiments. (A) Representative RNase probing data. Lanes 1 and 2 show the denaturing RNase T1 ladder and the alkaline hydrolysis ladder, respectively. Lanes 4–7 show the pattern of products resulting from treatment of 5′ 32P-labeled full-length B2 RNA with RNase T1 (unpaired GpN), RNase T2 (unpaired Ap>Np), RNase V1 (double-stranded RNA), and RNase 1 (unpaired NpN), respectively. The control in lane 3 shows the background pattern of bands for untreated B2 RNA. (B) Representative RNase probing data showing resolution of the 3′ end of B2 RNA. Lane 1 shows the denaturing RNase T1 ladder. Lanes 2, 3, 4, and 5 show digestion with RNase 1, RNase V1, RNase T1, and the undigested control, respectively. (C) Representative in-line probing data. Lanes 1 and 2 show the denaturing RNase T1 ladder and the alkaline hydrolysis ladder, respectively. Lane 4 shows the pattern of bands resulting from incubating 5′ 32P-labeled full-length B2 RNA under in-line probing conditions. The control in lane 3 shows the background pattern of bands for untreated B2 RNA. (D) Representative in-line probing data of B2 RNA(75–149). Lanes are as described for C. (E) Model for the secondary structure of B2 RNA. Nucleotides 1–155 of B2 RNA are shown. Nucleotides 156–178, which constitute an A-rich single-stranded 3′ tail, are not shown in the model. The data from multiple experiments are summarized on the model according to the legend. Symbols in parentheses indicate positions cleaved only weakly. Predominant features in the secondary structure are labeled as: stems 1–6 (S1–S6), bulges 1 and 2 (B1 and B2), loops 3–6 (L3–L6), and single-stranded regions 1–3 (SS1–SS3).
We used the data from the RNase digestion and in-line probing experiments to arrive at the secondary structure model for B2 RNA shown in Figure 2E. The RNA has three main regions: 1–72, 73–153, and 154–178. B2 RNA from 1 to 72 forms a long arm that is composed of three stems (S1–S3), two bulges (B1–B2), and an end loop (L3). The 73–153 region of the RNA is composed of three stem–loops (S4/L4–S6/L6) and three single-stranded stretches (SS1–SS3). The region from 154 to 178 is a 25 nt single-stranded A-rich tail that is not shown in the model (SS4). We previously found that this 3′ tail was not required for RNA polymerase II binding or transcriptional repression (Espinoza et al. 2004).
The data in Figure 2, A–D, support the structural model shown in Figure 2E. For example, the RNase T1 (cleaves unpaired GpN) data in Figure 2, A and B, can be readily compared to the denaturing RNase T1 ladders to reveal G residues that are not cleaved in the structured B2 RNA. These residues are found within each of the six stems in the model: G6, G7, G9, and G71 in S1; G14, G15, G57, and G59 in S2; G23, G24, and G40 in S3; G81, G82, G84, and G85 in S4; G125 in S5; and G136, G140, G146, and G148 in S6. This is also the case for the digestion patterns resulting from RNase T2 and RNase 1; the strong hits from these enzymes occur in single-stranded regions. We observed RNase V1 cleavage in all six stems in the structure. The in-line probing data also support the structural model in Figure 2E. The lines to the right of the gels in Figure 2, C and D, show the locations of the stems, bubbles, and single-stranded regions in the structural model; in general, the lack of cleavage correlates with the formation of stems.
RNA polymerase II protects residues in the 3′ region of B2 RNA from RNase digestion
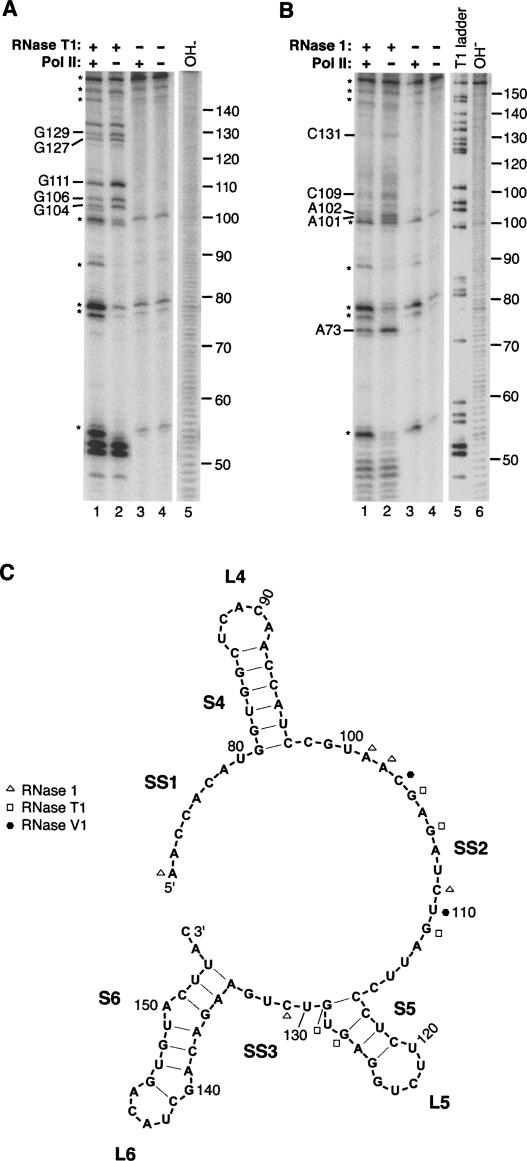
To begin to identify regions within full-length B2 RNA that bind to RNA polymerase II, we performed RNase footprinting experiments. Complexes between RNA polymerase II and full-length B2 RNA were formed and then briefly treated with an RNase (either RNase T1, 1, or V1). The cleavage patterns from these reactions were compared to cleavage patterns obtained from reactions lacking RNA polymerase II to reveal the residues that the polymerase protected from digestion. Representative data for RNase T1 and RNase 1 are shown in Figure 3, A and B, and the data are mapped on the 73–155 region of the B2 RNA secondary structure in Figure 3C. All of the protected residues (12 in total) were in the region from 73 to 131, with the majority of the protected residues (eight out of 12) falling in the single-stranded region SS2. RNA polymerase II protected residues 104, 106, 111, 127, and 129 from cleavage by RNase T1; residues 73, 101, 102, 109, and 131 from cleavage by RNase 1; and residues 103 and 110 from cleavage by RNase V1. We conclude that RNA polymerase II binds within the region from 73 to 131, which correlates well with the data in Figure 1 showing that B2 RNA(75–149) was sufficient for high-affinity binding to the polymerase.
FIGURE 3.
