Abstract
Analysis of the available crystal structures of the ribosome and of its subunits has revealed a new RNA motif that we call G-ribo. The motif consists of two double helices positioned side-by-side and connected by an unpaired region. The juxtaposition of the two helices is kept by a complex system of tertiary interactions spread over several layers of stacked nucleotides. In the center of this arrangement, the ribose of a nucleotide from one helix is specifically packed with the ribose and the minor-groove edge of a guanosine from the other helix. In total, we found eight G-ribo motifs in both ribosomal subunits. The location of these motifs suggests that at least some of them play an important role in the formation of the ribosome structure and/or in its function.
Keywords: loop 530, ribosomal RNA, ribosome, RNA motif, RNA structure
INTRODUCTION
An essential part of our knowledge on RNA structure has accumulated in the form of recurrent structural motifs. Recurrent motifs appear in the same or different molecules and have the same or very similar conformation (for review, see Batey et al. 1999; Moore 1999; Noller 2005). Here we present a new RNA structural motif named G-ribo, which has been found in eight different places within the ribosome. This motif represents a specific side-by-side arrangement of two double helices connected by an unpaired region. The juxtaposition of the helices is stabilized through a complex system of specific interactions, which spread over several layers of stacked nucleotides. The location of the identified cases of this motif within the ribosome suggests that at least some of them play an important role in the formation of the ribosome tertiary structure and/or in its function.
RESULTS
Definition of the G-ribo motif
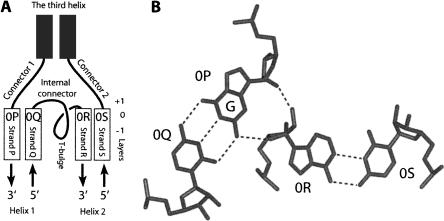
The G-ribo motif refers to a particular side-by-side arrangement of two double helices connected by a short unpaired region. To describe this arrangement, we use the following language. The above-mentioned connector region is called the “internal connector” (Fig. 1A). The helices flanking this connector on the 5′ and 3′ sides are referred to as Helices 1 and 2, respectively. The unpaired regions on the 5′ side of Helix 1 and on the 3′ side of Helix 2 are called Connectors 1 and 2, respectively. In some cases, there is a third helix between Connectors 1 and 2. In other arrangements, such a helix does not exist, so that Connectors 1 and 2 become one. The four strands composing the two helices are named “P,” “Q,” “R,” and “S.” For the nucleotides (nt) in these strands that compose the top base pairs of both helices, the number zero (0) is assigned. For other layers within the helices, the numbering propagates in the negative direction. For the nucleotides of connector regions that stack on top of the nucleotides of the zero layer, the letters are taken from the corresponding 0 nt, while the numbers are positive.
FIGURE 1.
(A) The definition of different elements of the G-ribo motif. (Rectangles) Helical strands; (curves) unpaired regions. Helix 1 consists of strands P and Q, while Helix 2 consists of strands R and S. The third helix exists only, in some cases, of the G-ribo motif. In all cases of the motif, the 3′ part of the internal connector makes a loop that we call the T-bulge (see the text and Fig. 3). The top base-pairs [0P;0Q] and [0R;0S] of Helices 1 and 2, respectively, form the zero layer. The positions of layers −1, 0, and +1 are shown on the right. (B) The juxtaposition of the two zero base pairs, [0P;0Q] and [0R;0S]. (Dashed lines) Hydrogen bonds within and between the base pairs. In this juxtaposition, the ribose of 0R interacts with the ribose and the base of 0P. To make this interaction possible, 0P should be guanosine.
The key element of the G-ribo motif pertains to a specific juxtaposition of top base-pairs [0P;0Q] and [0R;0S] of both helices, which form together a so-called zero layer (Fig. 1B). Within this layer, the ribose of nucleotide 0R interacts with the minor-groove edge of nucleotide 0P. Nucleotide 0P should be G, which would allow it to provide two chemical groups O2′ and NH2 for formation of the hydrogen bonds with O2′ and O4′ of nucleotide 0R, respectively. The described interaction between G in one helix and the ribose of a nucleotide in the other helix has given the name “G-ribo” to the whole arrangement.
