Abstract
Aptamers are small nucleic acid ligands that bind to their targets with specificity and high affinity. They are generated by a combinatorial technology, known as SELEX. This in vitro approach uses iterative cycles of enrichment and amplification to select binders from nucleic acid libraries of high complexity. Here we combine SELEX with the yeast three-hybrid system in order to select for RNA aptamers with in vivo binding activity. As a target molecule, we chose the RNA recognition motif-containing RNA-binding protein Rrm4 from the corn pathogen Ustilago maydis. Rrm4 is an ELAV-like protein containing three N-terminal RNA recognition motifs (RRMs). It has been implicated in microtubule-dependent RNA transport during pathogenic development. After 11 SELEX cycles, four aptamer classes were identified. These sequences were further screened for their in vivo binding activity applying the yeast three-hybrid system. Of the initial aptamer classes only members of two classes were capable of binding in vivo. Testing representatives of both classes against Rrm4 variants mutated in one of the three RRM domains revealed that these aptamers interacted with the third RRM. Thus, the yeast three-hybrid system is a useful extension to the SELEX protocol for the identification and characterization of aptamers with in vivo binding activity.
Keywords: SELEX, three-hybrid system, RNA recognition motif, RNA/protein interaction, RNA aptamer
INTRODUCTION
Aptamers are small DNA or RNA oligonucleotides that are selected to interact with a variety of different targets such as nucleic acids, polypeptides, sugar molecules, small organic compounds, as well as entire cells (http://aptamer.icmb.utexas.edu/; Homann and Göringer 1999; Göringer et al. 2003; Lee et al. 2004; Proske et al. 2005; Göringer et al. 2006; Ulrich 2006). Key usages are diagnostic and therapeutic applications where target proteins involved in human diseases are detected and inhibited by aptamers (Sullenger and Gilboa 2002; Nimjee et al. 2005). In addition, aptamers can be used to purify proteins or to investigate the sequence specificity of nucleic acid-binding proteins. The latter is important for the investigation of RNA-binding proteins, e.g., for the identification of potential target sites within a given genome in order to characterize the corresponding protein/RNA linkage map (Murphy et al. 2003; Ulrich 2006).
RNA aptamers are identified by the SELEX technology (Ellington and Szostak 1990; Tuerk and Gold 1990). SELEX uses iterative cycles of target binding and sequence enrichment to select for high-affinity ligands from combinatorial nucleic acid libraries with complexities of up to 1015 different molecules. Due to the iterative character of SELEX, even extremely rare sequences can be evolved from the initial library. However, for in vivo applications, the selected aptamers have to be evaluated on an individual basis.
The yeast three-hybrid system is a powerful technique to investigate interactions between RNA and proteins in vivo (SenGupta et al. 1996, 1999; Hook et al. 2005). It allows the identification of RNA and protein binding partners as well as the dissection of higher-order RNA/protein complexes (Bernstein et al. 2002). The system is based on the yeast two-hybrid system and consists of three components: the first hybrid contains the DNA-binding domain of LexA from Escherichia coli fused to the RNA-binding coat protein of phage MS2. The second hybrid is an RNA molecule that consists of the MS2-binding site and the RNA sequence of interest. The third hybrid is composed of the transcriptional activation domain of Gal4 from Saccharomyces cerevisiae and a given RNA-binding domain that interacts with the RNA of interest. The auxotrophic marker HIS3 and lacZ (encoding imidazole-glycerol-phosphate dehydratase from S. cerevisiae and β-galactosidase from E. coli) are used as reporter genes. Their promoter regions contain cognate lexA operator sequences that are recognized by the first hybrid (Bernstein et al. 2002). The strength of this approach is the detection of RNA/protein interactions in vivo. In addition, it offers a rapid and simple experimental routine for detecting reporter gene expression. It has been used for in vivo selection of RNA-binding sites (Bernstein et al. 2005), but the number of transformants, typically 107, limits the exploration of larger sequence space.
As a target molecule for the selection of aptamers we chose Rrm4 involved in pathogenic development of Ustilago maydis, the causative agent of corn smut disease (Becht et al. 2005). This RRM protein is essential for polar growth of the infectious hypha. Loss of Rrm4 abolishes formation of retraction septa at the distal pole and results in an increased number of bipolar growing cells (Becht et al. 2006). The protein is a component of particles that move along microtubules and is most likely involved in RNA transport from the nucleus to the cell poles. Rrm4 contains three N-terminal RRMs (RRM1–3) with characteristic spacing of embryonic lethal abnormal vision (ELAV)-type proteins (Soller and White 2004). A mutational analysis revealed that RRM1 is necessary for the determination of polarity during filamentation (Becht et al. 2006).
Here we followed the strategy to combine SELEX with the three-hybrid system (Cassiday and Maher 2001, 2003) to select RNA aptamers that interact with Rrm4 in vivo. Four aptamer classes were identified after 11 cycles of SELEX. Sequences from this cycle were tested for protein interaction in a yeast three-hybrid screen. Two out of four aptamer classes revealed in vivo binding to Rrm4 and members of both classes were shown to interact with the third RRM.
RESULTS
In vitro selection of Rrm4-binding RNA aptamers
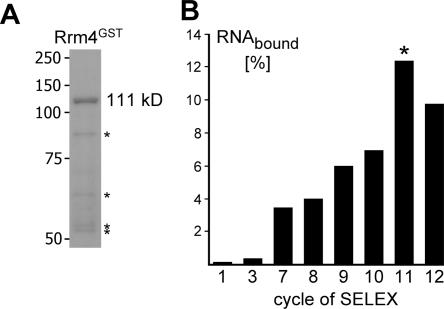
For in vitro selection we used a library of RNA molecules with an estimated complexity of 3 × 1014 unique sequences. The library contained a central domain of 40 randomized nucleotides (nt), flanked by primer binding sites of 21 and 24 nt. As a binding target we chose the Rrm4 protein from U. maydis. The polypeptide was expressed in E. coli as a C-terminal fusion protein with GST (111 kDa) and purified to near homogeneity by affinity chromatography (Rrm4GST; estimated grade of purity ∼85%) (Fig. 1A). Twelve cycles of selection and amplification were performed. Bound RNA was separated from unbound material by filter adsorption. To deplete aptamers that bind to filter material or the GST domain we performed counterselections in cycles 3–4 and 7–12 (see Materials and Methods). The enrichment of Rrm4-specific binders was monitored by determining the percentage of bound RNA in each cycle. An increase was observed that reached a plateau in cycle 11 (Fig. 1B). The RNA pools from cycles 7–12 were analyzed for binding to filter material and BrunoGST. Bruno, an ELAV-type protein from Drosophila melanogaster (Kim-Ha et al. 1995), was expressed as GST fusion protein and purified under identical conditions. In both cases binding never exceeded 2%, indicating that the observed increase was due to the selection of aptamers specific for the Rrm4 part of Rrm4GST.