RNA polymerase II protects specific nucleotides in the region from 73 to 131 of B2 RNA from RNase cleavage. (A) Representative data from RNase T1 footprinting. Protected residues are indicated to the left of the gel. Asterisks indicate reoccurring breakdown products of B2 RNA that are observed even in the absence of RNase T1. Lane 5 shows the alkali hydrolysis ladder. (B) Representative data from RNase 1 footprinting. Asterisks indicate reoccurring breakdown products of B2 RNA. Lanes 5 and 6 show the denaturing RNase T1 and the alkali hydrolysis ladders, respectively. (C) Footprinting data mapped onto the secondary structural model of B2 RNA. Shown is the region from 73 to 155, which contains all of the protected residues, although footprinting was performed with full-length B2 RNA. Data from experiments with RNase T1, RNase 1, and RNase V1 are summarized according to the legend.
Delimiting a region of B2 RNA that binds RNA polymerase II and blocks transcription in vitro
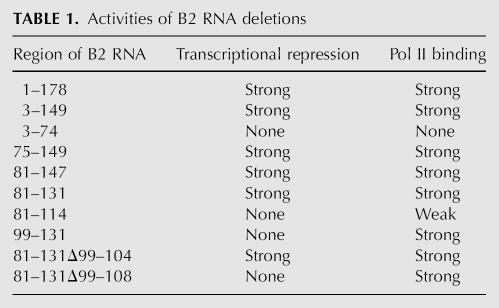
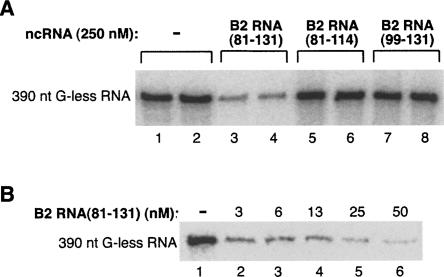
To further delimit regions of B2 RNA involved in repressing transcription, we generated a series of deletions and screened them for the ability to inhibit transcription in vitro (summarized in Table 1). We started with B2 RNA(75–149) and sequentially removed regions from the 5′ and 3′ ends. First, we removed the single-stranded region SS1 from the 5′ end to create B2 RNA(81–147), which repressed transcription. Next, we deleted the stem–loop S6/L6 and part of the single-stranded region SS3 to create B2 RNA(81–131), which also repressed transcription (Fig. 4A, cf. lanes 3,4 and lanes 1,2). Further deletion of either the stem–loop S4/L4 or the stem–loop S5/L5 resulted in B2 RNA(81–114) and B2 RNA(99–131), respectively, which were both incapable of repressing transcription even when added at a concentration of 250 nM (Fig. 4A, lanes 5–8). Hence, the minimal piece of B2 RNA identified that can repress RNA polymerase II transcription in vitro spans nucleotides 81–131 and consists of stem–loop S4/L4, single-stranded region SS2, and stem–loop S5/L5. To determine the potency of B2 RNA(81–131) as a transcriptional repressor, we titrated it into in the minimal transcription system. Figure 4B shows that transcription was inhibited by >50% at the lowest concentration of B2 RNA(81–131) tested (3 nM), and fitting the data resulted in an IC50 <2 nM.
TABLE 1.
Activities of B2 RNA deletions
FIGURE 4.
The region of B2 RNA from 81 to 131 is sufficient to repress RNA polymerase II transcription in vitro. (A) The region from 81 to 131 is the minimal identified piece of B2 RNA that functions as a transcriptional repressor in vitro. B2 RNA(81–131), B2 RNA(81–114), or B2 RNA(99–131) was added to in vitro reactions at a final concentration of 250 nM. Reactions were performed in duplicate. G-less transcripts (390 nt) are shown. (B) B2 RNA(81–131) potently represses RNA polymerase II transcription in vitro. B2 RNA(81–131) was titrated into in vitro transcription reactions at the concentrations indicated. G-less transcripts (390 nt) were visualized.
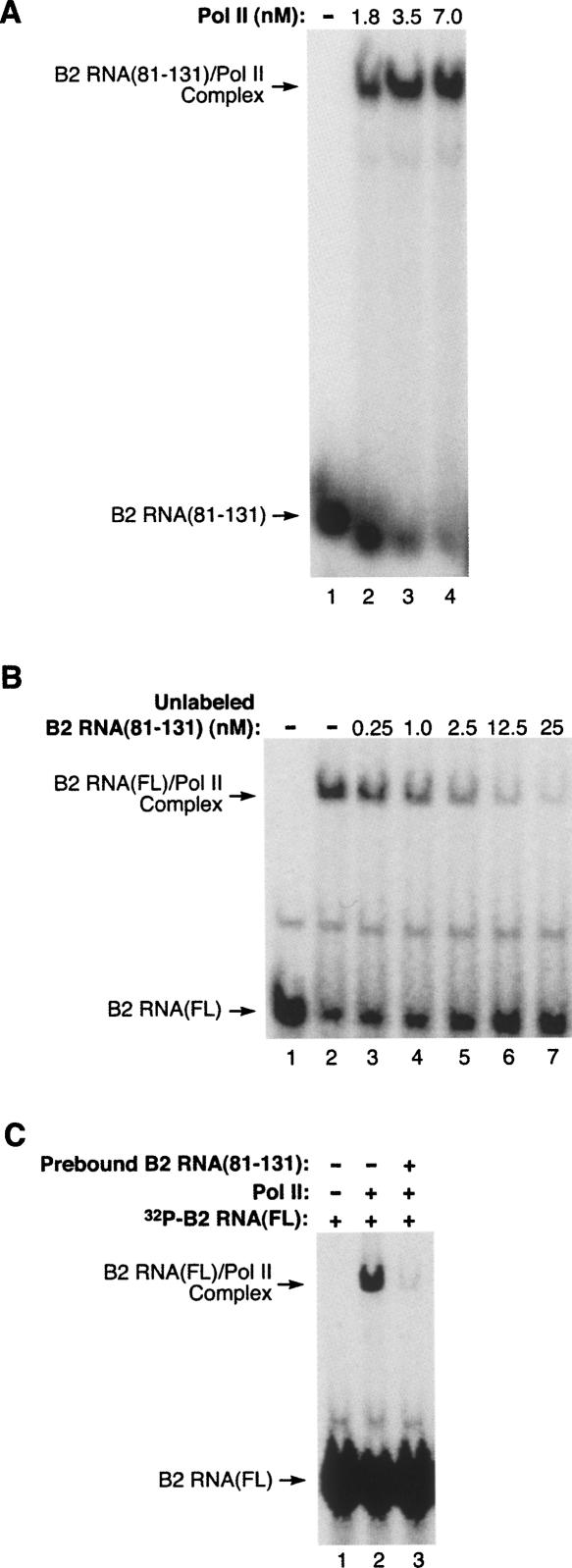
We next investigated the RNA polymerase II binding properties of B2 RNA(81–131). This region of B2 RNA bound the polymerase with high affinity as determined using EMSAs in which the concentration of the B2 RNA(81–131) was kept constant and the RNA polymerase II was titrated up (Fig. 5A). To determine the relative affinities of B2 RNA(81–131) and full-length B2 RNA, we performed competition assays in which increasing concentrations of unlabeled B2 RNA(81–131) were mixed with a constant concentration of 32P-labeled full-length B2 RNA. RNA polymerase II was added to the mixtures and complexes were resolved by EMSA. As shown in Figure 5B, unlabeled B2 RNA(81–131) competed well with 32P-labeled full-length B2 RNA; when added to reactions at a concentration equal to the full-length B2 RNA it caused ∼50% reduction in the amount of complex (Fig. 5B, lane 5). These results also indicate that B2 RNA(81–131) binds to the same, or an overlapping site on the polymerase as full-length B2 RNA. To extend this observation, we also performed a blocking experiment in which unlabeled B2 RNA(81–131) was prebound to RNA polymerase II, then an equal concentration of 32P-labeled full-length B2 RNA was added. As can be seen in Figure 5C, B2 RNA(81–131) completely blocked full-length B2 RNA from binding to the polymerase. Together the data in Figures 4 and 5 identify a 51-nt piece of B2 RNA that is fully functional for binding to RNA polymerase II and repressing transcription.