Identification of the G-ribo motif in the ribosome structure
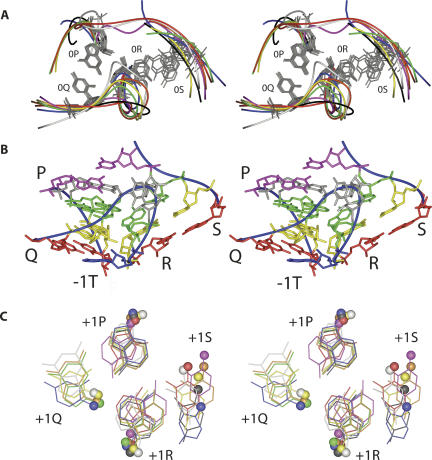
Within the ribosome structure, we found eight cases of the G-ribo motif, three in 16S rRNA and five in 23S rRNA (Figs. 2, 3). All these motifs are clearly seen in all available high-resolution crystal structures of the whole ribosome and of its subunits regardless of the organism (Ban et al. 2000; Schluenzen et al. 2000; Wimberly et al. 2000; Harms et al. 2001; Schuwirth et al. 2005; Korostelev et al. 2006; Selmer et al. 2006). At the same time, the detection of the motif was found to be sensitive to the resolution of the crystal structure because in the structures with a lower resolution (Schluenzen et al. 2000; Harms et al. 2001; Korostelev et al. 2006) almost none of the G-ribo motifs can be easily recognized. No example of the G-ribo motif was found in any RNA-containing entry of the PDB database not related to the ribosome. In all found cases of the motif collected from all structures, the arrangement of two, nucleotides 0P and 0R, is virtually identical. Their superposition provides for a RMSD of 0.5 Å (Fig. 2A), which also leads to a very similar juxtaposition of Helices 1 and 2. The angle between the axes of the helices is ∼60°, which makes the arrangement of the two base pairs at the zero layer essentially nonplanar.
FIGURE 2.
The tertiary structure of the G-ribo motifs in the E. coli ribosome (Schuwirth et al. 2005). (A) Superposition of all cases of the G-ribo motif. Helices 1 and 2 are on the left and right, respectively. For each case of the motif, the backbone is shown by the curve of a particular color. Only the nucleotides of the zero layer are shown explicitly. For three motifs S521 (white), L1024 (red), and L2383 (magenta), which do not have the third helix, the backbone continues from position 0S to 0P. For cases S861 (orange), A1047 (blue), L1309 (yellow), L1642 (green), and L2323 (black), which contain the third helix, the latter is not shown. The positions of 0P, 0Q, and 0R are well superimposed in all cases (for 0R, due to the variability of its identity, the superposition deals only with the backbone). The position of 0S is more flexible than those of 0P, 0Q, and 0R. (B) The tertiary structure of motif S861. The backbone (blue curve). The four strands P, Q, R, and S are indicated. Layers: −1 (red), 0 (yellow), and +1 (green). Nucleotide −1T (blue). Unpaired nucleotides of the levels above +1 (gray). Noncanonical base-pair G858–U828 (magenta) on top of the arrangement forms the first base pair of the third helix. (C) The superposition of the arrangements in the +1 layer in different G-ribo motifs. For each motif, the same color is used as in A. The C1′ atom of each nucleotide is shown as a ball. Nucleotides +1S in L1642 and +1P in L1024, which do not follow the common pattern, are not shown (see the text). In motifs L1024, L2323, and L2383, nucleotide +1Q does not exist (see Fig. 3). Despite the variations in the structure of different +1 arrangements, the positions of the glycosidic bonds of the equivalent nucleotides in different G-ribo motifs are rather close.
FIGURE 3.
