Abstract
A recently discovered class of gene regulatory RNAs, coined riboswitches, are commonly found in noncoding segments of bacterial and some eukaryotic mRNAs. Gene up- or down-regulation is triggered by binding of a small organic metabolite, which typically induces an RNA conformational change. Unique among these noncoding RNAs is the glmS catalytic riboswitch, or ribozyme, found in the 5′-untranslated region of the glmS gene in Gram-positive bacteria. It is activated by glucosamine-6-phosphate (GlcN6P), leading to site-specific backbone cleavage of the mRNA and subsequent repression of the glmS gene, responsible for cellular GlcN6P production. Recent biochemical and structural evidence suggests that the GlcN6P ligand acts as a coenzyme and participates in the cleavage reaction without inducing a conformational change. To better understand the role of GlcN6P in solution structural dynamics and function, we have separated the glmS riboswitch core from Bacillus subtilis into a trans-cleaving ribozyme and an externally cleaved substrate. We find that trans cleavage is rapidly activated by nearly 5000-fold to a rate of 4.4 min−1 upon addition of 10 mM GlcN6P, comparable to the cis-acting ribozyme. Fluorescence resonance energy transfer suggests that this ribozyme–substrate complex does not undergo a global conformational change upon ligand binding in solution. In addition, footprinting at nucleotide resolution using terbium(III) and RNase V1 indicates no significant changes in secondary and tertiary structure upon ligand binding. These findings suggest that the glmS ribozyme is fully folded in solution prior to binding its activating ligand, supporting recent observations in the crystalline state.
Keywords: catalytic RNA, conformational change, fluorescence resonance energy transfer, footprinting, ribozyme
INTRODUCTION
Over the past few years, it has been shown that noncoding RNAs can serve as molecular switches to regulate gene expression. These riboswitches are highly structured domains found in the 5′-untranslated region (5′-UTR) of mRNAs of Gram-positive bacteria, where it is thought that expression of at least 4% of all genes are riboswitch controlled (Winkler 2005), as well as in the 3′ UTRs and introns of certain eukaryotic mRNAs. A riboswitch with a novel, catalytic mode of action was recently discovered and termed the glmS ribozyme. It resides in the 5′ UTR of the mRNA of the glmS gene in at least 18 Gram-positive bacteria, including Bacillus subtilis and the pathogenic Bacillus anthracis and Bacillus cereus (Winkler et al. 2004). The glmS ribozyme self-cleaves upon specific binding of glucosamine-6-phosphate (GlcN6P), the metabolic product of the enzyme glutamine-fructose-6-phosphate amidotransferase (also referred to GlcN6P synthase), which is encoded by the glmS gene. Self-cleavage of the 5′ UTR correlates with down-regulation of glmS gene expression, providing for a negative feedback loop in response to metabolite accumulation (Winkler et al. 2004; Winkler 2005).
Recent studies focused on the role of GlcN6P in glmS ribozyme function (McCarthy et al. 2005; Jansen et al. 2006; Lim et al. 2006). Several structurally related amine-containing compounds, such as glucosamine (GlcN), serinol, serine, and even the buffer tris(hydroxymethyl) aminomethane, were found to partially activate the riboswitch. In addition, nucleotide analog interference mapping and suppression studies place the ligand close to the cleavage site (Jansen et al. 2006). These observations, along with enzymology data (McCarthy et al. 2005; Link et al. 2006), led to the hypothesis that GlcN6P functions as a coenzyme for general acid–base catalysis in self-cleavage rather than as an allosteric activator (McCarthy et al. 2005). Such a function is consistent with the observation that a trans-acting glmS ribozyme does not show changes in hydroxyl radical footprinting pattern upon ligand binding (Hampel and Tinsley 2006). Most recently, crystal structures of the reaction precursor of a glmS ribozyme from a thermophile in the absence and presence of a modified ligand have confirmed this hypothesis but have also revealed a different secondary structure than previously postulated, leading to a particularly compact and rigid double-nested pseudoknot structure (Fig. 1; Klein and Ferre-D'Amare 2006). This new crystallographic information now facilitates interpretation of solution-based structure probing data, while it will benefit, in turn, from solution-based techniques to verify its functional relevance (Nelson and Uhlenbeck 2006).
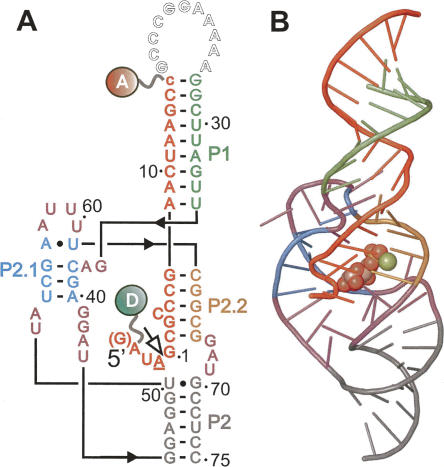
FIGURE 1.
Structure of the trans-acting glmS catalytic riboswitch from Bacillus subtilis as derived by homology modeling based on the crystal structure of the glmS riboswitch from Thermoanaerobacter tengcongensis (Klein and Ferre-D'Amare 2006). (A) Modeled secondary structure of the catalytic core ribozyme used in this study, color coded by structure element. The outlined nucleotides of the closing loop of helix P1 were removed to obtain a trans-acting ribozyme, the small “c” at the 3′ terminus was added to stabilize P1. The substrate strand is shown in red with the cleavage site indicated by an open arrow. For FRET studies “(G)” on the 5′ end was removed and a donor (“D,” fluorescein) and an acceptor (“A,” tetramethylrhodamine) fluorophore were attached to the substrate 5′ and 3′ termini, respectively, as indicated. To obtain a noncleavable substrate analog (ncS3) for structural studies of the precursor form, the 2′-hydroxyl of the underlined A-1 nucleotide was modified to 2′-methoxy. (B) Stick and ribbon depiction of the glmS precursor crystal structure from Thermoanaerobacter tengcongensis bound to glucose-6-phosphate (PDB ID 2H0Z; Klein and Ferre-D'Amare 2006) that shows only the elements contained in the trans-acting ribozyme studied here, using the same color code as in panel A. The Glc6P ligand is represented in space filling, a chelated Mg2+ ion is shown as a green sphere. Rendered using PyMol (DeLano 2002).