FIGURE 5.
B2 RNA(81–131) competes with full-length B2 RNA for binding RNA polymerase II. (A) B2 RNA(81–131) binds RNA polymerase II with high affinity. RNA polymerase II was titrated (at the indicated concentrations) into reactions containing 0.5 nM 32P-labeled B2 RNA(81–131). The positions of bands containing unbound B2 RNA(81–131) and the complex are indicated. (B) B2 RNA(81–131) efficiently competes with full-length B2 RNA for binding RNA polymerase II. 32P-labeled full-length B2 RNA (2.5 nM) was combined with unlabeled B2 RNA(81–131) at the concentrations indicated. RNA polymerase II was then added to a final concentration of 3 nM. Complexes were resolved by EMSA and visualized by phosphorimagery. (C) B2 RNA(81–131) blocks the binding of full-length B2 RNA to RNA polymerase II. Unlabeled B2 RNA(81–131) (2.5 nM) was preincubated with 2 nM RNA polymerase II prior to the addition of 32P-labeled full-length B2 RNA (2.5 nM). Controls lacking B2 RNA(81–131) and RNA polymerase II are also shown.
Binding to RNA polymerase II is not sufficient for transcriptional repression by an ncRNA
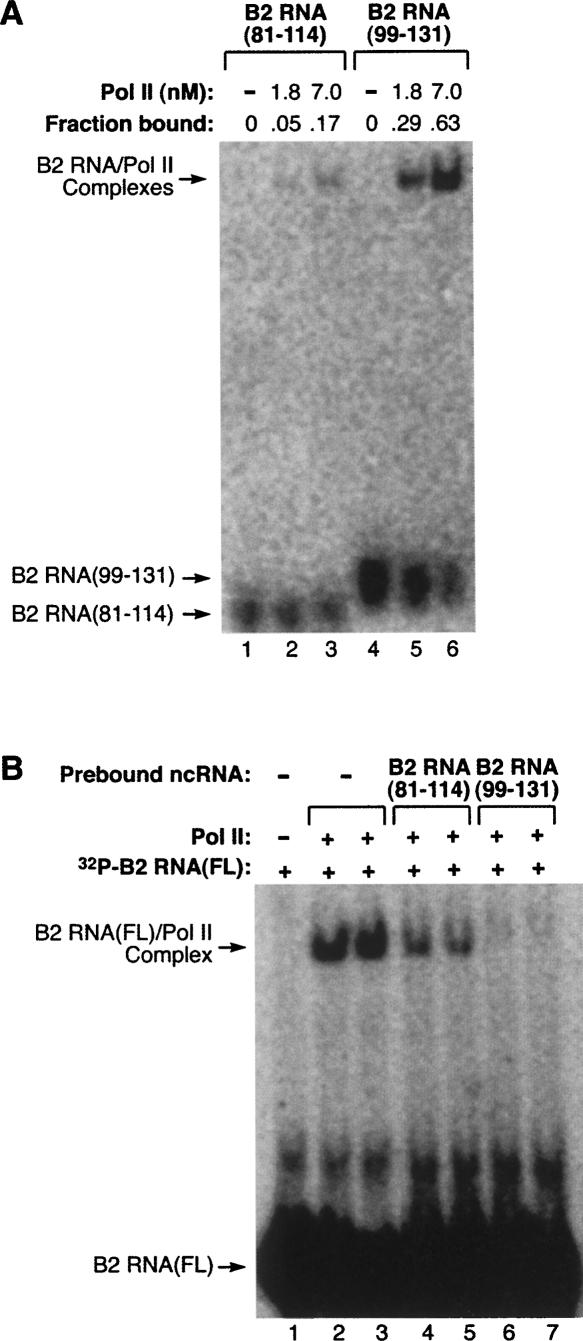
In Figure 4A we showed that two small pieces of B2 RNA (81–114 and 99–131) were unable to repress transcription. Both these pieces, however, are contained within the delimited region that binds the polymerase; therefore, it seemed possible that one or both of these pieces would bind RNA polymerase II. To test this we performed EMSAs and found that B2 RNA(99–131) bound RNA polymerase II with an affinity close to that of full-length B2 RNA, whereas B2 RNA(81–114) exhibited only weak binding (Fig. 6A). We next asked whether these RNAs could block full-length B2 RNA from binding the polymerase. Unlabeled B2 RNA(99–131) or B2 RNA(81–114) was incubated with RNA polymerase II, then 32P-labeled full-length B2 RNA was added and complexes were resolved by EMSA. As shown in Figure 6B, B2 RNA(99–131) fully blocked the binding of full-length B2 RNA to the polymerase (Fig. 6B, lanes 6 and 7), whereas B2 RNA(81–114) partially blocked full-length B2 RNA from binding (Fig. 6B, lanes 4 and 5). This indicates that B2 RNA(99–131) completely masks the region(s) on the polymerase to which full-length B2 RNA binds. Taken together, our data show that binding to RNA polymerase II is itself not sufficient for transcriptional repression.
FIGURE 6.
High-affinity binding to RNA polymerase II does not necessarily cause transcriptional repression. (A) B2 RNA(99–131) binds RNA polymerase II with high affinity, while B2 RNA(81–114) binds weakly. RNA polymerase II was titrated (at the indicated concentrations) into reactions containing 0.25 nM 32P-labeled RNA. The positions of bands containing unbound RNA and the complex are indicated. The fraction of the RNA bound is shown above each lane. (B) B2 RNA(99–131) blocks the binding of full-length B2 RNA to RNA polymerase II. 2.5 nM unlabeled B2 RNA(99–131) or B2 RNA(81–114) was preincubated with 2 nM RNA polymerase II before addition of 32P-labeled full-length B2 RNA (2.5 nM). Reactions were performed in duplicate. Lane 1 lacked RNA polymerase II.