FIGURE 1.
Course of the SELEX experiment using recombinant Rrm4GST as a target. (A) SDS-PAGE analysis of recombinant Rrm4GST preparations after staining with Coomassie brilliant blue. The full-length fusion protein (111 kDa) and putative degradation products (asterisks) are indicated. (B) Graphical representation of the percentages of bound RNA for different cycles of SELEX. DNA from the 11th cycle (asterisk) was cloned and 49 clones were sequenced.
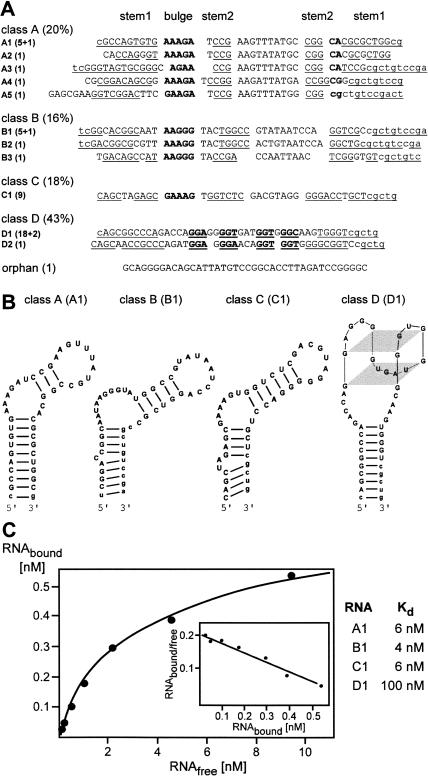
DNA templates from cycle 11 were cloned and 49 clones were sequenced. In total, 12 different sequences were identified (Fig. 2). These sequences were grouped into four classes and an orphan sequence on the basis of primary sequence and secondary structure predictions (Fig. 2A). Five members of class A (20% of the total sequences) exhibit high sequence identity to each other and are predicted to fold into a hairpin structure with a 8–14-nt stem, a highly conserved 10-nt AU-rich loop sequence (AAGUUUAUGC) and an asymmetric purine-rich internal bulge (AAAGA/CA) (Fig. 2B). Class B contains three members (16%), which can be folded into structures similar to class A members with loop sizes from 9 to 11 nt and different AU-rich loop sequences (Fig. 2B). Class C contains a single sequence that was found nine times (18%). It can be folded into a hairpin loop similar to classes A and B (Fig. 2B). However, the loop size is only 8 nt and its sequence lacks AU stretches. The two class D members (21 clones, 43%) are characterized by complementary sequence stretches of 10–11 nt plus four repeats of a GGN triplet sequence. These sequence motifs are likely to fold into a G-quartet structure (Homann et al. 2006), located on top of a helical domain as shown in Figure 2B. Finally, a single orphan sequence (Fig. 2A) was identified, which showed no similarity to any of the other sequences.
FIGURE 2.
Classification of in vitro selected aptamer sequences. (A) Four aptamer classes were identified (A to D, relative abundances in parenthesis). The designation of each aptamer is followed by its clone abundance in parentheses (+n, additional n sequences with single-point mutations) and the primary sequence of the randomized region (capital letters). Complementary sequences corresponding to stems 1 and 2 are underlined. Lower-case letters implicated in stem–loop formation indicate sequences derived from the constant primer binding sites. A purine-rich stretch in classes A–C, a conserved CA/G sequence in class A, and four GGN repeats in class D are printed in bold. (B) Secondary structure models for the most abundant member of classes A–D are shown. Models are based on structure predictions using Mfold and sequence similarities to known G-quartet structures (Zuker 2003; Homann et al. 2006). (C) Determination of equilibrium dissociation constants (Kd). Representative binding isotherm for the binding of aptamer B1 to Rrm4GST. The inset shows a Scatchard plot of the binding data with the slope of the curve representing −1/Kd.
Equilibrium dissociation constants (Kd) for the interaction of the most abundant aptamers A1, B1, C1, and D1 with recombinant Rrm4GST were determined by filter adsorption. A representative binding isotherm for aptamer B1 with corresponding Scatchard analysis is shown in Figure 2C. Aptamers A1, B1, and C1 bind to Rrm4 with Kd values between 4 and 6 nM and thus represent high-affinity RNA/protein interactions. In contrast, the binding of aptamer D1 was about 20-fold weaker and showed a Kd in the range of 100 nM.
In vivo selection of RNA aptamers interacting with Rrm4 using the yeast three-hybrid system
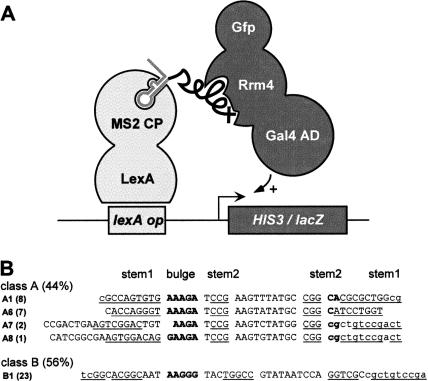
In order to identify aptamer sequences that interact with Rrm4 in vivo we applied the yeast three-hybrid system. Using a LEU2 gene-containing yeast vector, Rrm4 was expressed as fusion protein with the Gal4 activation domain and the green fluorescence protein (eGfp, enhanced version, Clontech) at the N and C terminus (Rrm4AD-G), respectively, (Fig. 3A). The resulting plasmid was transformed into yeast strain L40-coat (SenGupta et al. 1996) containing the two reporter genes lexA-op–lacZ as well as lexA-op–HIS3 and expressing the first hybrid LexA–MS2CP (Fig. 3A). A comparable fusion protein of Rrm4 and eGfp has been shown to be functional in U. maydis (Becht et al. 2006). Gfp fluorescence was used to verify, by microscopy, that Rrm4AD-G was expressed in the yeast nucleus (data not shown).
FIGURE 3.
Yeast three-hybrid selection of RNA aptamers in vivo. (A) Schematic representation of the yeast three-hybrid system. For simplicity only one MS2 protein and binding site are indicated (the MS2 protein binds as a dimer and the RNA hybrid contains two MS2 binding sites). The label “selex” symbolizes the presence of sequences from the 11th cycle of SELEX. CP, AD, and op are abbreviations for coat protein, activation domain, and operator, respectively. (B) Forty-one clones were sequenced and the aptamers were classified as described in Fig. 2A.
