Abstract
Programmed translational frameshift sites are sequences in mRNAs that promote frequent stochastic changes in translational reading frame allowing expression of alternative forms of protein products. The EST3 gene of Saccharomyces cerevisiae, encoding a subunit of telomerase, uses a programmed +1 frameshift site in its expression. We show that the site is complex, consisting of a heptameric sequence at which the frameshift occurs and a downstream 27-nucleotide stimulator sequence that increases frameshifting eightfold. The stimulator appears to be modular, composed of at least three separable domains. It increases frameshifting only when ribosomes pause at the frameshift site because of a limiting supply of a cognate aminoacyl-tRNA and not when pausing occurs at a nonsense codon. These data suggest that the EST3 stimulator may modulate access by aminoacyl-tRNAs to the ribosomal A site by interacting with several targets in a ribosome paused during elongation.
Keywords: context effect, recoding, translational accuracy, translational frameshifting
INTRODUCTION
The ribosome is a large macromolecular complex that orchestrates the process of protein synthesis from an mRNA template. The extension of a growing polypeptide chain occurs by sequential recruitment of aminoacyl-tRNAs (aa-tRNAs) to a binding site on the ribosome, the A site. Following this, the growing peptide chain is transferred to the aminoacyl-tRNA from a peptidyl-tRNA bound to the P site. The aa-tRNA binds in a ternary complex with elongation factor 1A and GTP (aa-tRNA•EF-1A•GTP). The translational machinery has evolved multiple mechanisms to reduce the frequency of errors during protein synthesis. The ribosome discriminates between correct (cognate) and incorrect (near or noncognate) aa-tRNAs by a process that involves both kinetic proofreading (Hopfield 1974; Ninio 1975) and discrimination based on induced fit (for review, see Rodnina et al. 2005). During kinetic proofreading, the more rapid dissociation of incorrect complexes during two successive selection steps operating before and after GTP hydrolysis by EF-1A select for cognate aa-tRNAs (for review, see Thompson 1988). More recent work showed that the rapid progression through the steps of aa-tRNA selection precludes maximal discrimination based on differences in aa-tRNA affinity for the ribosome (Gromadski and Rodnina 2004). The dominant effect on accuracy appears not to result from the kinetic effects of these affinity differences but rather on the ability of cognate aa-tRNAs to induce a change in ribosome structure that accelerates their acceptance by significantly increasing the rate of GTP hydrolysis (for review, see Rodnina et al. 2005).
Despite these mechanisms, translation is not completely accurate. Translation has evolved to maximize the conflicting demands of accuracy and translational output. For example, the intrinsic rate of GTP hydrolysis must be extremely fast so the ribosome can achieve an elongation rate of 10 per second while still allowing correct tRNAs to out compete the vast excess of incorrect tRNAs (Gromadski and Rodnina 2004). Rapid activation of the GTPase keeps the reaction far from equilibrium so that differences in stability have a small effect on accuracy. Thus, increasing protein output by maximizing GTP activation has the cost of failing to maximize accuracy (Gromadski and Rodnina 2004).
The error rate is also affected by mRNA sequence context (for review, see Parker 1989). The mechanism underlying these context effects is not understood, but could result from differences in the interaction between the mRNA and the ribosome's accuracy center. An extreme form of context effect is the existence of mRNA sequences, termed “recoding sites” (Gesteland et al. 1992). These sites cause translational errors at rates approaching as much as 0.5 errors per codon (Farabaugh 1996; Namy et al. 2004), several orders of magnitude more than the background rate, ∼5×10−4 per codon (Parker 1989). Recoding encompasses a variety of phenomena including programmed readthrough of nonsense codons, programmed frameshifting, or programmed bypassing of several codons (Farabaugh 1996; Namy et al. 2004). Programmed frameshifting can occur in either direction; typically, frameshifting involves shifts in the 5′ direction with respect to the mRNA (e.g., programmed −1 frameshifting) or the 3′ direction (e.g., programmed +1 frameshifting).
Programmed +1 frameshifting in Saccharomyces cerevisiae occurs at special heptameric sites composed of codons that occupy the ribosomal P and A sites during the frameshift. Frameshifting requires that a near-cognate tRNA occupy the P-site codon during the shift (Sundararajan et al. 1999). The near-cognate forms a weak wobble pair involving either a pyrimidine•pyrimidine or a purine•purine pair. We have hypothesized that the unusual pairing in the P site by disturbing the suite of interactions between the P-site codon•anticodon complex and the P site, may interfere with acceptance of even perfect cognate tRNAs in the A site (Stahl et al. 2002). This interference seems to eliminate the nearly absolute preference for in-frame cognate tRNAs, allowing the occasional acceptance of a +1 frame cognate tRNA to promote frameshifting (Stahl et al. 2002). The ability of the P-site tRNA to slip +1 on the mRNA can further increase the efficiency of frameshifting, for example, by tRNALeu UAG slipping from CUU to UUA (Belcourt and Farabaugh 1990; Sundararajan et al. 1999). Frameshifting can also occur without slippage, as when tRNAAla IGC binds to a P-site GCG codon (Farabaugh et al. 1993; Sundararajan et al. 1999). A second requirement for maximal frameshifting is slow recognition of the next in-frame codon, either because slow recognition of a sense codon by its cognate tRNA (a sense pause codon) or of a termination codon by peptide release factor (a nonsense pause codon).