The unstructured region of B2 RNA(81–131) is important for transcriptional repression
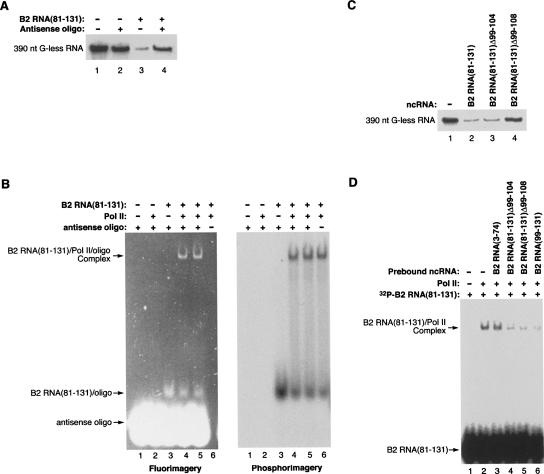
B2 RNA(81–131) consists of two stem–loops (S4/L4 and S5/L5) separated by a predominately single-stranded region (SS2). We have shown that both stem–loops are required for transcriptional repression. To test the function of SS2 in transcriptional repression, we annealed a complementary oligonucleotide to the 99–115 region of B2 RNA(81–131). As shown in Figure 7A, the antisense oligonucleotide attenuated transcriptional repression by B2 RNA(81–131). To ask whether the observed derepression caused by the annealed oligonucleotide was due to decreased binding to RNA polymerase II, we performed EMSAs with 32P-labeled B2 RNA(81–131), fluorescently labeled antisense oligonucleotide, and RNA polymerase II. Figure 7B shows both fluorimagery (left) and phosphorimagery (right) scans of the same gel. The two scans were aligned to determine the positions at which the oligonucleotide and B2 RNA(81–131) migrated. The oligonucleotide did not change the fraction of B2 RNA(81–131) that binds to RNA polymerase II (Fig. 7B, cf. lanes 4,5 and lane 6 in the phosphorimagery scan). Moreover, the oligonucleotide comigrated with the B2 RNA(81–131) in the shifted complex (Fig. 7B, lanes 4 and 5 in both scans), indicating that it remained annealed to B2 RNA(81–131) when the RNA bound to the polymerase. The experiments in Figure 7, A and B, show that either the unstructured nature of the single-stranded region in B2 RNA(81–131), or tertiary interactions involving this region, are important for transcriptional repression but not binding to RNA polymerase II.
FIGURE 7.
The single-stranded region of B2 RNA(81–131) is critical for transcriptional repression but not binding to RNA polymerase II. (A) An antisense oligonucleotide complementary to region 99–115 attenuates transcriptional repression by B2 RNA(81–131). Reactions contained either B2 RNA(81–131) (50 nM; lane 3), an antisense oligonucleotide (150 nM; lane 2), or the two annealed together (lane 4). G-less transcript (390 nt) is shown. (B) The antisense oligonucleotide does not affect binding to RNA polymerase II. Binding reactions contained RNA polymerase II (1.5 nM) incubated with either 32P-labeled B2 RNA(81–131) (0.2 nM; lane 6), fluorescently labeled antisense oligonucleotide (5 nM; lane 2), or the two annealed together (lanes 4,5, which are duplicate reactions). Bands were visualized with fluorimagery (left image) and phosphorimagery (right image). (C) Deleting residues 99–108 of B2 RNA(81–131) attenuates transcriptional repression. Each of the indicated ncRNAs was added to transcription reactions at a final concentration of 50 nM. G-less transcript (390 nt) is shown. (D) Deletions within the single-stranded region of B2 RNA(81–131) do not affect binding to RNA polymerase II. The indicated unlabeled ncRNAs (2.5 nM) were preincubated with 2 nM RNA polymerase II prior to the addition of 32P-labeled B2 RNA(81–131) (2.5 nM). Controls lacking unlabeled ncRNAs and RNA polymerase II are also shown.
To further test the role of SS2 in the function of B2 RNA(81–131), we made two deletions that removed nucleotides 99–104 (B2 RNA[81–131]Δ99–104) or 99–108 (B2 RNA[81–131]Δ99–108). When tested in our in vitro transcription system, B2 RNA(81–131)Δ99–104 repressed transcription as well as B2 RNA(81–131) (Fig. 7C). Therefore, the absolute length of SS2 (18 nt) is not important for transcriptional repression. By contrast, B2 RNA(81–131)Δ99–108 did not repress transcription (Fig. 7C), suggesting that SS2 must be at least 12 nt in length for repression to occur, or that the sequence from 105 to 108 is important for repression. To test whether the deletions affected binding to RNA polymerase II, we performed the blocking experiment shown in Figure 7D in which unlabeled ncRNAs were prebound to RNA polymerase II, then 32P-labeled B2 RNA(81–131) was added. Both B2 RNA(81–131)Δ99–104 and B2 RNA(81–131)Δ99–108 blocked the binding of B2 RNA(81–131) with an efficiency similar to B2 RNA(99–131), suggesting comparable affinities. As a negative control B2 RNA(3–73) did not block binding. Figure 7, C and D, shows that SS2 is important for transcriptional repression, and the region from 99 to 108 is not important for binding to RNA polymerase II.
B2 RNA deletions exert different effects on the assembly of preinitiation complexes and elongation complexes
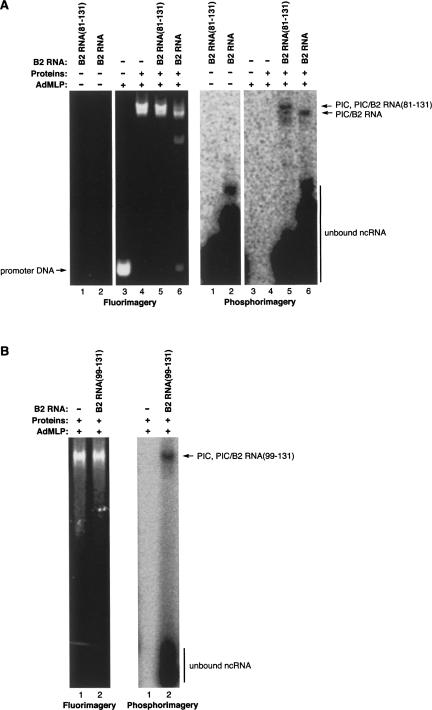
B2 RNA represses transcription in vitro by assembling into preinitiation complexes with RNA polymerase II, the general transcription factors, and promoter DNA, thus rendering the complexes inactive (Espinoza et al. 2004). Accordingly, we asked whether the minimal identified piece of B2 RNA that represses transcription could assemble into preinitiation complexes. To do so we monitored the formation of preinitiation complexes using EMSAs in which the AdMLP promoter DNA was fluorescently labeled and B2 RNA(81–131) or full-length B2 RNA (as a control) was 32P labeled at the 5′ end. Figure 8A shows both fluorimagery (left) and phosphorimagery (right) scans of the same gel; the two scans were aligned to determine the positions at which the promoter DNA and the ncRNAs migrated. Similar amounts of each of the ncRNAs comigrated with the promoter DNA in preinitiation complexes, showing that B2 RNA(81–131) assembled into preinitiation complexes as did the full-length B2 RNA. We next asked whether an ncRNA that binds RNA polymerase II but does not repress transcription could assemble into preinitiation complexes. As shown in Figure 8B, B2 RNA(99–131) assembled into preinitiation complexes. Therefore, preinitiation complexes can contain a bound ncRNA and be fully functional. We previously observed that preinitiation complexes containing full-length B2 RNA migrated faster in EMSAs than did preinitiation complexes lacking B2 RNA (Espinoza et al. 2004), which is evident in the experiment in Figure 8A (Fig. 8A, cf. lane 1 and lane 3 of the fluorimagery scan). Interestingly, B2 RNA(81–131) and B2 RNA(99–131) did not appreciably change the migration of preinitiation complexes. This suggests that the change in migration is dictated by the size or overall negative charge of the ncRNA.