To construct the hybrid RNA library we inserted DNA templates from the 11th SELEX cycle downstream from the MS2-binding sites of an URA3 gene-containing vector (pIIIMS2–4). For quality control, the library DNA was transformed into E. coli cells and the inserts of 39 randomly selected clones were sequenced. All four SELEX classes were represented with a relative abundance of 18% (class A), 36% (class B), 31% (class D), and 15% (class C) (data not shown). Next, the library DNA was transformed into yeast strain L40-coat (SenGupta et al. 1996) expressing Rrm4AD-G (see Materials and Methods), and 105 transformants were pooled and tested for HIS3 expression (growth on SC plates −his+5 mM 3-AT, see Materials and Methods). HIS3 and lacZ reporter gene expression could be confirmed for all 104 randomly selected histidine prototrophs. DNA inserts of 41 randomly selected clones were sequenced after amplification by whole-cell PCR. Sequence comparison revealed that members of classes A and B were selected exclusively (44% and 56%, respectively) including three new variants of class A (Fig. 3B; sequences A6–A8). The latter aptamers share the characteristic primary and secondary structure elements of class A including the highly conserved AU-rich 10-nt loop region (Fig. 2B). Thus, class A and B sequences were detected with roughly equal frequencies during SELEX and three-hybrid selection, whereas class C and D aptamers were completely absent from this selection although they account for >60% of the in vitro selected sequences. This might indicate that, whereas aptamers of classes A and B bind in vivo, those of the remaining classes C and D were nonfunctional.
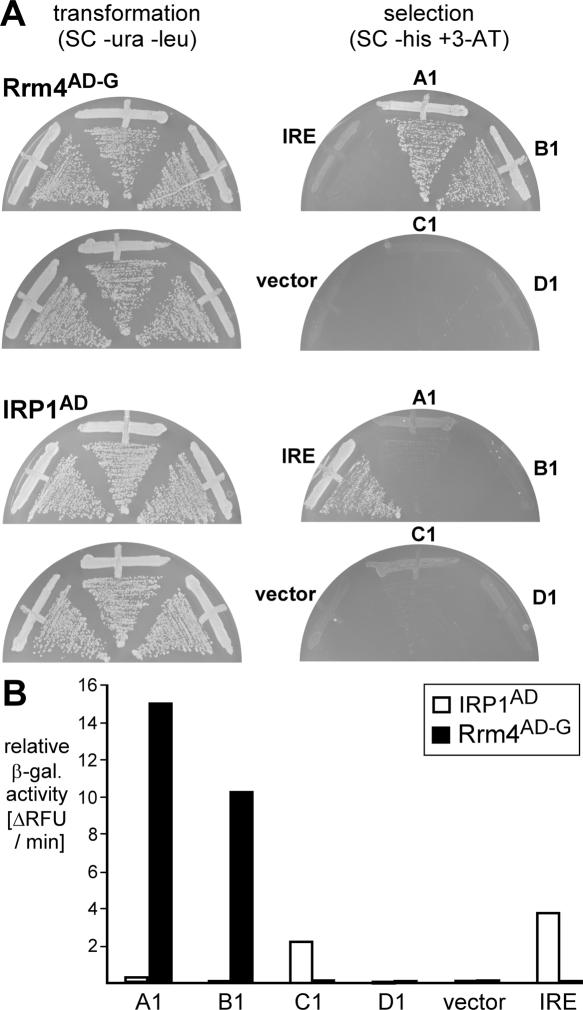
In order to verify this result, the most abundant member of each class was tested individually for its interaction with Rrm4AD-G in the three-hybrid assay. Plasmids encoding hybrid RNAs MS2-A1, MS2-B1, MS2-C1, and MS2-D1 were transformed into L40-coat expressing Rrm4AD-G. Only MS2-A1 and MS2-B1 were able to trigger HIS3 reporter gene expression, whereas MS2-C1 and MS2-D1 were inactive (Fig. 4A, top). The specificity of the aptamer-mediated activation was tested in a control experiment using the iron response protein (IRP1) from mammals that has been shown to bind the iron responsive element (IRE) in vivo (SenGupta et al. 1996). As expected, only MS2-IRE was able to trigger high HIS3 reporter gene expression (Fig. 4A, bottom). These results were further verified by quantifying β-galactosidase reporter gene activation using a fluorogenic substrate. In this assay, the change of relative fluorescence units (RFUs) over time was determined using whole-cell extracts of each transformant. Consistently, MS2-A1 and MS2-B1 were able to activate β-galactosidase expression in combination with Rrm4AD-G (Fig. 4B). In the case of IRP1AD, MS2-IRE, as well as MS2-C1, were able to elicit β-galactosidase expression, indicating a weak interaction of MS2-C1 with IRP1 (Fig. 4B). In summary, representatives from two aptamers classes, MS2-A1 and MS2-B1, interact specifically and efficiently with Rrm4 in vivo.
FIGURE 4.
Three-hybrid assay with representative members from all classes. (A) HIS3 expression was monitored as growth on selection plates (SC −his+5 mM 3-AT). Strain L40-coat expressed either Rrm4AD-G (top) or IRP1AD-G (bottom). The respective hybrid RNA is indicated next to the various plate sectors. (B) β-galactosidase expression was quantified using a fluorogenic substrate. The difference in relative fluorescence units (RFUs) per minute is given for strains expressing the different proteins (inlay) and hybrid RNAs (bottom) tested.