The sequences surrounding a recoding site stimulate translational errors by one of a variety of mechanisms. Some sequences locally reduce the rate of canonical translation; ribosomes commonly pause at the site of recoding with the pause presumed to provide sufficient time for a slow noncanonical event (frameshift, readthrough, or bypass) to occur. The pause-inducing sequences can comprise poorly recognized codons or secondary structures, such as hairpin loops or pseudoknots. Other sequences stimulate the noncanonical event. For example, programmed frameshift sites in bacteria include a Shine–Dalgarno (SD) site immediately upstream of the shift site (Larsen et al. 1995). The sequence appears to base pair with the anti-Shine–Dalgarno site near the 3′ end of the 16S rRNA. The distance between the SD and shift sites is such that it strains the ribosome effectively pulling the ribosome into the shifted frame (Weiss et al. 1988; Larsen et al. 1994).
Other context effects remain unexplained. For example, a 14-nucleotide (nt) sequence downstream of the retrotransposon Ty3 frameshift site in the yeast S. cerevisiae, termed the Ty3 stimulator, increases programmed +1 frameshifting by as much as 7.5-fold (Farabaugh et al. 1993). It functions as a primary mRNA sequence, suggesting that it might base pair (bp) with a region of the rRNA (Li et al. 2001). Our recent data are inconsistent with a model proposing base pairing between the Ty3 stimulator and the loop region of Helix 18 (unpublished observations).
We report here our analysis of another frameshift stimulator derived from the EST3 gene, which encodes a protein subunit of telomerase (Lingner et al. 1997; Hughes et al. 2000). The EST3 gene comprises a 5′ reading frame of 280 bp and a 3′ reading frame of 270 bp. We demonstrated that a heptameric sequence from the overlap between the open reading frames (ORFs), CUU-AGU-U, promotes +1 frameshifting at a rate of 8% by reading the CUU and GUU codons as Leu-Val, skipping the central A in the sequence (Vimaladithan and Farabaugh 1994). Morris and Lundblad (1997) demonstrated that mutations of this heptameric sequence predicted to block Est3 protein expression were lethal. Surprisingly, frameshifting is not essential since a mutation putting the EST3 gene into a single frame had no effect on growth (Morris and Lundblad 1997). Morris and Lundblad (1997) estimated the efficiency of the EST3 frameshift as high as 75%–90% by measuring ratio of the amount of the full-length frameshift product relative to the amount of a truncated form produced by termination at the UGA codon at the end of the 5′ ORF. Because there is no reason to suppose that the truncated product is stable, this estimate probably overestimates the frequency of frameshifting. However, the frequency appears to be far greater than we had previously measured, suggesting that the context surrounding the frameshift site might include a strong frameshift stimulator. We present an analysis that identifies a context region of at least 24 nt downstream of the shift site that stimulates frameshifting almost 10-fold. Like the Ty3 stimulator (Li et al. 2001), the EST3 stimulator effect requires a pause-inducing sense codon and has little effect on frameshifts dependent on termination codons. Saturation mutagenesis of the 30 nt downstream of the EST3 frameshift site shows the stimulator is a large and complex site, suggesting that its presumed ribosomal target(s) may be similarly large and complex.
RESULTS
Sequence requirements for frameshifting in EST3
The estimate of frameshift efficiency in EST3 by Morris and Lundblad (1997) may be inaccurate because it depends on quantifying the amount of a possibly highly unstable truncated protein. To obtain a more accurate estimate of EST3 frameshifting frequency we constructed a set of reporter plasmids by inserting the entire EST3 reading frame into a frameshift reporter plasmid (see Fig. 1). The plasmid, pANU-7 (Sundararajan et al. 1999), is a bacterial-yeast shuttle plasmid encoding a translational fusion of the S. cerevisiae HIS4 gene to the Escherichia coli lacZ gene. A short polylinker between the two genes allows the insertion of the fragment of interest. The reporter fuses the upstream HIS4 gene to the 5′ EST3 ORF and the 3′ ORF to the downstream lacZ gene. Expression of the lacZ product, β-galactosidase, requires +1 frameshifting within the EST3 insert. To determine the efficiency of frameshifting we compare the expression of this plasmid to a second plasmid in which changing the frameshift heptamer CTT-AGT-T to TTA-GTT fuses the two EST3 ORFs to encode Leu-Val without frameshifting. The frameshift efficiency is the ratio of expression of the frameshift to the frame fusion reporter, given as a percentage.
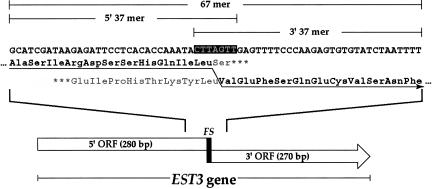
FIGURE 1.