FIGURE 8.
Pieces of B2 RNA that bind RNA polymerase II also assemble into preinitiation complexes. (A) B2 RNA(81–131) associates with preinitiation complexes, but does not alter their migration. Preinitiation complexes were assembled on fluorescently labeled promoter DNA in the absence (lane 4) and presence of 5′-32P-labeled B2 RNA(81–131) or full-length B2 RNA (lanes 5,6, respectively). Bands were visualized with fluorimagery (left image) and phosphorimagery (right image). (B) B2 RNA(99–131) associates with preinitiation complexes. Preinitiation complexes were assembled on fluorescently labeled promoter DNA in the absence (lane 1) and presence (lane 2) of 32P-labeled B2 RNA(99–131).
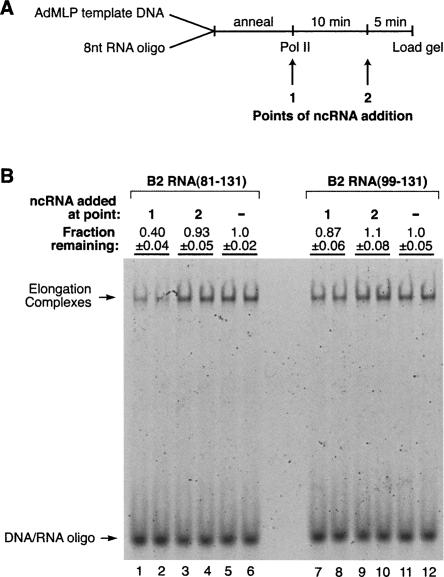
One mechanism by which B2 RNA could repress transcription is by blocking RNA polymerase II from properly associating with the DNA. As a first step in testing this model we asked whether B2 RNA(81–131) and B2 RNA(99–131) could block the artificial assembly of RNA polymerase II elongation complexes in the absence of general transcription factors. As diagrammed in Figure 9A, elongation complexes were assembled by incubating core RNA polymerase II with a fluorescently labeled template DNA strand annealed to a complementary 8-nt RNA oligonucleotide (Kireeva et al. 2000). Complexes were resolved on native gels and visualized by fluorimagery. B2 RNA(81–131) or B2 RNA(99–131) was added to assembly reactions at two different points: At point 1 the ncRNA was prebound to RNA polymerase II prior to assembling elongation complexes, and at point 2 the ncRNA was added to elongation complexes that had already been assembled. Therefore, this experiment does not ask how ncRNAs affect the step of transcript elongation, but rather how ncRNAs affect the binding of polymerase to promoter DNA and a short annealed RNA oligonucleotide.
FIGURE 9.
B2 RNA(81–131) blocks the assembly of elongation complexes, but B2 RNA(99–131) does not. (A) Schematic of the method used to assemble elongation complexes from RNA polymerase II, a fluorescently labeled template DNA strand, and a complementary 8 nt RNA oligo. ncRNA added at point 1 was preincubated with RNA polymerase II. See text for a complete description. (B) Elongation complexes were assembled as shown in A and described in the text. Reactions were performed in duplicate. The positions of elongation complexes and unbound nucleic acids are indicated. The amounts of complex remaining relative to reactions performed in the absence of added ncRNA are shown above the lanes (errors represent the range of the two measurements).
As shown in Figure 9B, B2 RNA(81–131) blocked the assembly of elongation complexes when it was prebound to RNA polymerase II (added at point 1; Fig. 9B, cf. lanes 1,2 and lanes 5,6). By contrast, B2 RNA(99–131) did not block assembly of elongation complexes when prebound to the polymerase (Fig. 9B, cf. lanes 7 and lanes 8–11 and lane 12). Neither ncRNA decreased the amount of elongation complex when added to reactions at point 2, after the polymerase had already engaged the template and RNA:DNA hybrid. Together our data suggest that a region of B2 RNA(81–131) occupies or overlaps with the site on RNA polymerase II to which the template DNA binds in elongation complexes. This is consistent with a model in which B2 RNA represses transcription by blocking RNA polymerase II from establishing the proper contacts with the promoter DNA.
DISCUSSION
Here we identified the regions in B2 RNA that are responsible for binding RNA polymerase II and repressing transcription. Biochemical assays were used to determine a model for the secondary structure of B2 RNA. RNase footprinting assays revealed that RNA polymerase II interacts with a distinct region in the 3′ half of B2 RNA. Deletion analysis identified a 51-nt piece of B2 RNA that binds RNA polymerase II and represses transcription with efficacy similar to that of the full-length molecule. This piece contains a large single-stranded region critical for transcriptional inhibition. Further deletion of the 51-nt RNA revealed a 33-nt piece of B2 RNA that tightly binds RNA polymerase II, but does not repress transcription in vitro; therefore, high-affinity binding to the polymerase does not always result in transcriptional repression. Both the 51-nt and 33-nt pieces incorporate into preinitiation complexes; however, only the 51-nt piece blocks the assembly of elongation complexes. These data are consistent with a model in which B2 RNA docks to RNA polymerase II, then the region responsible for transcriptional repression interferes with the interaction between the polymerase and template DNA.
We experimentally determined a model for the secondary structure of B2 RNA. The secondary structure has two large regions: 1–72 and 73–153. The 5′ region of B2 RNA consists of a long interrupted stem–loop, which is predicted using mfold (Mathews et al. 1999; Zuker 2003). This portion of B2 RNA is derived from the region of the B2 SINE that contains the A and B box promoter elements for RNA polymerase III transcription. The 73–153 region, which contains the polymerase binding and transcriptional repression functions, consists of a series of stem–loops interrupted by single-stranded sections. We observed that RNase V1, which normally cleaves double-stranded RNA, cleaved at positions C76, C103, and U110 within regions SS1 and SS2. It is possible these residues are involved in tertiary interactions. This also might explain the variability in the intensity of the in-line probing data in the SS2 region. The mouse genome contains ∼350,000 B2 SINEs, with some differing in sequence by only a single nucleotide, and others containing more significant sequence variations (Kramerov and Vassetzky 2005). It is possible that changes in the sequences, and hence structures, of the transcribed RNAs could dictate which B2 RNAs function as transcriptional repressors.