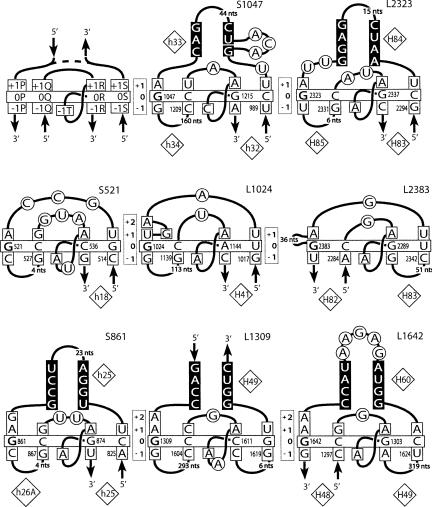
Secondary structures of the G-ribo motifs identified in the E. coli ribosome. (Upper left corner) A template with the named nucleotide positions of the layers from −1 to +1 is shown. (Dashed line) The possibility for a motif to have or not to have the third helix. In the name of a motif, “S” or “L” stands for the ribosomal subunit, small or large, in which the motif was found. The number in the name corresponds to that of nucleotide 0P in the standard E. coli numeration of rRNA. The double helices are arranged as in Figure 1A: the third helix is on top; Helices 1 and 2 are at bottom left and bottom right, respectively. (Vertically oriented rectangles) The positions of the layers. (Horizontally oriented rectangle) The base pairs at the zero layer. G in position 0P is bold. The nucleotides that stack to those of the zero layer are squared. The nucleotides of the third helix involved in base-pairing are shown on the black background. Other nucleotides are circled. The numbers of the helices in the standard 16S and 23S rRNA secondary structures are shown in diamonds.
Figure 3 demonstrates the nucleotide sequences of the identified cases of the G-ribo motif in the Escherichia coli ribosome. The cases are named “S” or “L” depending on the subunit in which they are found, followed by the number of the nucleotide occupying position 0P in 16S or 23S rRNA. In all these cases, base-pair [0P;0Q] is GC. It is also GC in all G-ribo motifs identified in the other structures except motif L2323 from the Thermus thermophilus ribosome, where it is GU (data not shown; Selmer et al. 2006). As to base-pair [0R;0S], in all cases it is either Watson–Crick (WC) or GU. Neither the replacement of GC as base-pair [0P;0Q] by GU, nor the mentioned variations in base-pair [0R;0S] affect the ability of the two base pairs to form the arrangement seen in Figure 1B.
Additional analysis of the available nucleotide sequences of 16S and 23S rRNA (Wuyts et al. 2004) showed a rather high level of conservation of base-pairs [0P;0Q] and [0R;0S] in different G-ribo motifs (Supplemental Table 1). In most identified G-ribo motifs, the presence of GC in base-pair [0P;0Q] exceeds 96%, and the most frequent alternative to the GC base pair is GU. Together, combinations GC and GU account for >98% of the cases of base-pair [0P;0Q] in all motifs but two. The exceptions pertain to motif S861 in eubacteria and to motif L1024 in archeabacteria. In these cases, combinations GC and GU account together for 94.2% and 91.9% of all cases of base-pair [0P;0Q], respectively. A relatively low presence of GC and GU in these cases could be due to problems with the alignment of the corresponding regions among different sequences of rRNA. Base-pair [0R;0S] is also very conserved. For all identified G-ribo motifs, either combination WC or GU was found in this position in >98% of the nucleotide sequences. Due to such a high level of conservation, the correct juxtaposition of base-pairs [0P;0Q] and [0R;0S] is possible in the overwhelming majority of the cases in all identified G-ribo motifs both in eubacteria and in archeabacteria.
Tetranucleotide arrangement at the +1 layer
In addition to the arrangement at the zero layer, all G-ribo motifs have common elements at layers +1 and −1. The central role in the +1 arrangement is played by nucleotide +1R, which belongs to the internal connector and stacks on top of nucleotide 0R. Compared to the position that +1R would have occupied in a regular A-conformation of Helix 2, this nucleotide is reoriented and displaced for several angstroms farther from Helix 1, seemingly because of a potential collision with base-pair [0P;0Q] (Supplemental Fig. 1). Such a displacement makes the distance between the C1′ atoms of +1R and 0R too long for a direct connection of the two nucleotides and necessitates the existence of at least one intervening nucleotide between them. In all identified cases of the G-ribo motif, nucleotide +1R is adenosine. Two more nucleotides of the +1 layer, +1P and +1S, are also characterized by predominant identities, although for each of them, there are exceptional G-ribo motifs in which these predominant identities are not conserved. Nucleotide +1P is adenosine in all motifs except L1024. Nucleotide +1S is uridine in all motifs except L1642.