We set out to apply biophysical structure probing techniques in combination with footprinting to the glmS ribozyme to better understand its structural dynamics in solution. To this end, we developed a trans-cleaving version of just the catalytic core of the B. subtilis glmS ribozyme, in which the substrate is bound and cleaved externally. We find that this trans-acting variant is rapidly activated by nearly 5000-fold to a rate of 4.4 min−1 upon addition of 10 mM GlcN6P, comparable to the cis-acting ribozyme. Using fluorescence resonance energy transfer (FRET) to monitor global structure, we show that this core trans-acting glmS catalytic riboswitch also does not undergo a global conformational change upon GlcN6P binding and maintains a substrate end-to-end distance of 52 Å, in agreement with the crystal structure, and therefore confirming its relevance for solution conditions. Nucleotide-resolution structural probing methods including terbium(III) and RNase V1-mediated footprinting further confirm that neither secondary nor tertiary structure significantly change upon ligand binding. Our data underscore that the glmS ribozyme is prefolded prior to binding and activation by ligand in both the crystalline and solution states, and they establish a short trans-acting variant as a suitable model for biophysical studies and bioanalytical applications.
RESULTS AND DISCUSSION
Trans cleavage by the minimal glmS catalytic riboswitch is activated 5000-fold upon addition of GlcN6P
Figure 1A shows the secondary structure of the trans-acting glmS ribozyme from B. subtilis, as modeled from the crystal structure of the glmS ribozyme from Thermoanaerobacter tengcongensis represented in Figure 1B. Early truncation analysis showed that the nucleotides downstream of C75 are nonessential for catalytic activity of the B. subtilis ribozyme (Winkler et al. 2004), although recent kinetic analyses indicate that a pseudoknot in this region enhances self-cleavage activity by stabilizing the core structure at limiting Mg2+ concentrations (Wilkinson and Been 2005; Roth et al. 2006). For our studies, in which we were particularly interested in the structural impact of ligand binding on the catalytic core, we therefore focused on the structurally presumably more labile catalytic core of the riboswitch. We further modified the riboswitch by removing the closing loop of helix P1, thus generating a two-strand trans-cleaving ribozyme termed G1 (Fig. 1; Barrick et al. 2004; Hampel and Tinsley 2006). This variant has the distinct advantage over a cis-acting (one-strand) construct in that chemical modifications may be more easily introduced, such as a 2′ O-methyl group at the cleavage site of the substrate strand to block catalysis and thus represent the catalytic precursor form. It also allows for a donor–acceptor fluorophore pair to be placed on the 5′ and 3′ termini, respectively, of the substrate strand for FRET-based distance measurements (Fig. 1A). To test the validity of such a construct, we sought to compare its catalytic activity with the previously characterized full-length, cis-acting glmS ribozyme (Winkler et al. 2004; McCarthy et al. 2005).
First, the ribozyme concentration dependence of cleavage by our trans-acting catalytic core construct was measured by varying the concentration of the ribozyme strand between 10 and 100 nM (Fig. 2A) under standard single-turnover (pre-steady-state) reaction conditions in the presence of 10 mM GlcN6P. The first-order rate constants were plotted as a function of ribozyme (Rz) concentration and fit with a binding equation, yielding a rate constant at saturation for the rate-limiting step of cleavage (k cleav) of 4.95 min−1 (Rz1/2 = 14 nM). This rate constant compares well with those reported for the cis-acting full-length glmS ribozyme under similar conditions, which typically range from 1.1 to 3.0 min−1 (Winkler et al. 2004; McCarthy et al. 2005; except for a recent study, conducted at elevated temperature [37°C] in the absence of monovalent ions, which found a small burst phase with further accelerated cleavage [at up to 15 min−1]; Wilkinson and Been 2005). Based on our titration, we chose 100 nM as our standard saturating trans-acting ribozyme concentration.
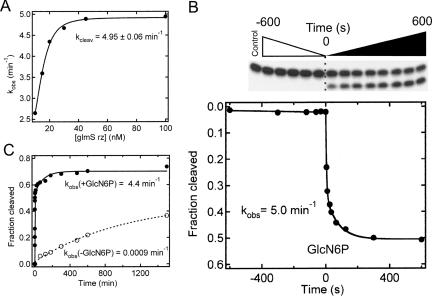
FIGURE 2.
Cleavage activity of the trans-acting glmS catalytic riboswitch. (A) Dependence of the cleavage time constant, k obs, on ribozyme concentration under standard buffer conditions at 25°C in the presence of 10 mM GlcN6P. The data were fit with a simple binding equation (Materials and Methods), yielding a kcleav (maximum cleavage rate at saturating ribozyme concentration) of 4.95 min−1 (Rz1/2 = 14 nM). (B) Rapid activation of the glmS ribozyme upon the addition of 10 mM GlcN6P carried out under standard buffer conditions and a ribozyme concentration of 100 nM. The dotted line signals the addition of GlcN6P. Data were fit with a single-exponential decrease function (—) to yield the reported rate constant k obs = 5.0 min−1. (C) Cleavage time course under standard buffer conditions with a ribozyme concentration of 100 nM with (•) or without (○) 10 mM GlcN6P. Data were fit with either a single- (---) or double-exponential (—) increase function to yield the (fast-phase) rate constants k obs reported in the graph; over such an extended time course, cleavage in the presence of GlcN6P showed a slow-phase rate constant of 0.011 ± 0.003 min−1, representing ∼25% of the total cleaved substrate. This rate constant is well separated from the fast-phase rate constant so that we were able to fit shorter time-scale reactions with single exponentials to obtain very similar rate constants, within error.
Next, under our standard single-turnover reaction conditions, we tested how rapidly the glmS ribozyme is activated by addition of GlcN6P. This was achieved by incubating the riboswitch in standard buffer (50 mM HEPES-KOH at pH 7.5, 200 mM KCl, and 10 mM MgCl2 at 25°C; Materials and Methods) for 10 min, followed by the addition of GlcN6P to a final concentration of 10 mM; reaction aliquots were analyzed throughout this period. While no detectable cleavage was observed over the initial 10 min in the absence of GlcN6P, addition of 10 mM GlcN6P resulted in ∼25% cleavage within 3 sec without any noticeable lag time, indicating that GlcN6P rapidly activates the riboswitch (Fig. 2B). The resulting curve was well fit to yield a first-order single-exponential rate constant of 5.0 min−1. This rapid activation, with no detectable lag phase, suggests that our trans-acting catalytic core construct forms its active state without the need for a preceding slow conformational search, similar to the cis-acting construct (Winkler et al. 2004) and consistent with the notion that it has a prefolded structure poised to bind ligand (Hampel and Tinsley 2006; Klein and Ferre-D'Amare 2006).