Mapping the Rrm4 interaction domain of the RNA aptamers in vivo
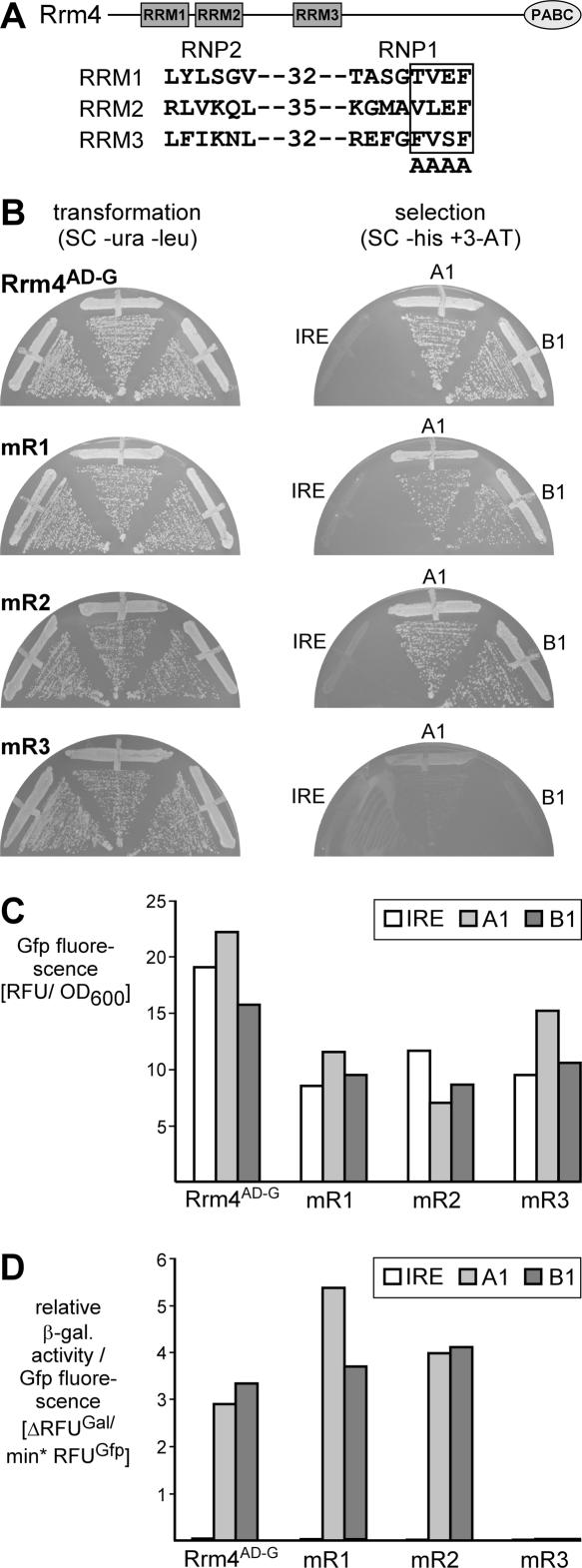
In order to identify the domains of Rrm4 involved in the interaction with aptamers A1 and B1, we tested Rrm4 variants carrying point mutations in the RNP1 region of RRM1, RRM2, and RRM3 (Rrm4-mR1AD-G, Rrm4-mR2AD-G, and Rrm4-mR3AD-G) (Fig. 5A). The same point mutations in RRM1 and RRM3 have been analyzed before in U. maydis (Becht et al. 2006). The corresponding plasmids were cotransformed with hybrid RNA-encoding plasmids expressing MS2-A1, MS2-B1, and MS2-IRE hybrid RNAs into yeast strain L40-coat. HIS3 expression (growth on SC plates −his+5 mM 3-AT) was detected in strains expressing hybrid RNAs MS2-A1 and MS2-B1 with Rrm4AD-G, Rrm4-mR1AD-G, and Rrm4-mR2AD-G but not with Rrm4-mR3AD-G, demonstrating that both aptamers interact with the third RRM domain (Fig. 5B). To control for the expression of Rrm4 mutants we determined Gfp fluorescence of the various Rrm4AD-G versions in combination with three different hybrid RNAs (RFUs per OD600). In all strains each variant was expressed, albeit Rrm4AD-G versions carrying point mutations were expressed with approximately twofold lower efficiency compared to wild type (Fig. 5C). To verify our results obtained during determining HIS3 expression, we measured β-galactosidase reporter gene expression using a fluorogenic substrate as described above. Since reporter gene expression is dependent on the amount of Rrm4AD-G versions (Hook et al. 2005), we calculated the increase of fluorescence over time relative to the amount of Gfp fluorescence (Fig. 5D; ΔRFUGal per min per RFUGfp). Consistently, β-galactosidase activity was not detectable when Rrm4-mR3AD-G was tested with MS2-A1 or MS2-B1 hybrid RNAs (Fig. 5D). Thus, the yeast three-hybrid system allowed rapid identification of the aptamer interaction domain of Rrm4.
FIGURE 5.
Identifying the aptamer interaction domain of Rrm4 in vivo. (A) Schematic representation of the Rrm4 domain architecture (top; RRM and C-terminal PABC domains are given as rectangles and oval, respectively) and the introduced point mutations (bottom). The boxed amino acids in the RNA contact region RNP1 were mutated to alanine. The numbers indicate the distance between RNP1 and RNP2. (B) HIS3 expression was monitored as growth on selection plates (SC −his+5 mM 3-AT). Strain L40-coat expressed different versions of Rrm4AD-G (Rrm4AD-G-mR1, Rrm4AD-G-mR2, and Rrm4AD-G-mR3 carry point mutations in RRM1, RRM2, and RRM3, respectively; labeled with mR1, mR2, and mR3 on the left). The respective hybrid RNA is indicated next to the various plate sectors (labeled on the right). (C) Expression of Rrm4AD-G versions (given at the bottom) was determined in strains expressing different hybrid RNAs (inlay) by quantifying Gfp fluorescence per OD600. (D) Relative β-galactosidase activity was determined using a fluorogenic substrate. To account for differences in protein expression, enzyme activity is given relative to Gfp fluorescence (ΔRFUGal per minute per RFUGfp). Different versions of Rrm4AD-G (bottom) were tested with various hybrid RNAs (inlay).
DISCUSSION
SELEX selects and amplifies RNA aptamers from a large pool of combinatorial sequences. The yeast three-hybrid system detects RNA/protein interactions in vivo. Here we combine these techniques (Cassiday and Maher 2001, 2003) to select and characterize RNA aptamers that interact with the RRM protein Rrm4 in vivo. After 11 cycles of SELEX, a single round of selection applying the three-hybrid system was sufficient to reduce the complexity of the SELEX sequences to ∼50% and identify a distinct subset of RNA aptamers that function in vivo. Secondary structure calculations of these selected aptamers revealed similarities consisting of two stem–loops, a purine-rich bulge and an AU-rich loop sequence (Fig. 2B). This related structure is in accordance with our observation that members of both classes interact with the same domain of the target Rrm4, i.e., the third RRM.
As outlined above RNA aptamers have the capability of binding to a variety of different targets. Therefore, the identification of RNA aptamers that interact with a known RNA-binding protein is no guarantee that the aptamers are directed against the RNA-binding domain of the target protein. Our approach to test different variants carrying point mutations in the RNA contact region of the RRMs allowed the identification of RRM3 as the interaction domain. Thus, applying the yeast three-hybrid system is also a straightforward approach to map the target “epitope” of RNA aptamers.