The particular positions and identities of nucleotides +1R, +1P, and +1S allow them to form a specific arrangement seen in Figure 2C and in Supplemental Figure 2. Normally, these nucleotides form a base triple consisting of two base pairs, [+1P;+1R] and [+1R;+1S]. Most often, adenosines +1P and +1R form a head-to-head base pair using their WC edges. However, in different motifs, any of the two adenosines, or even both at the same time, can flip around their glycosidic bonds, providing the Hoogsteen edges for the interaction with the other adenosine. Such a movement does not change the number of hydrogen bonds between the two bases, although it requires some rearrangement in the backbone conformation for the adjustment to a shorter distance between the glycosidic bonds. For different orientations of +1R, its base pair with uridine +1S would be either reverse Hoogsteen or reverse WC. A conversion from one base pair to the other would also need an adjustment of the positions of the glycosidic bonds. The +1 arrangement can also include nucleotide +1Q, which exists only in five G-ribo motifs and does not have a distinct identity. When +1Q is a purine, it forms a sheared base pair with adenosine +1P. If it is a pyrimidine, it provides atom O2 for a hydrogen bond with the amino group of the adenosine +1P. Regardless of the identity, +1Q always stays in about the same position with respect to adenosine +1P (Fig. 2C).
Analysis of the available nucleotide sequences of ribosomal RNA shows that the +1 arrangement described above is highly conserved both in eubacteria and archeabacteria. Thus, the adenosine identity of position +1R is preserved in all motifs at the level of 98.5% or higher (Supplemental Table 1). Also, in all G-ribo motifs except L1024, adenosine in position +1P is conserved at the level of 96.6% or higher. In all motifs except L1642 and S1047 (only in archeabacteria), the uridine identity of +1S is conserved at the level of 94.9% or higher.
In the exceptional motif L1024, position +1P is always occupied by uridine, which is involved in interactions not observed in any other G-ribo motif (Supplemental Fig. 2). However, nucleotides +1R and +1S stay at about the same places and form a base pair as in other G-ribo motifs. In motif L1642, the nucleotide in position +1S has neither a distinct identity nor a particular position within the 50S tertiary structure. At the same time, the other three nucleotides of the +1 layer—+1R, +1P, and +1Q—are positioned as in other G-ribo motifs. The same is probably true for motif S1047 in archeabacteria, in which position +1S does not have a distinct identity, while positions +1R and +1P are conserved as adenosines (Supplemental Table 1).
Despite some differences in the structure of the +1 layer, the conformation of the backbone and the positions of the glycosidic bonds of three nucleotides, +1R, +1P, and +1Q, in different G-ribo motifs are rather close (Fig. 2C). The position of the fourth nucleotide, +1S, varies more widely, which indicates its sensitivity to the variations in the position of adenosine +1R. The similarities between the structures at the +1 layer in different G-ribo motifs are based on the high conservation of nucleotides +1R, +1P, and +1S, which allows us to qualify these structures as variations of the same type of arrangement.
A-minor interaction at the −1 layer of Helix 1
Although nucleotides +1R and 0R stack on each other, they, as mentioned above, cannot be directly connected, which requires the presence of at least 1 nt between them. The part of the polynucleotide chain enclosed between +1R and 0R forms a bulge, which in most cases consists of only 1 nt, but can also contain 2 nt, as happens in S521 and L1309 (Fig. 3). For identification of nucleotides of this bulge, we use “T,” and the bulge is referred to as the T-bulge (Fig. 1A). The last nucleotide of the T-bulge, which at the same time is the last nucleotide of the internal connector, universally interacts with the −1 layer of Helix 1 and is thus identified as −1T.
The base of −1T interacts with the minor groove of base-pair [−1P;−1Q] (Fig. 2B; Supplemental Figs. 3, 4). In the known crystal structures, nucleotide −1T is almost exclusively adenosine, which allows it to make the A-minor interaction with base-pair [−1P;−1Q]. Also, there is always a hydrogen bond between O2′ of nucleotide −1Q and either N1 or N6 of adenosine −1T. Statistical analysis shows that for all motifs except S1047, both in eubacteria and archeabacteria, adenosine is the predominant identity of nucleotide −1T. However, depending on the motif, the level of conservation varies between 54% and 100% (Supplemental Table 1). Analysis of the structure of motif S1047 from E. coli, in which position −1T is occupied by cytidine, shows that even when −1T is not adenosine, its interaction with base-pair [−1P;−1Q] is similar to that of adenosine. In general, although position −1T displays a clear preference for adenosine, the integrity of the G-ribo motif does not seem to be critically dependent on the identity of −1T.