Structure of the EST3 gene and frameshift region. The diagram at the bottom shows the arrangement of the two open reading frames and the frameshift site (FS) within the overlap region. Above is the DNA sequence of the heptameric frameshift site (white letters on black) and the 30 nt both upstream and downstream. Below the DNA sequence is the encoded protein product in the zero (above) and +1 (below) reading frames with the amino acids encoded into the frameshift product in black and the arrow indicating decoding by frameshifting. The extent of three oligonucleotides referred to in Figure 2 (EST3 67 mer, 5′ 37 mer, and 3′ 37 mer are indicated above the DNA sequence.
The apparent frequency of frameshifting in EST3 based on this analysis is 47%, much less than previously estimated but much more than the frequency of frameshifting on the heptameric shift site outside the EST3 context (Fig. 2). The EST3 insert is 543 bp, and it was possible that the insert might have introduced a cryptic promoter and translation initiation site, allowing expression of lacZ without frameshifting. To eliminate this possibility, we introduced a nonsense mutation in the +1 frame at codon 97 (Q97*) immediately downstream of the frameshift site but upstream of either AUG in the second frame at which this putative cryptic initiation could occur (codons 120 and 136, which are the only AUGs downstream of the frameshift site). The first Met encoded in the lacZ region is downstream of a region required for β-galactosidase activity (Cigan et al. 1988). As shown in Figure 2, this nonsense mutation virtually eliminates expression of lacZ. This result disproves the cryptic initiation model, showing that ribosomes that translate lacZ initiate upstream of the frameshift site.
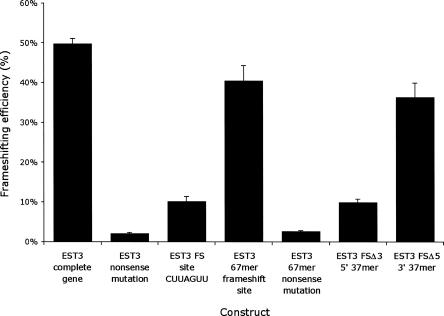
FIGURE 2.
Identifying the EST3 frameshift stimulator. Shown is the frameshift efficiency in percent (±SEM expressed as a percent) for the indicated constructs derived from the EST3 gene, described in the text.
The fact that frameshifting in the EST3 context is so much more efficient suggests that the gene includes one or more frameshift stimulating sequences. With a few exceptions, most context effects involve sequences very near the frameshift site. Since a ribosome binds to an mRNA region of about 30 nt (Steitz 1969), we tested whether a region including 30 nt upstream and downstream of the frameshift site would support a similar frequency of frameshifting. As shown in Figure 2, the ability of the 67mer region to promoted frameshifting was not significantly different from the effect of the entire EST3 gene. Although it is possible that there are more stimulatory elements in the gene, we chose to characterize this region since it appears to include most, if not all, of the stimulatory potential. To roughly map the position of stimulatory sites, we constructed two truncations of this 67mer, removing the 30 nt upstream (Δ5) or downstream (Δ3) of the frameshift site. The Δ5 deletion caused a very modest reduction in expression, but the Δ3 deletion eliminated the stimulatory effect entirely (Fig. 2), and inclusion of the Q97* mutation virtually eliminated expression. We concluded that the stimulatory site resides downstream of the frameshift signal.
Evidence for a modular structure of the EST3 stimulator
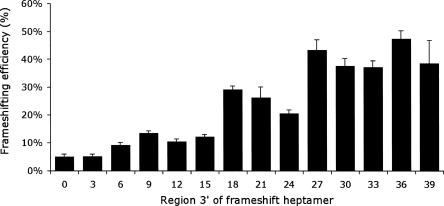
As a first step in characterizing the EST3 stimulator we constructed a set of 1 codon 3′ deletions of the 30-nt region downstream of the frameshift site (see Materials and Methods). Stimulator activity declined with increasing deletions in three discrete steps. Frameshift efficiency dropped from about 40% to 20% with deletion of the region from 27 nt to 24 nt downstream, to about 10% with deletion from 18 nt to 15 nt, and finally to about 5% with deletion from 6 nt to 3 nt (Fig. 3). To eliminate the idea that we had unwittingly destroyed a site located >27 nt downstream we made a series of constructs increasing the region to 33 nt, 36 nt, or 39 nt. Adding these nucleotides caused no further increase in frameshifting, suggesting that the stimulator is located entirely within the first 27 nucleotides 3′ of the frameshift heptamer.
FIGURE 3.
Deletion mapping of the EST3 stimulator. Each bar represents the frameshifting efficiency, as in Figure 2, for one of a series of reporters that includes increasing amounts of the region downstream of the EST3 frameshift site.
The discrete nature of the reduction of activity, with activity dropping in three well-defined steps, suggests that the EST3 stimulator might be modular. The data are consistent with there being three components to the region, each responsible for increasing frameshifting about twofold, resulting in a total effect of about eightfold.