The minimal identified region of B2 RNA that contains both binding and repression activities is 81–131, which consists of an 18-nt predominately single-stranded stretch (SS2) surrounded by two stem–loops (S4/L4 and S5/L5). The deletion of S4/L4 from B2 RNA(81–131) resulted in an RNA (B2 RNA[99–131]) that was able to bind the polymerase but not repress transcription. Therefore, stem–loop S4/L4 is required for transcriptional repression in the context of B2 RNA(81–131), and the minimal identified region sufficient for binding consists of SS2 and stem–loop S5/L5. This region contains all of the RNase footprints, with the exception of one located on residue 73. The majority of the RNase footprints occur in SS2, which is important for transcriptional repression. The data in Figure 7 suggest that both the length (minimum of 12 nt) and unstructured nature of SS2 are important for repression, although it remains possible that the sequence from 105 to 108 is important. We also show that both B2 RNA(81–131) and B2 RNA(99–131) can assemble into preinitiation complexes; however, only B2 RNA(81–131) represses transcription. Therefore, preinitiation complexes containing a bound ncRNA can indeed be active. This suggests that the site on RNA polymerase II, to which B2 RNA(99–131) binds, does not block essential interactions with TBP, TFIIB, TFIIF, or the promoter DNA. By contrast, the inhibitory regions of B2 RNA(81–131) likely block important contacts with either TBP, TFIIB, TFIIF, or the promoter DNA.
The observation that binding to RNA polymerase II does not necessarily result in transcriptional repression supports our working model that RNA polymerase II contains a high-affinity docking site to which ncRNAs bind and then regulate transcription by distinct mechanisms (Goodrich and Kugel 2006). This docking site is distinct from the site on polymerase targeted for transcriptional repression by B2 RNA. This model is also supported by our previous research in which we found that a synthetic RNA oligonucleotide composed of 20 guanosine residues (20rG-oligo) binds tightly to RNA polymerase II (Kugel and Goodrich 2002). Interestingly, the 20rG-oligo inhibits a specific step in early transcription that is different from the step inhibited by B2 RNA (Kugel and Goodrich 2002; Espinoza et al. 2004), although B2 RNA and the 20rG-oligo compete with one another for binding to RNA polymerase II (Espinoza et al. 2004). When the 20rG-oligo is shortened to 10 guanosines (10rG-oligo), it binds RNA polymerase II with a K D similar to that of the 20rG-oligo, but the 10rG-oligo is unable to repress transcription (Kugel and Goodrich 2002). Therefore the 20rG-oligo is analogous to full-length B2 RNA and the 10rG-oligo is analogous to B2 RNA(99–131). It remains to be determined whether other natural ncRNAs bind to this high-affinity docking site on RNA polymerase II to regulate transcription.
Once B2 RNA is docked to RNA polymerase II it may use a mechanism of inhibition similar to that of two other ncRNAs known to bind to an RNA polymerase: the FC aptamer, which binds yeast RNA polymerase II, and 6S RNA, which binds bacterial RNA polymerase. Both FC RNA and 6S RNA are thought to repress transcription by preventing the polymerase from correctly interacting with promoter DNA. A crystal structure of yeast RNA polymerase II bound to the FC aptamer revealed that the aptamer is located in the DNA binding cleft of RNA polymerase II and forms a double stem–loop in the site normally occupied by DNA (Kettenberger et al. 2006). Therefore, the FC aptamer is likely to interfere with the ability of yeast RNA polymerase II to form the proper contacts with the DNA in open complexes (Kettenberger et al. 2006). The 6S RNA secondary structure contains a double-stranded region and a central bulge that are critical for function (Barrick et al. 2005; Trotochaud and Wassarman 2005); deletions in this region abrogated both transcriptional repression and binding to E. coli RNA polymerase (Trotochaud and Wassarman 2005). Here we have found that deleting a portion of a single-stranded region of B2 RNA(81–131) also disrupted transcriptional repression, although it did not affect binding to RNA polymerase II. It was proposed that the central bulge in 6S RNA is suggestive of the open conformation of the promoter DNA during the early stages of transcription, and 6S RNA competes with the promoter for binding the RNA polymerase holoenzyme (Barrick et al. 2005; Trotochaud and Wassarman 2005). It was recently shown that 6S RNA engages the active site of E. coli RNA polymerase to form an open complex with the 6S RNA acting as a template (Wassarman and Saecker 2006). It remains to be determined whether B2 RNA also has this property.
Our data from experiments investigating elongation complex assembly (Fig. 9) suggest that contacts between RNA polymerase II and the template DNA are disrupted by B2 RNA(81–131). Assembly of elongation complexes was blocked by B2 RNA(81–131), but not B2 RNA(99–131). This suggests that transcriptional repression by B2 RNA(81–131) in preinitiation complexes involves disrupting important contacts between the polymerase and the promoter DNA. Although stable complexes containing both promoter DNA and B2 RNA(81–131) can be observed in native gels (Fig. 8), it is likely that polymerase–promoter contacts are disrupted in these complexes and the stability of the complexes arises from other interactions between RNA polymerase II and DNA-bound general transcription factors. This is consistent with our previous experiments showing that B2 RNA blocks all detectable RNA synthesis (Espinoza et al. 2004); therefore, it never allows an RNA:DNA hybrid to be established. Testing this model for the mechanism of repression by B2 RNA and determining precisely which polymerase–DNA contacts B2 RNA disrupts awaits further study.
MATERIALS AND METHODS
Plasmid construction
The construction of pUC–T7–B2, which encodes mouse B2 RNA, has been described elsewhere (Allen et al. 2004). The plasmids pUC–T7–B2(3–74), pUC–T7–B2(75–149), pUC–T7–B2(81–131), pUC–T7–B2(81–114), pUC–T7–B2(99–131), pUC–T7–B2(81–131)Δ99–104, and pUC–T7–B2(81–131)Δ99–108 were constructed using a similar approach. Briefly, sense and antisense oligonucleotides (Invitrogen) containing B2 SINE sequences and an upstream phage T7 promoter were annealed, ligated, and cloned into the Bam HI and Kpn I restriction sites of the pUC18 plasmid (Pharmacia Biotech). As a result of designing constructs that would be efficiently transcribed by T7 RNA polymerase, some ncRNAs contained an extra G and/or 1–3 U residue at the 5′ and 3′ ends, respectively.
Preparation of ncRNAs
32P-body labeled ncRNAs were generated by transcription with T7 RNA polymerase as described previously (Allen et al. 2004; Espinoza et al. 2004). To generate 5′-end labeled ncRNAs, the procedure was modified as follows. Transcription by T7 RNA polymerase was performed without labeled nucleotide. After the ncRNAs were gel purified, the 5′ phosphates were removed by treatment with calf intestinal phosphatase. The ncRNAs were phenol/chloroform extracted and ethanol precipitated, then 32P labeled at the 5′ end. The labeled ncRNAs were then gel purified. All ncRNAs were folded immediately before use by incubating samples at 90°C for 1 min in the buffer described for in vitro transcription, then transferring samples to 4°C.