Participation of riboses in the stabilization of the core of the G-ribo motif
Within the structure of the G-ribo motif, there are areas in which interactions involving backbone and riboses become especially important. One such interaction between guanosine 0P and the ribose of 0R has defined the G-ribo motif. Another area in which riboses play an important role encompasses nucleotides 0Q, +1Q, +1R, and −1T (Fig. 2B; Supplemental Fig. 4). In all motifs containing nucleotide +1Q, the base of +1R stacks to the ribose of +1Q. Also, in all cases, the base of −1T stacks to the ribose of 0Q. Interestingly, in motif L2323 from the T. thermophilus ribosome (Selmer et al. 2006), base-pair [0P;0Q] is GU. As mentioned above, this is the only case of this kind for which the structure is known. Compared to its position in the GC base pair, nucleotide 0Q in this GU base pair is shifted for several angstroms in the direction of the major groove. This displacement, however, is accompanied by the corresponding movement of nucleotide −1T in the same direction, which preserves the interaction between −1T and 0Q (data not shown). The importance of this interaction can explain the limited variability of base-pair [0P;0Q], discussed above, in which GU is the only acceptable exception from the standard GC pattern. Any other nucleotide combination in position [0P;0Q] would break either the interaction between 0P and 0R or that between 0Q and −1T.
In the same area of contact between nucleotides 0Q, +1Q, +1R, and −1T, several polar groups, mostly the O2′ groups of the riboses, become close to each other and can form hydrogen bonds. Although these hydrogen bonds do not follow a common pattern, the fact that they exist in all cases of the G-ribo motif can indicate their importance for the stability of the arrangement. While stacked to the ribose of 0Q, the −1T base effectively screens these hydrogen bonds from the solution. Such a screening would protect the donors and acceptors of these bonds from interaction with the solvent, thus additionally contributing to the stability of the region.
The consensus pattern of the G-ribo motif
Based on the analysis of the tertiary structures of different G-ribo motifs and of the available nucleotide sequences of ribosomal RNA, we can suggest a consensus sequence pattern that fits almost all cases (Fig. 4). This pattern includes two base pairs, four positions of individual nucleotides, and two connectors. It underlines the critical importance of the GC or GU identity for base-pair [0P;0Q] and of the WC or GU identity for base-pair [0R;0S]. Also, nucleotide +1R must be adenosine. Three more positions—+1P, +1S, and −1T—are also occupied by nucleotides of predominant identities, although in each of these positions exceptions are numerous. The preferred length of the internal connector is 4 nt, and, if the presence of the third helix is considered to be equivalent to the presence of two additional nucleotides, the preferred length of the region connecting positions 0S and 0P is 5 nt. This consensus pattern can be used to search for new candidates for G-ribo motifs in RNAs with unknown tertiary structure.
FIGURE 4.
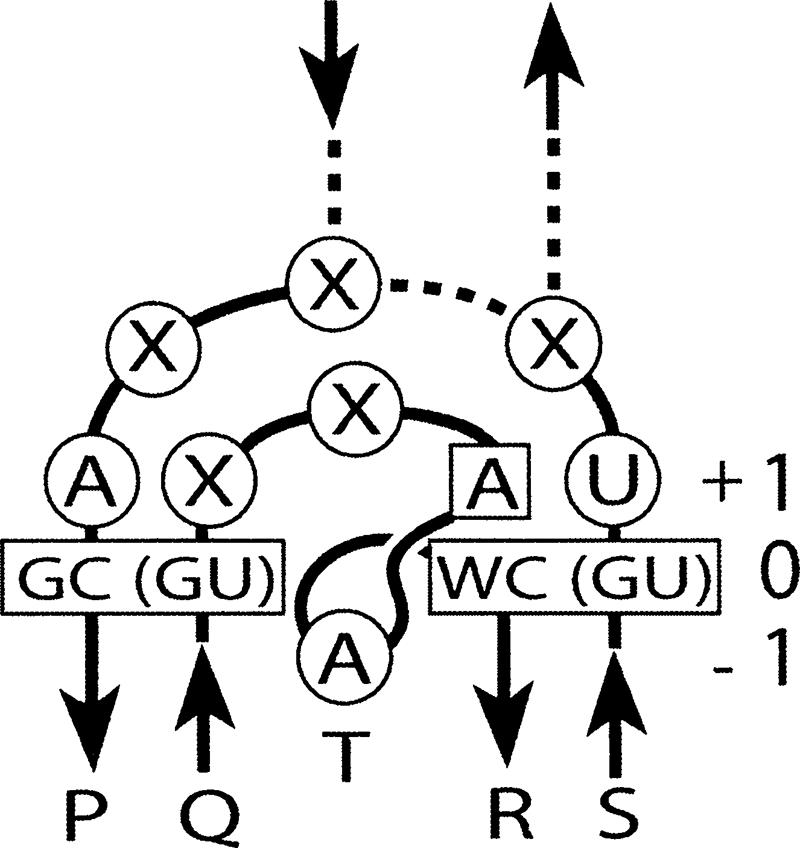