The cis-acting glmS ribozyme has previously been observed to cleave very slowly in the absence of GlcN6P (Winkler et al. 2004). To test whether this is the case for our trans-acting catalytic core construct and establish a baseline for potential biosensor applications, we measured cleavage activity in the absence and presence of 10 mM GlcN6P over an extended time course. Under these conditions, the riboswitch only cleaves at a very slow rate constant of 0.0009 min−1 in the absence of GlcN6P, while the presence of GlcN6P activates the riboswitch ∼5000-fold to a (biphasic) rate constant of 4.4 min−1 (Fig. 2B). These data further confirm that the ligand is integral to catalytic activity of the glmS riboswitch, and only slow, if site-specific, background cleavage occurs in its absence.
Cleavage of the glmS catalytic riboswitch has a strong dependence on GlcN6P concentration, ionic conditions, and pH
To further characterize the trans-acting core glmS ribozyme relative to its cis-acting full-length parent, we measured the dependence of cleavage activity on GlcN6P concentration under standard single-turnover conditions (100 nM ribozyme strand) with increasing GlcN6P concentration (Fig. 3A). The observed rate constant increases from 0.0009 to 5.26 min−1 as the GlcN6P concentration increases from 10 μM to 14 mM, yielding a GlcN6P half-titration point (GlcN6P1/2) of 4.8 ± 0.4 mM (with n = 1, i.e., no cooperativity of GlcN6P binding, consistent with the observed single GlcN6P binding site; Klein and Ferre-D'Amare 2006). The apparent GlcN6P affinity of our trans-acting catalytic core construct is thus ∼24-fold weaker than that reported for the cis-acting full-length glmS ribozyme (Winkler et al. 2004). This may be due to removal of the downstream pseudoknot, which is thought to play a role in stabilizing the overall RNA fold (Wilkinson and Been 2005) or due to loss of the capping loop on helix P1. Yet the activity of the trans-acting ribozyme still differentiates GlcN6P concentrations in the tens of micromolar range (Fig. 3A), making it a suitable biosensor candidate.
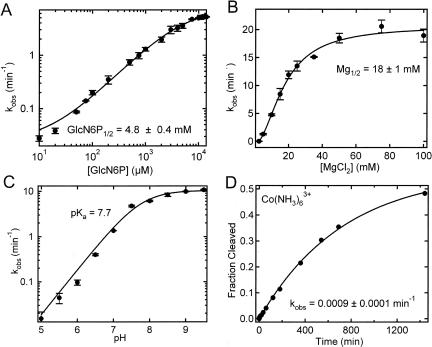
FIGURE 3.
Characteristics of glmS catalytic riboswitch cleavage in trans. (A) Dependence of cleavage rate, k obs, on GlcN6P concentration under standard buffer conditions at 25°C and a ribozyme concentration of 100 nM. Data were fit with a cooperative binding equation (Materials and Methods) to yield the reported dissociation constant (cooperativity constant n = 1). (B) Observed cleavage rate constants, k obs, as a function of Mg2+ concentration in 50 mM HEPES-KOH (pH 7.5) and 200 mM KCl at 25°C in the presence of 10 mM GlcN6P and a ribozyme concentration of 100 nM. The experimental data were fit with a binding equation to yield the reported apparent Mg2+ dissociation constant Mg1/2 = 18 mM (cooperativity constant n = 2). (C) pH dependence of the observed cleavage rate constants, k obs, under standard buffer conditions in the presence of 10 mM GlcN6P and 100 nM ribozyme. The data were fit as described in Materials and Methods (solid line) to yield a pKa of 7.7. (D) Cleavage time course of the glmS ribozyme in the presence of 1 mM Co(NH3)6 3+ in 50 mM HEPES-KOH (pH 7.5), 200 mM KCl, at 25°C, and a ribozyme concentration of 100 nM in the presence of 10 mM GlcN6P. Data were fit with a single-exponential increase function to yield the reported rate constant k obs.
Next, we measured the cleavage activity of our trans-acting catalytic core construct with increasing Mg2+ concentration under standard single-turnover conditions in the presence of 10 mM GlcN6P (Fig. 3B). The observed rate constant increases significantly from 0.009 to 18.7 min−1 as the Mg2+ concentration is raised from 2.5 to 100 mM, yielding a Mg2+ half-titration point (Mg1/2) of 18 ± 1 mM (cooperativity coefficient n = 2). This apparent Mg2+ binding affinity is ∼10-fold weaker than that of the cis-acting glmS riboswitch but retains some of its cooperativity (Winkler et al. 2004). This higher dependence on Mg2+ is consistent with the notion of a destabilized tertiary structure due to removal of the downstream pseudoknot (Wilkinson and Been 2005). The high activity of nearly 20 min−1 attained at saturating Mg2+ concentrations suggests that both folding into an active structure and reaction chemistry of the trans-acting glmS ribozyme are quite fast compared to other trans-acting ribozymes (Pereira et al. 2002; Zamel and Collins 2002; Rueda et al. 2004; Blount and Uhlenbeck 2005).
The pH dependency of a rate constant can help establish the role of functional groups in the catalytic mechanism of an RNA, which led to, for example, the proposal that GlcN6P promotes general acid–base catalysis in the cis-acting glmS ribozyme (McCarthy et al. 2005). We therefore tested the pH dependence of our trans-acting catalytic core ribozyme for comparison with the cis-acting version (Fig. 3C). Our G1 construct indeed displays a log-linear pH profile that saturates above a pH of 9, similar to the one of the cis-acting full-length glmS ribozyme. From the pH dependency we were able to extract a single pKa of 7.7. This value is in agreement with the pKa observed in a pH profile of the cis-acting glmS ribozyme using nonsaturating concentrations of GlcN6P, but is slightly lower than that of free GlcN6P (pKa = 8.2) (McCarthy et al. 2005). Thus, if GlcN6P indeed acts as a cofactor in the reaction, this slight shift toward neutrality in the context of the ribozyme's activity makes it a better suitable general acid or base during catalysis at a physiological pH of 7.4 (McCarthy et al. 2005).