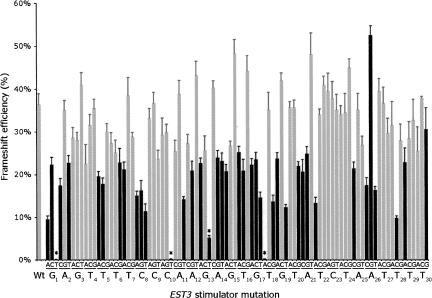
To provide a sense of the fine structure of the site we performed saturation point mutagenesis of the site, making the three possible changes at each position in the 30-nt region. We expected to find that many mutations had little effect on stimulator activity, and that some had a large effect, identifying nucleotides that made important contacts with a target in the ribosome. As shown in Figure 4, what we found is that over half of mutations cause an almost twofold decrease in stimulator activity, with frameshift efficiency falling from 40% to around 20%. Among these, a small number of mutations reduce frameshifting to around 10% (G1A, C8U, G17U, U18G, G19U, A21C, A26U, and U28C). Only one mutation (A26G) caused a significant increase in frameshifting to over 50%, although that reflected only a 25% increase in activity above wild type.
FIGURE 4.
Missense mutagenesis of the EST3 stimulator. Each bar represents β-galactosidase activity (±SEM) expressed from a reporter involving a wild-type (Wt) or missense mutant EST3 stimulator. Each wild-type nucleotide appears below the graph with a subscript indicating the position; the identity of each tested mutation appears just above in groups of 3. Black bars represent results judged by T-test as different from wild type (P ≤ 0.005). Asterisks indicate mutations that introduce an in frame nonsense codon, which strongly reduces expression of lacZ.
Point mutations causing significant decreases in frameshift efficiency are scattered throughout the 30 nt region and show little evidence of a modular structure to the stimulator. There is only one short region in which no mutations had a significant effect: C23-U24. The placement of these nucleotides does not correspond to an apparent division between modules in the 5′ deletion series but rather falls in the middle of the deletions that reduce stimulation twofold. Experiments are in progress to test if there are additive effects of mutations only when they fall in separate putative modules.
The spacing between the frameshift site and stimulator is critical
Sequences outside the site of the frameshift itself, like the EST3 stimulator, increase the efficiency of frameshifting in many characterized frameshift sites. Although their mechanism of action varies, all of these stimulatory sites share a requirement for strict spacing from the frameshift site. For example, the efficiency of most eukaryotic −1 simultaneous slippage frameshifts depends on the presence of a downstream mRNA pseudoknot; the distance between the −1 frameshift site and the pseudoknot is always very close to 6 nt (ten Dam et al. 1990; Farabaugh 1996). The distance is critical to frameshift stimulation since changing spacing by 1 nt reduces frameshift efficiency twofold, and larger changes have more severe effects (Brierley et al. 1989, 1992). Changing the spacing between the Ty3 frameshift site and its downstream stimulator had a more severe effect; inserting 1 nt abolished the stimulator effect (Li et al. 2001). We hypothesized that changing the spacing to the EST3 stimulator would have a similar effect on frameshift efficiency.
To test the effect of increased spacing, we inserted one codon between the frameshift site and the first nucleotide of the stimulator. This insertion reduced the frameshift efficiency from the wild-type level (36 ± 2.1%) to a level similar to the construct lacking the stimulator (5.3 ± 0.3%), or a reduction of 6.8-fold. This effect demonstrates that the stimulator requires precise spacing with respect to the frameshift site. The strict spacing requirement implies that the stimulator has a target that is located in space a precise distance from the frameshift site, which occupies the ribosomal A and P sites during the frameshift. Based on the solved structure of the mRNA path on the bacterial ribosome (Yusupova et al. 2001), we predict that the region of the stimulator up to 9 nt downstream of the frameshift site should interact with elements of the ribosomal entrance tunnel; this region roughly corresponds to the first apparent module of the stimulator described above. The remaining 15–18 nt downstream would appear to reside outside the ribosome but could interact with a structure on the solvent face of the small ribosomal subunit.
The EST3 stimulator increases programmed frameshifting at a variety of sites
The EST3 frameshift site involves a P-site codon that allows peptidyl-tRNA slippage (CUU) and a sense pause codon (AGU). We wondered if the EST3 stimulator would show a preference for this type of frameshift site or if it would generically affect any +1 frameshift site. For example, would the EST3 stimulator affect only sites, like the EST3 site, that work by peptidyl-tRNA slippage, or would it also affect frameshifting at those like the Ty3 frameshift site that frameshift without slippage? Would it require pausing at a poorly decoded sense codon, as in EST3, or would it affect frameshifting at poorly recognized termination codons?
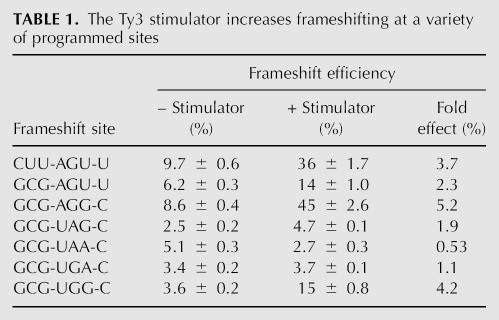
We replaced the P-site codon derived from the EST3 site (CUU) with the P-site codon of the nonslippage Ty3 site (GCG) by targeted mutagenesis (see Materials and Methods). A comparison of frameshifting on the normal EST3 site, CUU-AGU-U, with that on GCG-AGU-U shows that the stimulator significantly increases frameshifting on each site (Table 1). On the other hand, the stimulator showed a strong preference for sites involving sense pause codons. Sites using the pause codons AGU, AGG, or UGG showed increases from 2.3- to 5.2-fold, with a mean of 3.9-fold, whereas the stimulator changed frameshifting at three sites using UAA, UAG, or UGA as pause codons from 0.6- to 1.9-fold, with a mean of 1.2-fold. The Ty3 stimulator was also unable to increase frameshifting at nonsense pause codons (Li et al. 2001), implying that the two stimulators might use a shared mechanism.