RNase probing, in-line probing, and RNase footprinting
For RNase digestion, 5′-end labeled, folded B2 RNA was diluted into the buffer described for in vitro transcription. B2 RNA was treated for 30 sec at 30°C with individual RNases added to final concentrations of: RNase T1 (0.1 U/μL), RNase T2 (0.1 U/μL), RNase 1 (0.1 U/μL), or RNase V1 (0.01 U/μL). The positions of the RNase V1 cleavages were determined according to methods previously described (Brown and Bevilacqua 2005). For in-line probing 5′-end labeled, folded B2 RNA was incubated at 25°C for 24 h in 10 mM MgCl2, 100 mM KCl, and 50 mM Tris-HCl (pH 8.5). RNA products were phenol/chloroform extracted, ethanol precipitated, and resolved by either 8% or 12% denaturing PAGE. Bands were visualized using phosphorimagery.
For RNase footprinting, B2 RNA/RNA polymerase II complexes were formed by incubation at 30°C for 15 min. These complexes or free B2 RNA were treated for 30 sec at 30°C with RNase T1 (0.1 U/μL), RNase 1 (0.1 U/μL), or RNase V1 (0.01 U/μL). RNA products were phenol/chloroform extracted, ethanol precipitated, and resolved by either 8% or 12% denaturing PAGE. Bands were visualized using phosphorimagery.
In vitro transcription
Human TBP, TFIIB, TFIIF, and RNA polymerase II were prepared as described previously (Kugel and Goodrich 2003; Weaver et al. 2005). In vitro transcription assays were performed as described previously (Kugel and Goodrich 2003; Weaver et al. 2005). Specifically, 20 μL reactions were performed at 30°C in a buffer containing 10% glycerol, 10 mM Tris (pH 7.9), 50 mM KCl, 10 mM HEPES (pH 7.9), 4 mM MgCl2, 1 mM DTT, 25 μg/mL BSA, and 15 units/mL of RNA Guard (GE biosciences). Transcription factors and template DNA were used at the following final concentrations: 3.5 nM TBP, 10 nM TFIIB, 2 nM TFIIF, 1–3 nM RNA polymerase II, and 1–2 nM DNA template. The DNA template was a negatively supercoiled plasmid containing the AdMLP core promoter (−53 to +10) fused to a 380-bp G-less cassette (Goodrich and Tjian 1994). Purified human TFIIB, TFIIF, core RNA polymerase II, and ncRNA (when included) were incubated together at 30°C for 3 min. In a separate tube, TBP was incubated with the DNA template at 30°C. The contents of these two tubes were combined and allowed to incubate for 20 min at 30°C to allow preinitiation complexes to form. G-less transcription was initiated by addition of a nucleotide mix containing 625 μM ATP, 625 μM UTP, and 25 μM [α-32P] CTP (5 μCi per reaction). Transcription proceeded for 20 min at 30°C, at which point reactions were stopped with 100 μL of a solution containing 3.1 M ammonium acetate, 20 mM EDTA, 10 μg of carrier yeast RNA, and 15 μg proteinase K. Transcripts were ethanol precipitated, resolved by 6% denaturing PAGE, visualized by phosphorimagery, and quantitated using ImageQuant software.
Electrophoretic mobility shift assays (EMSAs)
EMSAs to resolve ncRNA/RNA polymerase II complexes were performed as previously described (Espinoza et al. 2004). 32P-body labeled ncRNAs and purified RNA polymerase II were incubated at 30°C for 10 min in the buffer described for in vitro transcription reactions. Reactions were subjected to electrophoresis at 150 V through 4% native polyacrylamide gels containing 0.5× TBE, 4% glycerol, and 5 mM magnesium acetate. Bands were visualized and quantitated by phosphorimagery. For the blocking experiments in Figures 5C, 6B, and 7D, unlabeled ncRNA was incubated with the polymerase for 10 min, then 32P-labeled ncRNA was added for 2–10 min and complexes were resolved as described above. Fraction bound was calculated as bound/(bound+free).
EMSAs to resolve preinitiation complexes were performed as previously described (Espinoza et al. 2004). Briefly, reactions were assembled as described for in vitro transcription, except plasmid DNA was replaced with a double-stranded DNA fragment containing the AdMLP from −40 to +20 that was fluorescently labeled with Alexa Fluor 647 (Molecular Probes). After allowing complexes to form, reactions were subjected to native PAGE as described above, using borosilicate gel plates (CBS Scientific). Bands were visualized using fluorimagery and phosphorimagery.
Assembly of elongation complexes was performed as described (Kireeva et al. 2000) with some modifications. Reactions were assembled in the buffer used for in vitro transcription reactions. The template strand of the AdMLP from −40 to +20 fluorescently labeled with Alexa Fluor 647 (0.67 μM; Molecular Probes) was annealed to an 8-nt RNA oligonucleotide complimentary to the +1 to +8 region (1.3 μM, Dharmacon) by slowly cooling from 45°C to 25°C. RNA polymerase II or RNA polymerase II/ncRNA complexes were added and reactions were incubated for 10 min at 30°C. Complexes were resolved on native gels as visualized as described above.
ACKNOWLEDGMENTS
We thank Aaron Hieb for providing plasmid pUC-T7-B2(75–149) and fluorescently labeled DNA oligonucleotides. This research was supported by Public Health Service Grant R01 GM068414 from the National Institutes of General Medical Sciences. C.A.E. was supported in part by NIH Predoctoral Training Grant T32 GM08759 and by an NRSA Fellowship (F31 GM070442) from the National Institutes of General Medical Sciences.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.310307.