Finally, we wanted to test whether Mg2+ is directly participating in catalytic activity of our G1 construct or primarily plays a structural role, as recently suggested for the cis-acting ribozyme (Klein and Ferre-D'Amare 2006; Roth et al. 2006). Therefore, we measured cleavage activity under buffer conditions of 50 mM HEPES-KOH (pH 7.5), 200 mM KCl, 2 mM EDTA, and 1 mM Co(NH3)6 3+ in the presence of 10 mM GlcN6P at 25°C. Cobalt(III) hexammine is a mimic of hydrated Mg2+ with largely exchange-inert ammonia ligands that allow it only to establish electrostatic outer-sphere contacts with RNA, but no inner-sphere contacts as necessary for any direct participation in reaction chemistry (Cowan 1993). Our G1 construct cleaves to a final extent of ∼50% in the presence of only Co(NH3)6 3+, albeit at a drastically slower rate of 0.0009 min−1 (Fig. 3D). This rate constant is considerably slower than that of the cis-acting ribozyme in Co(NH3)6 3+ (Roth et al. 2006) and is comparable to that found for the trans-acting ribozyme in the absence of GlcN6P in 10 mM Mg2+. These results suggest that while Mg2+ is not absolutely obligatory for the reaction it is more important for catalysis in a trans-acting core glmS ribozyme than it is for catalysis in the cis-acting full-length form.
Structurally similar metabolites are strongly discriminated against
One important characteristic of riboswitches is that they demonstrate remarkable specificity, even discriminating against metabolites that differ by only one functional group. To test whether our trans-acting catalytic core construct can be activated by metabolites structurally similar to GlcN6P, we compared cleavage under standard single-turnover conditions in the presence of GlcN6P, glucosamine (GlcN), or glucose-6-phosphate (Glc6P). The latter two metabolites differ from GlcN6P by removal of only one functional group; GlcN lacks the 6-phosphate functionality, while Glc6P lacks the 2-amino group. Addition of 10 mM GlcN activates the riboswitch, although to an ∼160-fold lower rate constant (0.03 min−1) than does GlcN6P. By contrast, Glc6P does not significantly activate the riboswitch (data not shown). These data show that our trans-acting catalytic core construct discriminates against structurally similar metabolites and has a specificity comparable to that previously reported for the cis-acting glmS ribozyme (Winkler et al. 2004; McCarthy et al. 2005).
Fluorescence resonance energy transfer analysis suggests that the global structure is formed prior to binding of the GlcN6P ligand
We utilized FRET between a donor–acceptor fluorophore pair to test whether a conformational change occurs upon binding of the GlcN6P ligand. We labeled the substrate strand of our trans-acting core glmS ribozyme with a 5′ fluorescein and 3′ tetramethylrhodamine donor–acceptor pair. This particular labeling scheme has the potential to detect distance changes along the central P1:P2.2 helical axis that encompasses the cleavage site and the GlcN6P binding site (Fig. 1; Klein and Ferre-D'Amare 2006). The distance between the labeling sites is expected from the crystal structure to be 54 Å, close to the Förster distance of the fluorescein–tetramethylrhodamine pair (∼55 Å), where distance sensitivity of FRET is at a maximum.
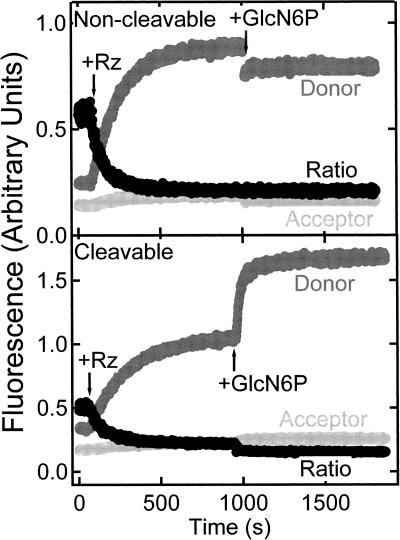
We first performed steady-state FRET experiments that allow for real-time observations of structural changes. When saturating concentrations of the ribozyme strand were added to either a labeled noncleavable substrate analog or cleavable substrate (Rz addition in Fig. 4), the acceptor-to-donor ratio decreased due to binding of the ribozyme, indicative of the expected FRET decrease upon extension of the substrate in the complex. After equilibration, addition of 10 mM GlcN6P to the noncleavable substrate analog–ribozyme (precursor) complex resulted in no further change in FRET efficiency (Fig. 4, top panel). By contrast, a significant change observed in an analogous control experiment with the cleavable substrate–ribozyme complex is due to cleavage and dissociation of the 5′ substrate sequence, which contains the donor fluorophore (Fig. 4, bottom panel). These data suggest that addition of GlcN6P does not cause a significant change in the fluorophore distance in the absence of cleavage and that the global fold of the trans-acting catalytic core construct is adopted prior to ligand binding. It should be noted that we cannot rule out that the 2′-methoxy modification at the cleavage site slightly modulates ligand affinity, which is why we used only high GlcN6P concentrations in our experiments, although it is reassuring that 2′-amino and 2′-deoxy modifications in the same substrate position are fully compatible with binding of the weaker Glc6P ligand to the crystalline glmS ribozyme from Thermoanaerobacter tengcongensis (Klein and Ferre-D'Amare 2006).
FIGURE 4.
Changes over time in donor fluorescence (fluorescein, dark gray), acceptor fluorescence (tetramethylrhodamine, light gray), and acceptor:donor fluorescence ratio (black) of 10 nM doubly labeled substrate (analog) strand upon addition of 100 nM glmS ribozyme (Rz), and subsequently 10 mM GlcN6P under standard buffer conditions of 50 mM HEPES-KOH (pH 7.5), 200 mM KCl, and 10 mM MgCl2 at 25°C, supplemented with 25 mM DTT as an oxygen scavenger.