The consensus structure of the G-ribo motif. Helices 1 and 2, as well as connector regions, are arranged as in Figures 1A and 3. (Enclosed in rectangles) Base-pairs [0P;0Q] and [0R;0S] as well as nucleotide +1R, whose identities are very restricted in the known cases of the G-ribo motif. (In circles) The preferable identities for nucleotides +1P, +1S, and −1T. (X) An unrestrained nucleotide identity. The preferred length of the internal connector is 4 nt. (Dashed lines) The alternative possibilities for a G-ribo motif to form the third helix or not. The length of the region connecting positions 0S and 0P is calculated as the sum of the lengths of Connectors 1 and 2 plus 2 nt, if the third helix exists. The preferred length of this region is 5 nt.
DISCUSSION
In this paper, we describe a new structural motif, called G-ribo, which has been found in eight places in the ribosomal RNA. The motif represents a particular juxtaposition of two double helices connected by an unpaired region. The G-ribo motif is characterized by a certain level of rigidity, as can be deduced from the superposability of all identified examples of the motif. The fixation of the juxtaposition of the two helices is achieved via formation of a complex network of contacts, which spread over three layers of stacked nucleotides. The central element of this system consists of two specifically arranged nucleotides, 0P and 0R, from the top base pairs of both helices. The ribose and the base of 0P interact with the ribose of 0R. 0P must be guanosine, while the identity of 0R may not be important. The particular juxtaposition of these nucleotides has been given the name “G-ribo” to the whole arrangement and has served for identification of all cases of the motif. It has been surprising, however, that the structures identified with use of a relatively simple definition of the motif demonstrate a more complex similarity that spreads over three layers of stacked nucleotides. The fact that all identified structures contain the same or very similar elements that have not been included in the definition of the motif signifies the importance of these elements for the motif's integrity and/or function.
The analysis of the identified cases of the G-ribo motif allows us to suggest a chain of cause–effect relationships between different elements of the structure, which can explain how the formation of the G-ribo juxtaposition of the two zero base pairs has determined the formation of other common interactions at the levels between −1 and +1. Everything seems to originate from the fact that the G-ribo juxtaposition of the base pairs at the zero layer does not allow Helix 2 to continue in the regular A-conformation to the +1 level without a collision with Helix 1 (Supplemental Fig. 1). As a result, +1R becomes displaced to a very special position in which it still stacks to 0R but cannot be directly connected to it. This particular position of +1R together with the conserved identities of +1R, +1P, and +1S help the four nucleotides of the +1 level to form a specific arrangement that would stabilize the given juxtaposition of the two helices. Then, the inability of +1R to be directly connected to 0R necessitates the appearance of at least one unpaired nucleotide, −1T, between 0R and +1R. Nucleotide −1T interacts simultaneously with the ribose of nucleotide 0Q and with base-pair [−1P;−1Q] of Helix 1, providing an additional stabilizing effect for the whole arrangement. To make these interactions possible, the ribose of 0Q should be positioned in a very particular way, which explains the almost universal GC identity of base-pair [0P;0Q]. The only acceptable exception from the standard GC pattern is GU, in which 0Q is still able to maintain the interaction with −1T.