REFERENCES
- Allen, T.A., Von Kaenel, S., Goodrich, J.A., Kugel, J.F. The SINE-encoded mouse B2 RNA represses mRNA transcription in response to heat shock. Nat. Struct. Mol. Biol. 2004;11:816–821. doi: 10.1038/nsmb813. [DOI] [PubMed] [Google Scholar]
- Bachvarova, R. Small B2 RNAs in mouse oocytes, embryos, and somatic tissues. Dev. Biol. 1988;130:513–523. doi: 10.1016/0012-1606(88)90346-6. [DOI] [PubMed] [Google Scholar]
- Barrick, J.E., Sudarsan, N., Weinberg, Z., Ruzzo, W.L., Breaker, R.R. 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA. 2005;11:774–784. doi: 10.1261/rna.7286705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown, T.S., Bevilacqua, P.C. Method for assigning double-stranded RNA structures. Biotechniques. 2005;38:368–372. doi: 10.2144/05383BM04. [DOI] [PubMed] [Google Scholar]
- Espinoza, C.A., Allen, T.A., Hieb, A.R., Kugel, J.F., Goodrich, J.A. B2 RNA binds directly to RNA polymerase II to repress transcript synthesis. Nat. Struct. Mol. Biol. 2004;11:822–829. doi: 10.1038/nsmb812. [DOI] [PubMed] [Google Scholar]
- Findly, R.C., Pederson, T. Regulated transcription of the genes for actin and heat-shock proteins in cultured Drosophila cells. J. Cell Biol. 1981;88:323–328. doi: 10.1083/jcb.88.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornace A.J., Jr., Mitchell, J.B. Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986;14:5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, D.S., Lis, J.T. In vivo interactions of RNA polymerase II with genes of Drosophila melanogaster . Mol. Cell. Biol. 1985;5:2009–2018. doi: 10.1128/mcb.5.8.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, J.A., Kugel, J.F. Non-coding-RNA regulators of RNA polymerase II transcription. Nat. Rev. Mol. Cell. Biol. 2006;7:612–616. doi: 10.1038/nrm1946. [DOI] [PubMed] [Google Scholar]
- Goodrich, J.A., Tjian, R. Transcription factors IIE and IIH and ATP hydrolysis direct promoter clearance by RNA polymerase II. Cell. 1994;77:145–156. doi: 10.1016/0092-8674(94)90242-9. [DOI] [PubMed] [Google Scholar]
- Kettenberger, H., Eisenfuhr, A., Brueckner, F., Theis, M., Famulok, M., Cramer, P. Structure of an RNA polymerase II-RNA inhibitor complex elucidates transcription regulation by noncoding RNAs. Nat. Struct. Mol. Biol. 2006;13:44–48. doi: 10.1038/nsmb1032. [DOI] [PubMed] [Google Scholar]
- Kireeva, M.L., Komissarova, N., Waugh, D.S., Kashlev, M. The 8-nucleotide-long RNA:DNA hybrid is a primary stability determinant of the RNA polymerase II elongation complex. J. Biol. Chem. 2000;275:6530–6536. doi: 10.1074/jbc.275.9.6530. [DOI] [PubMed] [Google Scholar]
- Kramerov, D.A., Vassetzky, N.S. Short retroposons in eukaryotic genomes. Int. Rev. Cytol. 2005;247:165–221. doi: 10.1016/S0074-7696(05)47004-7. [DOI] [PubMed] [Google Scholar]
- Kramerov, D.A., Tillib, S.V., Shumyatsky, G.P., Georgiev, G.P. The most abundant nascent poly(A)+ RNAs are transcribed by RNA polymerase III in murine tumor cells. Nucleic Acids Res. 1990;18:4499–4506. doi: 10.1093/nar/18.15.4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel, J.F., Goodrich, J.A. Translocation after synthesis of a four-nucleotide RNA commits RNA polymerase II to promoter escape. Mol. Cell. Biol. 2002;22:762–773. doi: 10.1128/MCB.22.3.762-773.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel, J.F., Goodrich, J.A. In vitro studies of the early steps of RNA synthesis by human RNA polymerase II. Methods Enzymol. 2003;370:687–701. doi: 10.1016/S0076-6879(03)70056-1. [DOI] [PubMed] [Google Scholar]
- Li, T., Spearow, J., Rubin, C.M., Schmid, C.W. Physiological stresses increase mouse short interspersed element (SINE) RNA expression in vivo. Gene. 1999;239:367–372. doi: 10.1016/s0378-1119(99)00384-4. [DOI] [PubMed] [Google Scholar]
- Liu, W.M., Chu, W.M., Choudary, P.V., Schmid, C.W. Cell stress and translational inhibitors transiently increase the abundance of mammalian SINE transcripts. Nucleic Acids Res. 1995;23:1758–1765. doi: 10.1093/nar/23.10.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik, S., Roeder, R.G. Transcriptional regulation through mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci. 2000;25:277–283. doi: 10.1016/s0968-0004(00)01596-6. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., Turner, D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Naar, A.M., Lemon, B.D., Tjian, R. Transcriptional coactivator complexes. Annu. Rev. Biochem. 2001;70:475–501. doi: 10.1146/annurev.biochem.70.1.475. [DOI] [PubMed] [Google Scholar]
- Narlikar, G.J., Fan, H.Y., Kingston, R.E. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- O'Brien, T., Lis, J.T. Rapid changes in Drosophila transcription after an instantaneous heat shock. Mol. Cell. Biol. 1993;13:3456–3463. doi: 10.1128/mcb.13.6.3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides, G., Lagrange, T., Reinberg, D. The general transcription factors of RNA polymerase II. Genes & Dev. 1996;10:2657–2683. doi: 10.1101/gad.10.21.2657. [DOI] [PubMed] [Google Scholar]
- Rudin, C.M., Thompson, C.B. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- Singh, K., Carey, M., Saragosti, S., Botchan, M. Expression of enhanced levels of small RNA polymerase III transcripts encoded by the B2 repeats in simian virus 40-transformed mouse cells. Nature. 1985;314:553–556. doi: 10.1038/314553a0. [DOI] [PubMed] [Google Scholar]
- Sonna, L.A., Gaffin, S.L., Pratt, R.E., Cullivan, M.L., Angel, K.C., Lilly, C.M. Effect of acute heat shock on gene expression by human peripheral blood mononuclear cells. J. Appl. Physiol. 2002;92:2208–2220. doi: 10.1152/japplphysiol.01002.2001. [DOI] [PubMed] [Google Scholar]
- Soukup, G.A., Breaker, R.R. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz, G., Altuvia, S., Wassarman, K.M. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- Thomas, M., Chedin, S., Carles, C., Riva, M., Famulok, M., Sentenac, A. Selective targeting and inhibition of yeast RNA polymerase II by RNA aptamers. J. Biol. Chem. 1997;272:27980–27986. doi: 10.1074/jbc.272.44.27980. [DOI] [PubMed] [Google Scholar]
- Trotochaud, A.E., Wassarman, K.M. A highly conserved 6S RNA structure is required for regulation of transcription. Nat. Struct. Mol. Biol. 2005;12:313–319. doi: 10.1038/nsmb917. [DOI] [PubMed] [Google Scholar]
- Wassarman, K.M., Saecker, R.M. Synthesis-mediated release of a small RNA inhibitor of RNA polymerase. Science. 2006;314:1601–1603. doi: 10.1126/science.1134830. [DOI] [PubMed] [Google Scholar]
- Wassarman, K.M., Storz, G. 6S RNA regulates E. coli RNA polymerase activity. Cell. 2000;101:613–623. doi: 10.1016/s0092-8674(00)80873-9. [DOI] [PubMed] [Google Scholar]
- Weaver, J.R., Kugel, J.F., Goodrich, J.A. The sequence at specific positions in the early transcribed region sets the rate of transcript synthesis by RNA polymerase II in vitro. J. Biol. Chem. 2005;280:39860–39869. doi: 10.1074/jbc.M509376200. [DOI] [PubMed] [Google Scholar]
- White, R.J., Stott, D., Rigby, P.W. Regulation of RNA polymerase III transcription in response to F9 embryonal carcinoma stem cell differentiation. Cell. 1989;59:1081–1092. doi: 10.1016/0092-8674(89)90764-2. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]