To obtain a more quantitative measure of the fluorophore distance, we employed time-resolved FRET (trFRET) to measure the donor–acceptor distances in the FRET-labeled glmS catalytic riboswitch as previously described (Pereira et al. 2002; Rueda et al. 2003). Time-resolved donor decay curves of the precursor complex were recorded in the absence and presence of GlcN6P, each of them singly labeled with donor as well as doubly labeled with donor and acceptor. We found that the donor–acceptor distance for the complex in the absence of GlcN6P ligand is 52 Å (with a full width at half maximum, [FWHM] of the distance distribution of 21 Å) and remains at 52 Å upon addition of 10 mM GlcN6P (FWHM of 19 Å), within error of the ligand-free measurement and in excellent agreement with the crystal structure that yields a distance between the labeling sites of 54 Å (Fig. 1). In both cases a single distance distribution fit the trFRET data well, as judged by the residuals and reduced χ2 values (<1.2). Finally, the fluorescence anisotropies of 0.10 for fluorescein and 0.23 for tetramethylrhodamine show no significant change upon addition of any of the three ligands, GlcN6P, GlcN, or Glc6P, to the precursor complex, indicating no detectable change in rotational mobility of the fluorophores upon ligand binding and justifying the assumption of isotropic fluorophore orientation for our distance analysis (Pereira et al. 2002; Rueda et al. 2003). Taken together, these data further support the notion that the trans-acting catalytic core glmS ribozyme does not undergo a global structural change along the P1/P2.2 helical axis upon binding of GlcN6P.
The footprinting pattern of the trans-acting glmS catalytic riboswitch at nucleotide resolution is not modulated upon binding of ligand
Footprinting techniques are often used to examine local or global structural changes of RNA at nucleotide resolution. There are many different types of footprinting assays that can give valuable information on changes in secondary and tertiary structure. For example, to identify single-stranded or non-Watson–Crick base-paired regions, higher (millimolar) concentrations of terbium(III) have previously been employed that slowly cut the RNA phosphodiester backbone in a largely sequence-independent manner. Double-stranded regions and regions involved in tertiary interactions are less accessible to solvent and thus less likely to be cut (Hargittai and Musier-Forsyth 2000; Walter et al. 2000; Harris and Walter 2005). In contrast, RNase V1 specifically cuts regions of RNA that are double stranded or are involved in base-stacking interactions, also largely independent of sequence. We therefore set out to utilize Tb3+ and RNase V1 as complementary footprinting agents to probe and compare the structures of the trans-acting glmS ribozyme precursor in the absence and presence of ligand.
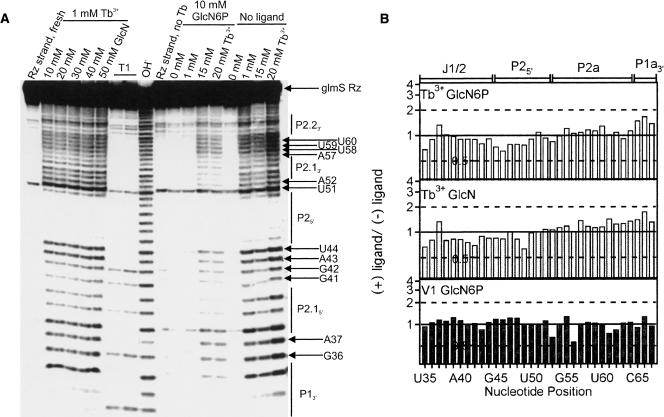
To this end, trace amounts of 5′-32P-radiolabeled ribozyme strand were preannealed with a saturating excess of an unlabeled noncleavable substrate analog under standard buffer conditions, followed by addition of 10 mM GlcN6P (see Materials and Methods). Next, increasing concentrations of terbium(III) were added to initiate slow backbone scission at 25°C over the course of 2 h (under these conditions, only a small fraction [<5%] of RNA is cut, thus avoiding secondary hits). We found that the RNA becomes strongly globally protected from Tb3+ scission in the presence of 10 mM GlcN6P. This protection is presumably due to sequestration of Tb3+ by the negatively charged phosphate group on GlcN6P, as it is not observed in the presence of GlcN, which lacks that phosphate group (Fig. 5A). In order to ensure that we indeed induce backbone scission in the presence of GlcN6P, we increased the concentration of TbCl3 in the reaction to 15 or 20 mM and compared these footprinting patterns to that obtained from 1 mM Tb3+ in the absence of GlcN6P. As a control, we performed terbium(III)-mediated footprinting in the presence of 1 mM Tb3+ and increasing concentrations of GlcN, which also activates the trans-acting glmS ribozyme (Fig. 4) and is known to bind to the cis-acting form (Jansen et al. 2006). Under all conditions we found evidence for formation of the proposed secondary structure, as helices P2.2, P2, P2.1, and P1 are protected relative to the formally single-stranded regions such as the G36–A37, G41–U44, U51–A52 regions and the capping loop of helix P2.1 (Fig. 5A). Remarkably, the footprinting patterns appear essentially the same in the absence and presence of GlcN6P or GlcN (Fig. 5A).
FIGURE 5.
Footprinting of the trans-acting glmS ribozyme. (A) Terbium(III)-mediated footprinting of a 5′-32P-labeled glmS ribozyme strand upon incubation with Tb3+ for 2 h in 50 mM HEPES-KOH (pH 7.5), 200 mM KCl, and 10 mM MgCl2 at 25°C. From left to right are shown, as indicated; freshly labeled ribozyme strand without further incubation, footprint with 1 mM Tb3+ and increasing concentration of GlcN, RNase T1 digests, alkali (OH−) ladder, ribozyme strand incubated in buffer without Tb3+, footprint with increasing Tb3+ concentrations in the presence of GlcN6P, and footprint with increasing Tb3+ concentrations in the absence of ligand. (B) Histogram plots showing relative change in scission intensity from with ligand to without ligand, expressed as a ratio between the normalized intensities of scission (Π values, see Materials and Methods). The top panel represents terbium(III)-induced backbone scission in the presence (at 20 mM Tb3+) or absence (at 1 mM Tb3+) of 10 mM GlcN6P, the middle panel represents terbium(III)-induced backbone scission in the presence or absence of 20 mM GlcN (both at 1 mM Tb3+), and the bottom panel represents backbone scission by RNase V1 (0.0025 units) in the presence or absence of 10 mM GlcN6P. A ratio of 1 (continuous horizontal line) signifies no change in scission intensity.
To better quantify the scission patterns and detect more subtle differences we calculated the relative percentages of RNA cut at each resolved nucleotide of the ribozyme strand in the presence and absence of GlcN6P and GlcN. Figure 5B shows the ratios of the relative extent of scission (Π values) in the absence and presence of ligand for all resolved positions. A ratio of unity signifies no change, while we consider ratios of 0.5–2 as the same within error. The terbium(III)-mediated footprinting patterns in the presence and absence of ligand are indeed identical within error (Fig. 5B, GlcN6P: top panel, GlcN: middle panel), providing further evidence that the trans-acting catalytic core ribozyme is folded prior to binding of the ligand.