Rrm4 has been implicated as functioning during polar growth of the infectious hyphae in U. maydis. Since it binds RNA in vivo, and as it moves bidirectionally along microtubules toward both ends of the tip cell, it has been hypothesized that Rrm4 is involved in mRNA transport from the nucleus to the poles (Becht et al. 2006). Currently, we are applying an in vivo UV cross-linking approach to identify target RNAs (Ule et al. 2005). The identified RNA aptamers will be helpful in guiding our search for the identification of Rrm4 binding sites within candidate sequences.
Why does Rrm4 discriminate between the four SELEX classes in the yeast three-hybrid system? The most plausible explanation at least for the observation that Rrm4 interacts with members of classes A and B but not with members of class D is the difference in binding affinities. Members of classes A and B bind with high affinity (Kd values of ∼6 and 4 nM, respectively), whereas the member of class D exhibits ∼20-fold lower affinity (Kd of ∼100 nM). This assumption is supported by recent work systemically investigating the relationship between reporter gene expression in the yeast three-hybrid system and in vitro affinity of an RNA/protein interaction (Hook et al. 2005). High-affinity interactions of FBF-1 from Caenorhabditis elegans with its wild-type binding sequence FBEa (Kd of ∼10 nM) can be detected on selection plates (SC −his) even in the presence of 100 mM 3-AT. When testing low-affinity interactions such as the binding of FBF-1 to a different binding site PME (Kd of ∼80 nM), optimal growth on selection plates is detected only in the absence of 3-AT (Hook et al. 2005). This is consistent with our observation that members of class D exhibiting weak interaction with Rrm4 (Kd of ∼100 nM) do not grow on our selection plates containing 5 mM 3-AT. It remains to be solved why Rrm4 interaction with the class C aptamer is not detectable in the three-hybrid system although the binding affinity in vitro is high (Kd of ∼6 nM). Possible explanations are that this particular RNA aptamer sequence is expressed poorly as a hybrid RNA with the tandem MS2-binding sequences. Alternatively, the aptamer binding site in Rrm4 could be masked in the fusion protein, for example, by the activation domain. However, it is also conceivable that this particular aptamer is unable to fold correctly in vivo. The discrimination of the different aptamers for in vivo binding supports the dual approach combining SELEX and the three-hybrid system. It would have been impossible to predict the correct binding behavior of the various aptamers in vivo solely based on the in vitro binding data.
In previous studies the yeast three-hybrid system was used to select from a random pool in vivo binding sequences for human mRNA export factor Tap or the PUF protein FBF-1 from Caenorhabditis elegans (Kang et al. 1999; Bernstein et al. 2005). These studies were successful for the selection of new variants of known binding sequences. However, both studies were limited in exploring the sequence space by the number of yeast transformants, typically 107. The combination of in vitro and in vivo techniques has the potential to overcome this obstacle. This has been demonstrated by combining SELEX with the three-hybrid system to identify an RNA aptamer in vivo that functions as an RNA decoy for the transcription factor NF-kB (Cassiday and Maher 2001, 2003). Aptamers with improved in vivo activity in the three-hybrid system were selected by testing mutagenized versions of the original RNA aptamer or assaying binding candidates after a few cycles of SELEX under increased selection pressure for HIS3 expression (using higher 3-AT concentrations) (Cassiday and Maher 2003). In the latter case an additional in vivo binder could be identified that might have been lost after excessive in vitro cycles. Here we demonstrate that the combination of SELEX with the three-hybrid system is a useful strategy not only for selection but also for classification of in vivo RNA aptamers. In addition the application of the three-hybrid system allows rapid mapping of the aptamer binding site of the interaction partner.
Future extensions of this approach could be the combination of rapid non-SELEX selection methods such as nonequilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) (Berezovski et al. 2006) with the yeast three-hybrid system or the application of SELEX with the yeast one-hybrid system (Li and Herskowitz 1993) to identify DNA-binding sites of transcription factors.
Combination of SELEX and the three-hybrid system might become particularly attractive for drug discovery, RNA diagnostics, and therapeutics (Cassiday and Maher 2003). Since both techniques can be automated (Cox et al. 2002; Eulberg et al. 2005; Stelzl et al. 2005), high-throughput applications also are feasible.
MATERIALS AND METHODS
Strains, plasmids, and plasmid constructions
The yeast strain L40-coat with the following genotype was used: MATa, ura3–52, leu2–3,112, his3Δ200, trp1Δ1, ade2, LexA–MS2 coat (TRP1), LYS2∷lexA-op–HIS3, and ura3∷lexA-op–lacZ (SenGupta et al. 1996). For cloning purposes and protein expression we used E. coli K-12 derivatives Top10 (Invitrogen) and BL21(DE3)ΔpLys (Stratagene). The plasmids used were pCR2.1-TOPO (Invitrogen) for cloning, pGEX-2T (GE healthcare) for protein expression, and pIIIMS2–1 as well as pACTII for the three-hybrid analysis (SenGupta et al. 1996). Plasmids were constructed as follows. In the case of pRrm4GST-pGEX (pUMa429), a 3.1-kb BamHI/EcoRI fragment from pRrm4AD-G-ACTIIA (pUMa427) was inserted into pGEX-2T. The resulting plasmid encodes the complete rrm4 ORF (Becht et al. 2005) fused to the coding region of the enhanced fluorescence protein (eGfp, Clontech) via the short amino acid linker AANAAT. Plasmid pBrunoGST-pGEX (pUMa435) contains a 2-kb BamHI/EcoRI fragment from pBruno-ACTII encoding the complete Bruno ORF in pGEX-2T. In the case of pACTIIA (pUMa399), a 2.1-kb BsrGI/EagI fragment from pIIIAMS2–2 (Bernstein et al. 2002) encoding