TABLE 1.
The Ty3 stimulator increases frameshifting at a variety of programmed sites
DISCUSSION
The mRNA context surrounding programmed frameshift sites frequently includes sequence elements that stimulate frameshift efficiency. These stimulatory sequences can form a secondary structure that blocks the ribosome's forward progress, as do the pseudoknots that follow programmed −1 frameshift sites in eukaryotes (Brierley and Pennell 2001). Other sequences directly interact with a target in the rRNA to promote frameshifting, as do the Shine–Dalgarno sites found upstream of prokaryotic frameshift sites (Larsen et al. 1995). Here we identify a stimulator sequence downstream of the EST3 programmed +1 translational frameshift site.
The EST3 stimulator increases +1 frameshifting up to eightfold, including sites that function by either peptidyl-tRNA slippage or out-of-frame recruitment of aa-tRNA. The stimulator affects frameshifts dependent on pausing at sense codons but not those using termination codons. Recognition of termination codons appears to use a mechanism of codon recognition unrelated to aa-tRNA selection, although the detailed mechanism remains unclear (Bertram et al. 2000; Nakamura et al. 2000; Inagaki et al. 2002). Eukaryotic peptide release factor-1 (eRF1) appears to mimic the overall structure of tRNA (Song et al. 2000). That apparent mimicry does not extend to the detailed structure of the anticodon (Inagaki et al. 2002) so it is unlikely that the ribosome contributes to termination codon recognition in the way it governs tRNA recognition. Given that the EST3 stimulator can distinguish in-frame sense and nonsense codons, it seems likely that it functions at the step of aa-tRNA recruitment, presumably by interfering with cognate in-frame recognition, since that would cause more efficient frameshifting. It shares the ability to distinguish between sense and nonsense codons with a stimulator sequence from the Ty3 retrotransposon (Li et al. 2001), although the two sequences have no primary sequence similarity. Previously, we proposed that the Ty3 stimulator might function by base pairing with a conserved sequence of Helix 18 of 18S rRNA (Li et al. 2001). Data from a partial set of stimulator missense mutations were consistent with the model, but the results of a complete set of missense mutations are inconsistent with this model (C. Guarraia, L. Norris, A. Raman, and P. J. Farabaugh, unpubl.). For the EST3 stimulator, we have found no evidence for extensive complementarity with the rRNA (data not shown). In the absence of a specific model, it is difficult to comment on whether the EST3 stimulator mutagenesis data are inconsistent with mRNA•rRNA pairing. As a rough test, we would expect transition mutations (G↔A, U↔C) to tend to be more compatible with Watson–Crick pairing than transversions (purine↔pyrimidine); the correlation between type of mutation and phenotype is random (data not shown), which fails to support the model of Watson–Crick pairing. Given the result with the Ty3 stimulator we believe that a model not involving base pairing is more probable.
Yusupova et al. (2001) have mapped out the path of the mRNA on a bacterial ribosome. Cryoelectron microscopic imaging suggests that a central, highly conserved region of the ribosome is quite similar between prokaryotes and eukaryotes (Spahn et al. 2001), suggesting that we can predict how the mRNA would interact with the yeast ribosome using a bacterial structure, which shows that mRNA up to 9 nt downstream of the A site lie within the ribosomal entrance tunnel. This tunnel is made up of a layer of rRNA, covering from 1 nt to 4 nt downstream, and a layer consisting of the ribosomal proteins rpS3, rpS4, and rpS5, covering from 5 nt to 9 nt downstream. The fact that the EST3 stimulator is 27 nt in length suggests that most of it must reside outside the ribosome during frameshifting unless the frameshifting process drastically alters the pathway of the mRNA. The 3′ deletion series suggests that the EST3 stimulator may consist of three distinct domains: one upstream of position 6, one upstream of position 18, and one upstream of position 27. The first two of these are close enough to reside wholly or partially within the entrance tunnel. The third probably lies outside it. We have tested the possibility that the EST3 stimulator might function by forming a secondary structure. We have analyzed the potential for structure formation using the programs Mfold (Zuker 2003), which looks for hairpin loops, and pknotsRG (Reeder and Giegerich 2004), which looks for pseudoknots. We find no evidence that the 30-nt minimial sequence can form either structure (data not shown).