All eight identified G-ribo motifs are highly conserved in the ribosomal RNA of eubacteria and archeabacteria, which indicates their importance for the ribosome structure and function. For some motifs, a particular functional role has already been known or can be suggested based on their position in the ribosome. For example, motif S521 caps the so-called loop 530. This loop is known to form a part of the decoding center, being involved in the accuracy control (O'Connor et al. 1997; Ogle et al. 2001). It also participates in the binding of the initiation factor IF1 (Moazed et al. 1995; Carter et al. 2001). Similarly, Helix 34, which stays as Helix 1 in S1047, is known to form a part of the decoding center (O'Connor et al. 1997; Ogle et al. 2001). Therefore, the involvement of the G-ribo motif S1047 in the proper positioning of the proximal part of this helix could also be essential for the decoding process. Another motif, L2323, is involved in the formation of the central protuberance. Helix 84 of 23S rRNA, which stays as the third helix of L2323, interacts with protein L5, which, in turn, interacts with protein S13 from the small ribosomal subunit. Both L5 and S13 contact the P-site tRNA (Yusupov et al. 2001). They are also known to be important for the subunit association during the initiation of translation (Correll et al. 1999; Cukras and Green 2005) and for the ratchet-like movement of the ribosomal subunits during the translocation (Valle et al. 2003). Thus, motif L2323, due to its closeness to proteins L5 and S13, may be involved both in the subunit association and in the translocation.
It is probable that other G-ribo motifs also play particular, yet unknown, functional roles. The elucidation of these roles and the understanding of how the details of the structure of the G-ribo motif determine its functionality in each particular case will be a matter of further analysis.
SUPPLEMENTAL DATA
Supplemental material is available at http://www.esi.umontreal.ca/chteinbe/publications/supplement.html.
ACKNOWLEDGMENTS
The authors are grateful to Drs. Lea Brakier-Gingras and Pascal Legaut for important discussions. S.V.S. acknowledges a grant from CIHR and fellowships from CIHR and FRSQ.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.387107.
REFERENCES
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- Batey, R.T., Rambo, R.P., Doudna, J.A. Tertiary motifs in RNA structure and folding. Angew. Chem. Int. Ed. Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Carter, A.P., Clemons W.M., Jr., Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., Ramakrishnan, V. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Wool, I.G., Munishkin, A. The two faces of the Escherichia coli 23 S rRNA sarcin/ricin domain: The structure at 1.11 Å resolution. J. Mol. Biol. 1999;292:275–287. doi: 10.1006/jmbi.1999.3072. [DOI] [PubMed] [Google Scholar]
- Cukras, A.R., Green, R. Multiple effects of S13 in modulating the strength of intersubunit interactions in the ribosome during translation. J. Mol. Biol. 2005;349:47–59. doi: 10.1016/j.jmb.2005.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., Yonath, A. High-resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell. 2001;107:679–688. doi: 10.1016/s0092-8674(01)00546-3. [DOI] [PubMed] [Google Scholar]
- Korostelev, A., Trakhanov, S., Laurberg, M., Noller, H.F. Crystal structure of a 70S ribosome–tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–1077. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- Moazed, D., Samaha, R.R., Gualerzi, C., Noller, H.F. Specific protection of 16 S rRNA by translational initiation factors. J. Mol. Biol. 1995;248:207–210. doi: 10.1016/s0022-2836(95)80042-5. [DOI] [PubMed] [Google Scholar]
- Moore, P.B. Structural motifs in RNA. Annu. Rev. Biochem. 1999;68:287–300. doi: 10.1146/annurev.biochem.68.1.287. [DOI] [PubMed] [Google Scholar]
- Noller, H.F. RNA structure: Reading the ribosome. Science. 2005;309:1508–1514. doi: 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- O'Connor, M., Thomas, C.L., Zimmermann, R.A., Dahlberg, A.E. Decoding fidelity at the ribosomal A and P sites: Influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 1997;25:1185–1193. doi: 10.1093/nar/25.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons W.M., Jr., Tarry, M.J., Carter, A.P., Ramakrishnan, V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F., et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- Schuwirth, B.S., Borovinskaya, M.A., Hau, C.W., Zhang, W., Vila-Sanjurjo, A., Holton, J.M., Cate, J.H. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- Selmer, M., Dunham, C.M., Murphy, F.V.T., Weixlbaumer, A., Petry, S., Kelley, A.C., Weir, J.R., Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- Valle, M., Zavialov, A., Sengupta, J., Rawat, U., Ehrenberg, M., Frank, J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., Brodersen, D.E., Clemons W.M., Jr., Morgan-Warren, R.J., Carter, A.P., Vonrhein, C., Hartsch, T., Ramakrishnan, V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- Wuyts, J., Perriere, G., Van De Peer, Y. The European ribosomal RNA database. Nucleic Acids Res. 2004;32:D101–D103. doi: 10.1093/nar/gkh065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., Noller, H.F. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]