Ade2p was inserted into pACTII. In the case of pRrm4AD-G-ACTIIA (pUMa427), a 3.1-kb NcoI/AscI fragment from pRrm4G-NatR (Becht et al. 2006) was inserted into pACTIIA using oligonucleotide linkers. Plasmids pRrm4-mR1AD-G-ACTIIA (pUMa581), pRrm4-mR2AD-G-ACTIIA (pUMa594), and pRrm4-mR3AD-G-ACTIIA (pUMa526) are comparable to pRrm4AD-G-ACTIIA but contain the rrm4 ORF with mutations in the RNP1 regions of RRM1, RRM2, and RRM3 (Fig. 5A). Mutations mR1, mR2, and mR3 were introduced using oligonucleotide primers oMF750 5′-TATCTAGAGTCTGCGCCGCGGCCGCGCCGCTGGCTGTCTGGCCC-3′, oMF764 5′-CGGCGCGCTGGGCGTCTTGTTCAGAGTAGGCCGCCGCGGCGGCCATACCCTTGAAGCC-3′, and oMF755 5′-GAGAATTCGGGCCGCGGCTGCCACCACTCCTGACGAAGCGC-3′. In the case of pIRP1AD-ACTIIA (pUMa412), a 3-kb MluI/BamHI fragment from pAD-IRP1 (Bernstein et al. 2002) was inserted into pACTIIA. The plasmid pIIIMS2–4 (pUMa486), which was used for the construction of the RNA hybrid library, was generated by annealing oligonucleotides oMF762 5′-CCGGAATCGATGCTTAATTAAGGCGCGC-3′ with oMF763 5′-CCGGGCGCGCCTTAATTAAGCATCGATT-3′ and inserting the double-stranded product into the XmaI site of pIIIMS2–1 (Bernstein et al. 2002). Thereby, the multiple-cloning site contains the restriction endonuclease cleavage sites for ClaI, PacI, and AscI, allowing directional cloning of ClaI/AscI inserts. In the case of pIRE-IIIMS2–4 (pUMa492), oligonucleotides oMF636 5′-AAATCGATCCCGAGGATCTGTATCTTGCTTCAACAGTGTTTGGACGGAACAGACCCGGATCGATAA-3′ and oMF637 5′-TTATCGATCCGGGTCTGTTCCG-TCCAAACACTGTTGAAGCAAGATACAGATCCTCGGGATCGATTT-3′ were annealed, cleaved with ClaI, and inserted into pIIIMS2–4. Plasmids pA1-IIIMS2–4 (pUMa550), pB1-IIIMS2–4 (pUMa546), pC1-IIIMS2–4 (pUMa866), and pD1-IIIMS2–4 (pUMa867) correspond to E. coli transformants that were generated during quality control of the library. All plasmid constructs were verified by DNA sequencing.
Protein expression in E. coli
Recombinant Rrm4GST, BrunoGST, and GST were expressed as follows. BL21(DE3)ΔpLys cells harboring plasmids pRrm4GST-pGEX, pBrunoGST-pGEX, or pGEX-2T were grown in dYT medium containing ampicillin (100 μg/mL) and chloramphenicol (25 μg/mL) at 37°C. At an OD600 of 0.8, cells were induced with 0.2 mM IPTG for 16 h at 22°C; 600 mL cell suspension were harvested, washed, and resuspended in 20 mL buffer A (50 mM Tris/HCl pH 8.0, 150 mM NaCl, 5 mM DTT, protease inhibitor cocktail Complete, Roche). Cells were sonicated and insoluble cell debris was removed by centrifugation (4°C, 45 min, 50,000g). The supernatant was incubated with 3 mL of GSH-sepharose beads (GE healthcare) for 5 h at 4°C. Beads were washed on the column (five times) with buffer A. Elution of GST fusion proteins was performed using 20 mM reduced glutathione, which was subsequently removed by dialysis against buffer A for 20 h at 4°C. Finally, the protein concentration was determined (Bradford 1976).
In vitro selection (SELEX)
The single-stranded DNA starting library (5′-GAATTCAGTCGGACAGCG(N)40GATGG-ACGAATATCGTCTCCC-3′) was synthesized and amplified as described (Homann and Göringer 1999). It contained a central sequence stretch of 40 randomized nucleotides flanked by constant regions C1 and C2. In the initial cycle, 100 μg of the library were transcribed in transcription buffer (20 mM NaxHyPO4 pH 7.9, 8 mM MgCl2, 20 mM DTT, 4 mM spermidine) using 500 U T7 RNA polymerase (Stratagene), 160 U RNasin (Promega), and 1 mM NTPs for 1.5 h at 37°C in a volume of 1.2 mL. In subsequent cycles, 20–50 μg PCR product were used as template for in vitro transcription. The reaction was terminated upon addition of MgCl2 (final concentration of 50 mM) and DNase I (1 h 37°C; 1 U DNase I/μg DNA, Roche). RNAs were extracted with phenol, precipitated with ethanol and redissolved in binding buffer (25 mM Tris/HCl pH 7.5, 5 mM MgCl2, 1 mM DTT, 100 mM NaCl, 4 mM KCl, 5 mM glucose). A final purification was achieved by size exclusion chromatography in the same buffer. Before each cycle of selection, all RNAs were pre-incubated at 37°C for 30 min. For the first cycle, 250 μg RNA library were incubated with Rrm4GST (750 nM) in 1 mL binding buffer for 30 min at RT. In later cycles, the selection stringency was increased stepwise by decreasing the amount of Rrm4GST (from 750 nM in cycle 1 to 4nM in cycles 11–12), decreasing the amount of RNA (from 250 μg in cycle 1 to 12.5 μg in cycles 9–12), and increasing the reaction volume from 1 to 8 mL (cycles 11–12). Nonspecific competitor RNA (total RNA from U. maydis) was added in cycle 4 (10 μg/μg pool RNA) and in cycles 9–12 (100 μg/μg pool RNA). RNA sequences with affinity for the filter material or the GST moiety of Rrm4GST were depleted from the pool by counterselection in cycles 3–4 and 7–12. This was achieved by passing the in vitro transcribed RNA through a filter prior to binding and by pre-incubation with GST using an RNA/GST ratio of 1:10.
Bound RNAs were partitioned from unbound material by filter adsorption to nitrocellulose/cellulose acetate mixed bed filters. Samples were filtered using a mild vacuum and filters were washed with binding buffer (at least fivefold excess to the initial binding volume). Selected RNAs were recovered by phenol extraction followed by ethanol precipitation. RNA was reverse-transcribed using 150 U MuLV-RT (USB) and 10 μg primer oMF731 5′-GGTCGAGAATTCAGTCGGACAGCG-3′ for 45 min at 42°C followed by 5 min at 48°C. The resulting DNA products were amplified by PCR with oligonucleotides oMF731 and oMF730 5′-GCGAAGCTTTAATACGACTCACTATAGGGAGACGATATTCGTCCATC-3′. The resulting DNA was used as a template for the next cycle. DNA from cycle 11 was inserted into pCR2.1-TOPO and inserts were sequenced by dideoxy terminator sequencing.
Classification of aptamer sequences and structure determination
The aptamer sequences were aligned using Clone Manager Professional Suite version 8 using the Multi-Way alignment type. The identification of sequence motifs and the final alignment were performed by visual inspection and with the help of secondary structure calculations using Mfold (Zuker 2003).