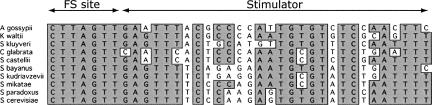
The fact that the EST3 gene evolved such a complex stimulator suggests that efficient frameshifting may be essential for its function. Our recent work shows that frameshifting evolved in EST3 >150 million years ago (mya), since the divergence of the genuses Kluyveromyces and Saccharomyces (Farabaugh et al. 2006). We found the same frameshift site in the EST3 gene from genome sequences of 10 budding yeast. The least-related homolog employing frameshift came from Ashbya gossypii (Dietrich et al. 2004). Ashbya's nearest relative not showing evidence of frameshifting was Kluyveromyces lactis, which diverged from the other budding yeasts about 150 mya (Langkjaer et al. 2003). The stimulator region has diverged during this period (Fig. 5; Farabaugh et al. 2006), so analysis of the pattern of sequence conservation could provide clues to identify essential sequence elements. As shown in Figure 4, two regions (the first 6 nt and the last 14 nt of the 27 nt minimal stimulator) are highly conserved, but the central region (7–13) is poorly conserved. The extent of conservation in the region is exceptional. There are 103 of 628 completely conserved residues in the entire EST3 gene, but 12 of 27 are conserved within the stimulator, threefold more than expected on a random basis. A comparison of the alignment with the 5′ deletion data show that the three putative domains roughly correspond to the regions of maximum sequence conservation: the first domain to the conserved region 1–6, and domains 2 and 3 roughly to the upstream and downstream halves of the second conserved region. The conservation also compares well with the missense mutagenesis. Of the 13 missense mutations that reduced the stimulator's effect more than twofold, only two alter an imperfectly conserved residue (U18 and A21).
FIGURE 5.
Alignment of budding yeast EST3 stimulators. The sequence of the frameshift sites and downstream stimulator sequences from 10 budding yeast species. Identity is indicated by shading.
We conclude that the EST3 gene has evolved over the last 150 million years to conserve the high level of frameshifting dependent on the downstream stimulator sequence. The similarities between the EST3 stimulator and the better-characterized Ty3 stimulator suggest that both sequences function by interfering with in-frame cognate recognition in the A site to block continued translation in the unshifted reading frame.
MATERIALS AND METHODS
Strains, DNA transformation, and β-galactosidase assays
The Escherichia coli strain used was DH5α (F− Φ80dlacZΔM15 Δ[lacZYA-argF]U169 deoR recA1 endA1 hsdR17[rk− mk+] phoA supE44 λ- thi-1 gyrA96 relA1) (Hanahan 1983). Bacterial transformation was performed using DH5α made chemically competent using the Z-competent protocol (Zymo Research). Transformants were grown on LB medium with 100 μg/mL ampicillin.
The Saccharomyces cerevisiae strain used was FY1679–18b, MATα ura3–52 leu2Δ1 trp1Δ63 his3Δ200 (Friel et al. 2003). Yeasts were transformed using the method LiAc method of Ito et al. (1983). Transformants were grown on Yeast Nitrogen Base (Difco) medium supplemented with 2% glucose and the amino acids histidine, leucine, and tryptophan (Burke et al. 2000). Activity of β-galactosidase in S. cerevisiae was assayed as previously described (Farabaugh et al. 1989).
Plasmid construction
The plasmid pANU7 (Sundararajan et al. 1999) encodes the first 33 codons of the S. cerevisiae HIS4 gene, which encodes three enzymes of histidine biosynthesis. The HIS4 region is fused through a short polylinker, which includes BamHI and KpnI restriction sites, with a reporter gene, the E. coli lacZ gene encoding β-galactosidase. The plasmid also contains replication origins for bacteria and yeast, as well as the bla gene, which confers ampicillin resistance in bacteria, and the S. cerevisiae URA3 gene, which complements the uracil prototrophy of the FY1679–18b yeast strain. The plasmid was subjected to QuikChange mutagenesis (Stratagene) to replace the BamHI restriction site with an NheI restriction site. The resulting plasmid is pDT254.
The EST3 coding region, lacking the termination codon, was isolated from S. cerevisiae genomic DNA using colony PCR (Burke et al. 2000). The primers were designed to add NheI and KpnI restriction sites at the 5′ and 3′ ends of the gene, respectively. The PCR product was then digested with NheI and KpnI and the fragment encompassing the EST3 coding region was ligated into a similarly digested pDT254. The resulting fragment fuses the upstream ORF of EST3 to the upstream HIS4 region and the downstream EST3 ORF to the downstream lacZ gene. The structure of the plasmid was confirmed by sequencing of the entire EST3 coding region.
Reporters were created in which segments of the EST3 gene encompassing various amounts of the frameshift region were inserted between the NheI and KpnI sites in pDT254. These reporters were created by ligating into NheI/KpnI-digested plasmid any of several double-stranded DNA fragments created by annealing complementary DNA oligonucleotides (purchased from Integrated DNA Technologies). In some cases, the oligonucleotides only partly overlapped, leaving large 5′ extensions. DNA polymerase was used to create fully double-stranded fragments that were then digested with NheI and KpnI before ligation into pDT254.
Constructs with and without context contain either the EST3 stimulator sequence (Fig. 1) or the sequence TCTAGGGCAAGAAGATCTAGGGCAAGAAGAA, respectively. This sequence comprises two copies of a mutant form of the Ty3 stimulator previously shown to eliminate any effect on +1 programmed frameshifting in yeast (Raman et al. 2006). Duplication of the mutant Ty3 stimulator created a nonframeshift stimulating mRNA sequence approximately the size of the EST3 stimulator.