To complement our terbium(III)-mediated footprinting, we performed RNase V1 footprinting in the absence and presence of 10 mM GlcN6P. The reaction was prepared in standard buffer (see Materials and Methods) essentially as described above except that 0.2 mg/mL of tRNA carrier and increasing concentrations of RNase V1 were added in place of Tb3+ and the sample was reacted for just 10 min. We observed RNase V1 backbone scission above background at 0.00125 units and saturation at 0.0025 units (data not shown). The overall cleavage pattern was again consistent with the secondary structure of the glmS ribozyme in that expected double-stranded regions were more strongly cut than single-stranded regions. Consistent with our results with terbium(III), addition of GlcN6P does not change the RNase V1 cleavage pattern. A plot of the ratio of the relative percentage cut in the presence and absence of GlcN6P for all resolved positions illustrates that the footprinting patterns in the presence and absence of GlcN6P are identical within error, with all ratios around unity (Fig. 5B, bottom panel). Our footprinting results therefore further support the notion that the glmS ribozyme precursor does not undergo structural changes upon addition of GlcN6P, which is consistent with the model of a glmS riboswitch that is prefolded in solution prior to binding and activation by ligand, as suggested in solution for the extended B. subtilis glmS ribozyme with a downstream pseudoknot by hydroxyl radical footprinting (Hampel and Tinsley 2006) and for the Thermoanaerobacter tengcongensis glmS ribozyme by X-ray crystallography (Klein and Ferre-D'Amare 2006).
CONCLUSIONS
Our results further support the notion that the glmS ribozyme(–substrate complex) is prefolded to recognize and bind its GlcN6P ligand in solution, and that catalytic activation occurs via a mechanism, presumably involving participation in reaction chemistry, which is distinct from the allostery found in other riboswitches. In addition, we have characterized a previously undescribed, particularly small, trans-acting form of the glmS ribozyme (Fig. 1), which we find to be activated ∼5000-fold by its specific ligand GlcN6P (Fig. 2C). Addition of comparable amounts of GlcN, lacking the 6-phosphate moiety, is able to activate the riboswitch by only 30-fold, and Glc6P, lacking the 2-amino functionality, only results in background activity. This high-level specificity of our trans-activating glmS ribozyme, together with its rapid response time and high dynamic range, compares well with most aptazymes, which were engineered and/or in vitro evolved for applications as biosensor components (Breaker 2002). The fact that our glmS ribozyme construct utilizes an external substrate enables multiple-turnover assays (i.e., amplified product formation; data not shown) and reloading with substrate, virtues sought in biosensor applications. Only future studies will reveal how suitable the glmS ribozyme is for such applications and whether its effective activation mechanism can be engineered into other aptazymes.
MATERIALS AND METHODS
Preparation of RNA oligonucleotides
The substrate strand, termed gS (see Fig. 1 for sequence), was purchased from the Howard Hughes Medical Institute Biopolymer/Keck Foundation Biotechnology Resource RNA Laboratory at the Yale University School of Medicine. The RNA contained 2′-protection groups and was deprotected as recommended by the manufacturer and purified as previously described (Harris et al. 2002; Pereira et al. 2002). For FRET measurements, gS was modified on the 5′ and 3′ ends with fluorescein (donor) and tetramethylrhodamine (acceptor), respectively (FgST), as also previously described (Walter 2001, 2002). To obtain the chemically blocked, noncleavable substrate analog for structural studies of the precursor form, the substrate was additionally modified with a 2′-methoxy group at the cleavage site (ncFgST; ncgS is the unlabeled form). RNA concentrations were calculated from their absorption at 260 nm and corrected for the additional absorption of fluorescein and tetramethylrhodamine by using the relations A260/A492 = 0.3 and A260/A554 = 0.49, respectively. The ribozyme strand was generated by run-off transcription from a double-stranded, PCR-amplified template that encoded an upstream T7 promoter. Transcriptions were purified as previously described (Harris et al. 2004), and the RNA concentration was calculated as described above.
Cleavage reactions
Cleavage activity of the riboswitch was determined using the two-strand glmS ribozyme construct depicted in Figure 1. A 3′-32P-labeled substrate was prepared by ligation with [32P]pCp using T4 RNA ligase, followed by desalting using a CentriSep spin column (Princeton Separations). All cleavage reactions were conducted under single-turnover (pre-steady-state) conditions. Standard buffer was 50 mM HEPES-KOH (pH 7.5), 200 mM KCl, and 10 mM MgCl2, unless otherwise stated. In general, ribozyme and substrate strands were annealed separately in standard buffer by heating to 70°C for 2 min and cooling to room temperature over the course of 5 min. After preincubation for 15 min at 25°C, a trace (<1 nM) amount of 3′-32P-labeled substrate in standard buffer was added to the ribozyme strand. After combining ribozyme and substrate solutions and incubating for 2 min, an aliquot of glucosamine-6-phosphate was added to a final concentration of 10 mM (unless otherwise stated), thereby diluting the ribozyme to a final concentration of 100 nM. Aliquots (5 μL) were taken at appropriate time intervals and the reactions quenched with 10 μL of 80% formamide, 0.025% bromophenol blue, 50 mM EDTA, and 7 M urea. The radiolabeled 3′ cleavage product was separated and analyzed as previously described (Pereira et al. 2002). Time traces of product formation were fit, unless otherwise noted, with the single-exponential first-order rate equation y = y 0 + A(1−e −τ/t), employing Marquardt–Levenberg nonlinear regression (Igor Pro 5.03), where A is the amplitude and τ−1 is the pseudo-first-order rate constant k obs. The ribozyme dependency of cleavage was fit with a noncooperative binding equation, while the Mg2+ and GlcN6P dependencies of cleavage were fit with a cooperative binding equation, yielding the cleavage rate constant k cleav under standard conditions, the ribozyme and metal ion half-titration points Rz1/2 and Mg1/2, respectively, and the cooperativity coefficient n as described (Pereira et al. 2002). For the GlcN6P concentration dependence, n = 1 gave the best fit. The pH dependency was fit with equation k obs = k max/(1 + 10pKa1−pH + 10pH−pKa2) as previously described (Harris et al. 2002; Pereira et al. 2002; Tinsley et al. 2003).