Determination of equilibrium dissociation constants
The binding affinity of RNA aptamers to recombinant Rrm4GST was measured by filter adsorption. Uniformly (32P)-labeled RNA aptamers were incubated with Rrm4 (15 nM) for 20 min at RT in 50 μL binding buffer (25 mM Tris/HCl pH 7.5, 5 mM MgCl2, 2 mM DTT, 100 mM NaCl, 5 mM KCl, 100 ng/μL BSA, 70 ng/μL tRNAyeast). Samples were filtered through nitrocellulose/cellulose acetate mixed bed filters and washed with 2 mL binding buffer. The percentages of bound RNA were determined by scintillation counting. Equilibrium dissociation constants (Kd) were derived from experiments determining the bound percentage of labeled aptamer (0.1 nM) at increasing concentrations of unlabeled RNA (0.6–300 nM) in the presence of a constant Rrm4GST concentration (15 nM). Kd values were calculated based on the Scatchard equation: r/[A] = n/Kd−r/Kd, with the slope of the plot representing −1/Kd ([A] = [free RNA], r = [bound RNA], and n = number of binding sites).
Yeast three-hybrid analysis
Construction of the yeast three-hybrid RNA library was done as follows. cDNA of the 11th SELEX cycle was PCR-amplified using primer oMF767 5′-AACATCGATGAGACGATATTCGTCCATC-3′ and oMF768 5′-TATGGCGCGCCAGAATTCAGTCGGACAGCG-3′ introducing ClaI and AscI restriction endonuclease sites 5′ and 3′ to the constant regions C1 and C2, respectively. PCR products were cleaved (ClaI/AscI) and inserted into the hybrid RNA encoding vector pIIIMS2–4. About 4 × 104 transformants were pooled and plasmid DNA was isolated. For quality control, 1 μg of the library DNA was transformed into E. coli and 39 randomly chosen clones were sequenced after whole-cell PCR using standard procedures.
Subsequently, 10 μg of library DNA were transformed into strain L40-coat (Gietz and Woods 2002) that contained plasmid pRrm4AD-G-ACTIIA, and transformants were incubated on synthetic complete plates lacking leucine and uracil (SC −leu–ura; 2 d at 28°C) to select for the presence of both plasmids. About 105 transformants were pooled and incubated on SC plates lacking histidine and containing 5 mM 3-aminotriazole (SC −his+5 mM 3-AT; an inhibitor of His3p). One-hundred-four randomly chosen clones were verified to express both reporter genes according to growth on plates (SC −his+5 mM 3-AT) and β-galactosidase activity, which was determined using standard filter assays with X-gal as the chromogenic substrate (Bernstein et al. 2002). Forty-one randomly chosen clones were subjected to whole-cell PCR analysis with oligonucleotide primers oMF713 5′-AAATTGGAAGTTGGATATGG-3′ and oMF725 5′-CATGGTACGCTGTGGTGC-3′ using standard procedures. The resulting PCR products were sequenced.
To test representative aptamers of each class individually, RNA (pA1-IIIMS2–4, pB1-IIIMS2–4, pC1-IIIMS2–4, pD1-IIIMS2–4, and pIRE-MS2–4) and protein-encoding plasmids (pRrm4AD-G-ACTIIA and pIRP1AD-G-ACTIIA) were cotransformed in strain L40-coat and incubated on SC −ura–leu plates (2 d at 28°C). At least three transformants were assayed on plates (SC −his+5 mM 3-AT; 3 d at 28°C).
To test different mutant versions of Rrm4 for RNA binding in vivo, RNA (pA1-IIIMS2–4, pB1-IIIMS2–4, and pIRE-MS2–4) and protein-encoding plasmids (pRrm4AD-G-ACTIIA, pRrm4-mR1AD-G-ACTIIA, pRrm4-mR2AD-G-ACTIIA, and pRrm4-mR3AD-G-ACTIIA) were cotransformed in strain L40-coat and assayed for reporter gene expression as mentioned.
Fluorimetric measurements of β-galactosidase activity
A densely grown culture of cells (SC −ura–leu, 28°C, shaking with 200 rpm) was diluted (optical density, OD600 of ∼0.2). After ∼4 h of incubation (OD600 of ∼0.6–0.8) ∼2 × 108 cells (107 cells/mL represent OD600 = 1) were harvested by centrifugation (room temperature, RT, 1000g, 5 min). The pellet was washed with 1 mL chilled reaction buffer (0.1 M NaxHyPO4 pH 7.3; 1 mM MgCl2) and resuspended in 150 μL chilled reaction buffer containing 45 mM β-mercaptoethanol. Upon adding of 0.3 g acid-washed glassbeads, cells were rigorously shaken (4°C, 1400 rpm, 5 min). After two rounds of centrifugation (4°C, 16,000g, 5 and 20 min each), the protein concentration of the supernatant was determined (Bradford 1976). Fifty nanograms of total protein extract were assayed in the presence of 100 μL CUG working solution (3-carboxyumbelliferyl β-D-galactopyranoside, FluoReporter quantification kit; Invitrogen) using a monochromator fluorescence reader (Tecan). Relative fluorescence units were determined at 25°C with excitation and emission wavelengths of 385 and 445 nm, respectively (bandwidth of 12.5 nm each). Measurements were performed in 1-min intervals (in total 30 min). Relative β-galactosidase activity was expressed as RFUs per minute. Negative values due to very low enzyme activity were considered to be zero. Two independent experiments were performed and representative results are shown.
To control expression of mutant Rrm4AD-G variants, ∼3 × 105 cells were harvested by centrifugation (RT, 1000g, 5 min) and resuspended in 200 μL H2O. Gfp fluorescence was measured with excitation and emission wavelengths of 485 nm and 520 nm, respectively (bandwidth of 7.5 nm each). The resulting RFUs were normalized to OD600. To account for expression differences of Rrm4AD-G variants, we normalized the relative β-galactosidase activity to the Gfp fluorescence. Two independent experiments were performed and representative results are shown.
ACKNOWLEDGMENTS
We thank Dr. A. Brachmann and the laboratory members for valuable discussions and critically reading of the manuscript. We thank Janine Koepke for initial work on Rrm4 mutants in yeast and Julia Hohenner for excellent technical assistance. We are grateful to Drs. M. Wickens and P. MacDonald for the generous gift of the three-hybrid system and a Bruno expression plasmid, respectively. This work was supported by funds from Bayer CropScience and DFG Fe448/3 to M.F. as well as by an International Research Scholar grant from the Howard Hughes Medical Institute (HHMI) and DFG SFB579 to H.U.G.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.334307.