ACKNOWLEDGMENTS
This work was supported by a grant to P.J.F. from the National Institutes of Health, R01 GM029480. D.T. was supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health, F31 GM073575.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.412707.
REFERENCES
- Belcourt, M.F., Farabaugh, P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: tRNAs induce slippage on a 7 nucleotide minimal site. Cell. 1990;62:339–352. doi: 10.1016/0092-8674(90)90371-K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram, G., Bell, H.A., Ritchie, D.W., Fullerton, G., Stansfield, I. Terminating eukaryote translation: Domain 1 of release factor eRF1 functions in stop codon recognition. RNA. 2000;6:1236–1247. doi: 10.1017/s1355838200000777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I., Pennell, S. Structure and function of the stimulatory RNAs involved in programmed eukaryotic-1 ribosomal frameshifting. Cold Spring Harb. Symp. Quant. Biol. 2001;66:233–248. doi: 10.1101/sqb.2001.66.233. [DOI] [PubMed] [Google Scholar]
- Brierley, I., Digard, P., Inglis, S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell. 1989;57:537–547. doi: 10.1016/0092-8674(89)90124-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley, I., Jenner, A.J., Inglis, S.C. Mutational analysis of the “slippery-sequence” component of a coronavirus ribosomal frameshifting signal. J. Mol. Biol. 1992;227:463–479. doi: 10.1016/0022-2836(92)90901-U. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke, D., Dawson, D., Steans, T. Methods in yeast genetics. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2000. [Google Scholar]
- Cigan, A.M., Pabich, E.K., Donahue, T.F. Mutational analysis of the HIS4 translational initiator region in Saccharomyces cerevisiae . Mol. Cell. Biol. 1988;8:2964–2975. doi: 10.1128/mcb.8.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, F.S., Voegeli, S., Brachat, S., Lerch, A., Gates, K., Steiner, S., Mohr, C., Pohlmann, R., Luedi, P., Choi, S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Farabaugh, P., Liao, X.-B., Belcourt, M., Zhao, H., Kapakos, J., Clare, J. Enhancer and silencerlike sites within the transcribed portion of a Ty2 transposable element of Saccharomyces cerevisiae . Mol. Cell. Biol. 1989;9:4824–4834. doi: 10.1128/mcb.9.11.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J. Programmed translational frameshifting. Microbiol. Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J., Zhao, H., Vimaladithan, A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell. 1993;74:93–103. doi: 10.1016/0092-8674(93)90297-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farabaugh, P.J., Kramer, E., Vallabhaneni, H., Raman, A. Evolution of +1 programmed frameshifting signals and frameshift-regulating tRNAs in the order Saccharomycetales. J. Mol. Evol. 2006;63:545–561. doi: 10.1007/s00239-005-0311-0. [DOI] [PubMed] [Google Scholar]
- Friel, D., Vandenbol, M., Haissam Jijakli, M. Cloning and sequence analysis of the TRP1 gene encoding the phosphoribosyl anthranilate isomerase from Pichia anomala (strain K) Yeast. 2003;20:1331–1337. doi: 10.1002/yea.1033. [DOI] [PubMed] [Google Scholar]
- Gesteland, R., Weiss, R., Atkins, J. Recoding: Reprogrammed genetic decoding. Science. 1992;257:1640–1641. doi: 10.1126/science.1529352. [DOI] [PubMed] [Google Scholar]
- Gromadski, K.B., Rodnina, M.V. Kinetic determinants of high-fidelity tRNA discrimination on the ribosome. Mol. Cell. 2004;13:191–200. doi: 10.1016/s1097-2765(04)00005-x. [DOI] [PubMed] [Google Scholar]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hopfield, J. Kinetic proofreading: A new mechanism for reducing errors in biosynthetic processes requiring high specificity. Proc. Natl. Acad. Sci. 1974;71:4135–4139. doi: 10.1073/pnas.71.10.4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, T.R., Evans, S.K., Weilbaecher, R.G., Lundblad, V. The Est3 protein is a subunit of yeast telomerase. Curr. Biol. 2000;10:809–812. doi: 10.1016/s0960-9822(00)00562-5. [DOI] [PubMed] [Google Scholar]
- Inagaki, Y., Blouin, C., Doolittle, W.F., Roger, A.J. Convergence and constraint in eukaryotic release factor 1 (eRF1) domain 1: The evolution of stop codon specificity. Nucleic Acids Res. 2002;30:532–544. doi: 10.1093/nar/30.2.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., Kimura, A. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langkjaer, R.B., Cliften, P.F., Johnston, M., Piskur, J. Yeast genome duplication was followed by asynchronous differentiation of duplicated genes. Nature. 2003;421:848–852. doi: 10.1038/nature01419. [DOI] [PubMed] [Google Scholar]