To study rapid activation of the glmS ribozyme by GlcN6P, separate solutions of 100 nM ribozyme (final concentration) and trace amounts of 5′-32P-labeled glmS substrate were prepared in standard buffer as described above. After preincubation for 15 min at 25°C, the ribozyme and substrate were mixed together and allowed to incubate for 10 min while 5 μL aliquots were removed at appropriate time intervals, quenched, and analyzed as described above. To initiate the reaction after 10 min, a final concentration of 10 mM GlcN6P was added to the ribozyme–substrate solution and reaction aliquots were quenched and analyzed as described above.
Steady-state FRET measurements
Steady-state FRET measurements of the glmS ribozyme doubly labeled with fluorescein and tetramethylrhodamine were performed on our AB2 spectrofluorometer in a manner similar to that of previously described experiments (Pereira et al. 2002). Typically, stock solutions for a final concentration of 10 nM doubly labeled noncleavable substrate analog ncFgST or cleavable substrate FgST and 100 nM ribozyme were incubated separately in standard buffer supplemented with 25 mM dithiothreitol as a radical quencher at 25°C for at least 15 min. The substrate (145 μL) was then transferred to a 150 μL cuvette and the ribozyme was manually added to a saturating 10-fold excess to form the glmS riboswitch. After complex formation, as indicated by reaching a fluorescence plateau, GlcN6P was added to a final concentration of 10 mM to initiate the reaction. Throughout these additions fluorescein was excited at 490 nm (4 nm bandwidth), and fluorescence emission was recorded simultaneously at the fluorescein (520 nm, 8 nm bandwidth) and tetramethylrhodamine (585 nm, 8 nm bandwidth) wavelengths, by shifting the emission monochromator back and forth. A FRET ratio was calculated as F 585/F 520.
Time-resolved FRET measurements
The global structure of the glmS ribozyme in the presence and absence of 10 mM GlcN6P was studied by trFRET. Preannealed complexes (75 μL, 1 μM noncleavable substrate analog ncFgST, and a saturating excess of 3 μM ribozyme strand) were incubated at 25°C for at least 15 min in standard buffer supplemented with 25 mM dithiothreitol, prior to collecting time-resolved donor emission profiles using time-correlated single-photon counting. Donor–acceptor distances were measured and analyzed following previously described procedures yielding a three-dimensional Gaussian distribution in distance R (Pereira et al. 2002; Rueda et al. 2003). In all cases, a single-distance distribution gave a good fit, as judged by low, reduced χ2 values (<1.2) and evenly distributed residuals. To extract absolute distances, a value of 55 Å for the Förster distance R 0 of fluorescein and tetramethylrhodamine was used (Pereira et al. 2002), and a value of 2/3 was assumed for the orientation factor, based on the relatively high mobility of the fluorophores as evident from their moderate fluorescence anisotropies.
Fluorescence anisotropy measurements
Binding of ligand to the trans-acting glmS ribozyme was analyzed using fluorescence anisotropy measurements performed on a Fusion Universal Microplate Analyzer (Packard Instrument Company). For detection of fluorescein, fluorescence was excited with polarized light at 485 ± 10 nm and emission was collected at 535 ± 12 nm through a polarizer. For detection of tetramethylrhodamine, fluorescence was excited with polarized light at 535 ± 10 nm and emission was collected at 580 ± 10 nm through a polarizer. A preannealed, noncleavable substrate analog–ribozyme complex (100 μL, with a 10 nM ncFgST noncleavable substrate strand and a 100 nM ribozyme strand) was incubated at 25°C for at least 15 min in standard buffer. Anisotropy values were automatically measured 10 times either in the presence or absence of 10 mM GlcN6P, 10 mM GlcN, or 10 mM Glc6P on two different samples and averaged. The factory G value (0.9) was used for all assays for correction.
Terbium(III)-mediated and RNase V1 footprinting
To observe the slow backbone scission mediated by the aqueous Tb3+ derived species Tb(OH)(aq)2+ and RNA footprinting mediated by the double-strand specific RNase V1, a purified ribozyme strand was 5′-32P-phosphorylated with T4 polynucleotide kinase and [γ-32P]ATP, repurified as described above. The labeled RNA strand (250,000 cpm per 10 μL reaction volume) was preannealed with a final concentration of 500 nM noncleavable substrate analog ncgS, denatured at 70°C for 2 min, and slowly cooled to room temperature over 5 min. After cooling, 0.2 mg/mL of tRNA carrier were added to the RNase V1 reactions. The sample was split into two halves, and GlcN6P was added to only one half to a final (saturating) concentration of 10 mM (unless otherwise stated). Terbium(III)-mediated scission was initiated by mixing with 2 μL of an appropriate serial dilution of TbCl3 to achieve the desired final Tb3+ concentration (typically, 1–20 mM) and incubated at 25°C for 2 h. RNase V1 degradation was initiated by mixing 2 μL of the RNA solution with RNase V1 to achieve the desired final nuclease concentration (0.00125–0.125 units per reaction) and incubated at 25°C for 10 min. Reaction mixtures were stopped by addition of an equal volume of 50 mM EDTA (pH 8.0) and ethanol precipitation at −20°C. The precipitated RNA was redissolved in urea loading buffer (80% formamide, 0.025% xylene cyanol, 0.025% bromophenol blue, 50 mM EDTA), and analyzed on a wedged 20% polyacrylamide, 7 M urea, sequencing gel, alongside sequencing ladders from partial digestion with G-specific RNase T1 under denaturing conditions and from alkaline hydrolysis. Product bands were visualized as described above and analyzed as previously described in the literature (Walter et al. 2000; Jeong et al. 2003; Harris et al. 2004; Harris and Walter 2005).
ACKNOWLEDGMENTS
We thank Ken Hampel for sharing preliminary data, Adrian Ferre-D'Amare for sending his coordinates on the day they were published, Robert Kennedy for use of his Microplate analyzer, and all members of the Walter laboratory for stimulating discussions and thoughtful suggestions.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.341807.