REFERENCES
- Becht, P., Vollmeister, E., Feldbrügge, M. A role for RNA-binding proteins implicated in pathogenic development of Ustilago maydis . Eukaryot. Cell. 2005;4:121–133. doi: 10.1128/EC.4.1.121-133.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becht, P., König, J., Feldbrügge, M. The RNA-binding protein Rrm4 is essential for polarity in Ustilago maydis and shuttles along microtubules. J. Cell Sci. 2006;119:4964–4973. doi: 10.1242/jcs.03287. [DOI] [PubMed] [Google Scholar]
- Berezovski, M., Musheev, M., Drabovich, A., Krylov, S.N. Non-SELEX selection of aptamers. J. Am. Chem. Soc. 2006;128:1410–1411. doi: 10.1021/ja056943j. [DOI] [PubMed] [Google Scholar]
- Bernstein, D.S., Buter, N., Stumpf, C., Wickens, M. Analyzing mRNA-protein complexes using a yeast three-hybrid system. Methods. 2002;26:123–141. doi: 10.1016/S1046-2023(02)00015-4. [DOI] [PubMed] [Google Scholar]
- Bernstein, D., Hook, B., Hajarnavis, A., Opperman, L., Wickens, M. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA. 2005;11:447–458. doi: 10.1261/rna.7255805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cassiday, L.A., Maher, L.J. In vivo recognition of an RNA aptamer by its transcription factor target. Biochemistry. 2001;40:2433–2438. doi: 10.1021/bi002376v. [DOI] [PubMed] [Google Scholar]
- Cassiday, L.A., Maher, L.J. Yeast genetic selections to optimize RNA decoys for transcription factor NF-κB. Proc. Natl. Acad. Sci. 2003;100:3930–3935. doi: 10.1073/pnas.0736013100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J.C., Hayhurst, A., Hesselberth, J., Bayer, T.S., Georgiou, G., Ellington, A.D. Automated selection of aptamers against protein targets translated in vitro: From gene to aptamer. Nucleic Acids Res. 2002;30:e108. doi: 10.1093/nar/gnf107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington, A.D., Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Eulberg, D., Buchner, K., Maasch, C., Klussmann, S. Development of an automated in vitro selection protocol to obtain RNA-based aptamers: Identification of a biostable substance P antagonist. Nucleic Acids Res. 2005;33:e45. doi: 10.1093/nar/gni044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz, R.D., Woods, R.A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Göringer, H.U., Homann, M., Lorger, M. In vitro selection of high-affinity nucleic acid ligands to parasite target molecules. Int. J. Parasitol. 2003;33:1309–1317. doi: 10.1016/s0020-7519(03)00197-8. [DOI] [PubMed] [Google Scholar]
- Göringer, H.U., Homann, M., Zacharias, M., Adler, A. RNA aptamers as potential pharmaceuticals against infections with African trypanosomes. Handb. Exp. Pharmacol. 2006;173:375–393. doi: 10.1007/3-540-27262-3_19. [DOI] [PubMed] [Google Scholar]
- Homann, M., Göringer, H.U. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999;27:2006–2014. doi: 10.1093/nar/27.9.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homann, M., Lorger, M., Engstler, M., Zacharias, M., Göringer, H.U. Serum-stable RNA aptamers to an invariant surface domain of live African trypanosomes. Comb. Chem. High Throughput Screen. 2006;9:491–499. doi: 10.2174/138620706777935324. [DOI] [PubMed] [Google Scholar]
- Hook, B., Bernstein, D., Zhang, B., Wickens, M. RNA–protein interactions in the yeast three-hybrid system: Affinity, sensitivity, and enhanced library screening. RNA. 2005;11:227–233. doi: 10.1261/rna.7202705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, Y., Bogerd, H.P., Yang, J., Cullen, B.R. Analysis of the RNA binding specificity of the human Tap protein, a constitutive transport element-specific nuclear RNA export factor. Virology. 1999;262:200–209. doi: 10.1006/viro.1999.9906. [DOI] [PubMed] [Google Scholar]
- Kim-Ha, J., Kerr, K., MacDonald, P.M. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell. 1995;81:403–412. doi: 10.1016/0092-8674(95)90393-3. [DOI] [PubMed] [Google Scholar]
- Lee, J.F., Hesselberth, J.R., Meyers, L.A., Ellington, A.D. Aptamer database. Nucleic Acids Res. 2004;32:D95–D100. doi: 10.1093/nar/gkh094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.J., Herskowitz, I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Murphy, M.B., Fuller, S.T., Richardson, P.M., Doyle, S.A. An improved method for the in vitro evolution of aptamers and applications in protein detection and purification. Nucleic Acids Res. 2003;31:e110. doi: 10.1093/nar/gng110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimjee, S.M., Rusconi, C.P., Sullenger, B.A. Aptamers: An emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Proske, D., Blank, M., Buhmann, R., Resch, A. Aptamers: Basic research, drug development, and clinical applications. Appl. Microbiol. Biotechnol. 2005;69:367–374. doi: 10.1007/s00253-005-0193-5. [DOI] [PubMed] [Google Scholar]
- SenGupta, D.J., Zhang, B., Kraemer, B., Pochart, P., Fields, S., Wickens, M. A three-hybrid system to detect RNA–protein interactions in vivo. Proc. Natl. Acad. Sci. 1996;93:8496–8501. doi: 10.1073/pnas.93.16.8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta, D.J., Wickens, M., Fields, S. Identification of RNAs that bind to a specific protein using the yeast three-hybrid system. RNA. 1999;5:596–601. doi: 10.1017/s1355838299002113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soller, M., White, K. ELAV. Curr. Biol. 2004;14:R53. doi: 10.1016/j.cub.2003.12.041. [DOI] [PubMed] [Google Scholar]
- Stelzl, U., Worm, U., Lalowski, M., Haenig, C., Brembeck, F.H., Goehler, H., Stroedicke, M., Zenkner, M., Schoenherr, A., Koeppen, S., et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Sullenger, B.A., Gilboa, E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Ule, J., Jensen, K., Mele, A., Darnell, R.B. CLIP: A method for identifying protein–RNA interaction sites in living cells. Methods. 2005;37:376–386. doi: 10.1016/j.ymeth.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Ulrich, H. RNA aptamers: From basic science toward therapy. Handb. Exp. Pharmacol. 2006;173:305–326. doi: 10.1007/3-540-27262-3_15. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]