- Larsen, B., Peden, J., Matsufuji, S., Matsufuji, T., Brady, K., Maldonado, R., Wills, N.M., Fayet, O., Atkins, J.F., Gesteland, R.F. Upstream stimulators for recoding. Biochem. Cell Biol. 1995;73:1123–1129. doi: 10.1139/o95-121. [DOI] [PubMed] [Google Scholar]
- Larsen, B., Wills, N.M., Gesteland, R.F., Atkins, J.F. rRNA–mRNA base pairing stimulates a programmed −1 ribosomal frameshift. J. Bacteriol. 1994;176:6842–6851. doi: 10.1128/jb.176.22.6842-6851.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z., Stahl, G., Farabaugh, P.J. Programmed +1 frameshifting stimulated by complementarity between a downstream mRNA sequence and an error-correcting region of rRNA. RNA. 2001;7:275–284. doi: 10.1017/s135583820100190x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingner, J., Cech, T.R., Hughes, T.R., Lundblad, V. Three ever shorter telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc. Natl. Acad. Sci. 1997;94:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris, D.K., Lundblad, V. Programmed translational frameshifting in a gene required for yeast telomere replication. Curr. Biol. 1997;7:969–976. doi: 10.1016/s0960-9822(06)00416-7. [DOI] [PubMed] [Google Scholar]
- Nakamura, Y., Ito, K., Ehrenberg, M. Mimicry grasps reality in translation termination. Cell. 2000;101:349–352. doi: 10.1016/s0092-8674(00)80845-4. [DOI] [PubMed] [Google Scholar]
- Namy, O., Rousset, J.P., Napthine, S., Brierley, I. Reprogrammed genetic decoding in cellular gene expression. Mol. Cell. 2004;13:157–168. doi: 10.1016/s1097-2765(04)00031-0. [DOI] [PubMed] [Google Scholar]
- Ninio, J. Kinetic amplification of enzyme discrimination. Biochimie. 1975;57:587–595. doi: 10.1016/s0300-9084(75)80139-8. [DOI] [PubMed] [Google Scholar]
- Parker, J. Errors and alternatives in reading the universal genetic code. Microbiol. Rev. 1989;53:273–298. doi: 10.1128/mr.53.3.273-298.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman, A., Guarraia, C., Taliaferro, D., Stahl, G., Farabaugh, P.J. An mRNA sequence derived from a programmed frameshifting signal decreases codon discrimination during translation initiation. RNA. 2006;12:1154–1160. doi: 10.1261/rna.13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder, J., Giegerich, R. Design, implementation, and evaluation of a practical pseudoknot folding algorithm based on thermodynamics. BMC Bioinformatics. 2004;5:104. doi: 10.1186/1471-2105-5-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina, M.V., Gromadski, K.B., Kothe, U., Wieden, H.J. Recognition and selection of tRNA in translation. FEBS Lett. 2005;579:938–942. doi: 10.1016/j.febslet.2004.11.048. [DOI] [PubMed] [Google Scholar]
- Song, H., Mugnier, P., Das, A.K., Webb, H.M., Evans, D.R., Tuite, M.F., Hemmings, B.A., Barford, D. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- Spahn, C.M., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., Frank, J. Structure of the 80S ribosome from Saccharomyces cerevisiae–tRNA–ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Stahl, G., McCarty, G.P., Farabaugh, P.J. Ribosome structure: Revisiting the connection between translational accuracy and unconventional decoding. Trends Biochem. Sci. 2002;27:178–183. doi: 10.1016/S0968-0004(02)02064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steitz, J.A. Polypeptide chain initiation: Nucleotide sequences of the three ribosomal binding sites in bacteriophage R17 RNA. Nature. 1969;224:957–964. doi: 10.1038/224957a0. [DOI] [PubMed] [Google Scholar]
- Sundararajan, A., Michaud, W.A., Qian, Q., Stahl, G., Farabaugh, P.J. Near-cognate peptidyl-tRNAs promote +1 programmed translational frameshifting in yeast. Mol. Cell. 1999;4:1005–1015. doi: 10.1016/s1097-2765(00)80229-4. [DOI] [PubMed] [Google Scholar]
- ten Dam, E., Pleij, C., Bosch, L. RNA pseudoknots: Translational frameshifting and readthrough of viral RNAs. Virus Genes. 1990;4:121–136. doi: 10.1007/BF00678404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, R. EFTu provides an internal kinetic standard for translational accuracy. Trends Biochem. Sci. 1988;13:91–93. doi: 10.1016/0968-0004(88)90047-3. [DOI] [PubMed] [Google Scholar]
- Vimaladithan, A., Farabaugh, P.J. Special peptidyl–tRNA molecules promote translational frameshifting without slippage. Mol. Cell. Biol. 1994;14:8107–8116. doi: 10.1128/mcb.14.12.8107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss, R., Dunn, D., Dahlberg, A., Atkins, J., Gesteland, R. Reading frame switch caused by base-pair formation between the 3′ end of 16S rRNA and the mRNA during elongation of protein synthesis in Escherichia coli . EMBO J. 1988;7:1503–1507. doi: 10.1002/j.1460-2075.1988.tb02969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova, G.Z., Yusupov, M.M., Cate, J.H., Noller, H.F. The path of messenger RNA through the ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]