REFERENCES
- Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., et al. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, K.F., Uhlenbeck, O.C. The structure–function dilemma of the hammerhead ribozyme. Annu. Rev. Biophys. Biomol. Struct. 2005;34:415–440. doi: 10.1146/annurev.biophys.34.122004.184428. [DOI] [PubMed] [Google Scholar]
- Breaker, R.R. Engineered allosteric ribozymes as biosensor components. Curr. Opin. Biotechnol. 2002;13:31–39. doi: 10.1016/s0958-1669(02)00281-1. [DOI] [PubMed] [Google Scholar]
- Cowan, J.A. Metallobiochemistry of RNA. Co(NH3)6 3+ as a probe for Mg2+(aq) binding sites. J. Inorg. Biochem. 1993;49:171–175. doi: 10.1016/0162-0134(93)80002-q. [DOI] [PubMed] [Google Scholar]
- DeLano, W. The PyMOL molecular graphics system. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- Hampel, K.J., Tinsley, M.M. Evidence for preorganization of the glmS ribozyme ligand binding pocket. Biochemistry. 2006;45:7861–7871. doi: 10.1021/bi060337z. [DOI] [PubMed] [Google Scholar]
- Hargittai, M.R., Musier-Forsyth, K. Use of terbium as a probe of tRNA tertiary structure and folding. RNA. 2000;6:1672–1680. doi: 10.1017/s135583820000128x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, D.A., Walter, N.G. Terbium(III) footprinting as a probe of RNA structure and metal-binding sites. In: Hartmann R.K., et al., editors. Handbook of RNA biochemistry. Wiley-VCH; Weinheim, Germany: 2005. pp. 205–213. [Google Scholar]
- Harris, D.A., Rueda, D., Walter, N.G. Local conformational changes in the catalytic core of the trans-acting hepatitis delta virus ribozyme accompany catalysis. Biochemistry. 2002;41:12051–12061. doi: 10.1021/bi026101m. [DOI] [PubMed] [Google Scholar]
- Harris, D.A., Tinsley, R.A., Walter, N.G. Terbium-mediated footprinting probes a catalytic conformational switch in the antigenomic hepatitis delta virus ribozyme. J. Mol. Biol. 2004;341:389–403. doi: 10.1016/j.jmb.2004.05.074. [DOI] [PubMed] [Google Scholar]
- Jansen, J.A., McCarthy, T.J., Soukup, G.A., Soukup, J.K. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat. Struct. Mol. Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- Jeong, S., Sefcikova, J., Tinsley, R.A., Rueda, D., Walter, N.G. Trans-acting hepatitis delta virus ribozyme: Catalytic core and global structure are dependent on the 5′ substrate sequence. Biochemistry. 2003;42:7727–7740. doi: 10.1021/bi034627g. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Ferre-D'Amare, A.R. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- Lim, J., Grove, B.C., Roth, A., Breaker, R.R. Characteristics of ligand recognition by a glmS self-cleaving ribozyme. Angew. Chem. Int. Ed. Engl. 2006;45:6689–6693. doi: 10.1002/anie.200602534. [DOI] [PubMed] [Google Scholar]
- Link, K.H., Guo, L., Breaker, R.R. Examination of the structural and functional versatility of glmS ribozymes by using in vitro selection. Nucleic Acids Res. 2006;34:4968–4975. doi: 10.1093/nar/gkl643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy, T.J., Plog, M.A., Floy, S.A., Jansen, J.A., Soukup, J.K., Soukup, G.A. Ligand requirements for glmS ribozyme self-cleavage. Chem. Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- Nelson, J.A., Uhlenbeck, O.C. When to believe what you see. Mol. Cell. 2006;23:447–450. doi: 10.1016/j.molcel.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Pereira, M.J., Harris, D.A., Rueda, D., Walter, N.G. Reaction pathway of the trans-acting hepatitis delta virus ribozyme: A conformational change accompanies catalysis. Biochemistry. 2002;41:730–740. doi: 10.1021/bi011963t. [DOI] [PubMed] [Google Scholar]
- Roth, A., Nahvi, A., Lee, M., Jona, I., Breaker, R.R. Characteristics of the glmS ribozyme suggest only structural roles for divalent metal ions. RNA. 2006;12:607–619. doi: 10.1261/rna.2266506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rueda, D., Wick, K., McDowell, S.E., Walter, N.G. Diffusely bound Mg2+ ions slightly reorient stems I and II of the hammerhead ribozyme to increase the probability of formation of the catalytic core. Biochemistry. 2003;42:9924–9936. doi: 10.1021/bi0347757. [DOI] [PubMed] [Google Scholar]
- Rueda, D., Bokinsky, G., Rhodes, M.M., Rust, M.J., Zhuang, X., Walter, N.G. Single-molecule enzymology of RNA: Essential functional groups impact catalysis from a distance. Proc. Natl. Acad. Sci. 2004;101:10066–10071. doi: 10.1073/pnas.0403575101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley, R.A., Harris, D.A., Walter, N.G. Significant kinetic solvent isotope effects in folding of the catalytic RNA from the hepatitis delta virus. J. Am. Chem. Soc. 2003;125:13972–13973. doi: 10.1021/ja037870b. [DOI] [PubMed] [Google Scholar]
- Walter, N.G. Structural dynamics of catalytic RNA highlighted by fluorescence resonance energy transfer. Methods. 2001;25:19–30. doi: 10.1006/meth.2001.1212. [DOI] [PubMed] [Google Scholar]
- Walter, N.G. Probing RNA structural dynamics and function by fluorescence resonance energy transfer (FRET) Curr. Protocols Nucleic Acid Chem. 2002;11.10:11.10.11–11.10.23. doi: 10.1002/0471142700.nc1110s11. [DOI] [PubMed] [Google Scholar]
- Walter, N.G., Yang, N., Burke, J.M. Probing nonselective cation binding in the hairpin ribozyme with Tb(III) J. Mol. Biol. 2000;298:539–555. doi: 10.1006/jmbi.2000.3691. [DOI] [PubMed] [Google Scholar]
- Wilkinson, S.R., Been, M.D. A pseudoknot in the 3′ noncore region of the glmS ribozyme enhances self-cleavage activity. RNA. 2005;11:1788–1794. doi: 10.1261/rna.2203605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler, W.C. Metabolic monitoring by bacterial mRNAs. Arch. Microbiol. 2005;183:151–159. doi: 10.1007/s00203-005-0758-9. [DOI] [PubMed] [Google Scholar]
- Winkler, W.C., Nahvi, A., Roth, A., Collins, J.A., Breaker, R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Zamel, R., Collins, R.A. Rearrangement of substrate secondary structure facilitates binding to the Neurospora VS ribozyme. J. Mol. Biol. 2002;324:903–915. doi: 10.1016/s0022-2836(02)01151-8. [DOI] [PubMed] [Google Scholar]