Abstract
Prostaglandin J2 (PGJ2) and its metabolites Δ12-PGJ2 and 15-deoxy-Δ12,14-PGJ2 (15d-PGJ2) are naturally occurring derivatives of prostaglandin D2 that have been suggested to exert antiinflammatory effects in vivo. 15d-PGJ2 is a high-affinity ligand for the peroxisome proliferator-activated receptor γ (PPARγ) and has been demonstrated to inhibit the induction of inflammatory response genes, including inducible NO synthase and tumor necrosis factor α, in a PPARγ-dependent manner. We report here that 15d-PGJ2 potently inhibits NF-κB-dependent transcription by two additional PPARγ-independent mechanisms. Several lines of evidence suggest that 15d-PGJ2 directly inhibits NF-κB-dependent gene expression through covalent modifications of critical cysteine residues in IκB kinase and the DNA-binding domains of NF-κB subunits. These mechanisms act in combination to inhibit transactivation of the NF-κB target gene cyclooxygenase 2. Direct inhibition of NF-κB signaling by 15d-PGJ2 may contribute to negative regulation of prostaglandin biosynthesis and inflammation, suggesting additional approaches to the development of antiinflammatory drugs.
Prostaglandin J2 (PGJ2) and its metabolites are naturally occurring derivatives of prostaglandin D2 (PGD2). The pathway for formation of these compounds involves sequential conversion of PGD2 to PGJ2, Δ12-PGJ2, and 15-deoxy-Δ12,14-PGJ2 (15d-PGJ2) (1). The last of these metabolites, 15d-PGJ2, is a high-affinity ligand for peroxisome proliferator-activated receptor γ (PPARγ) (2, 3). 15d-PGJ2 represses several genes in activated macrophages, including the inducible NO synthase (iNOS) and tumor necrosis factor α (TNFα) genes, and this repression is at least partly dependent on PPARγ expression (4–6). 15d-PGJ2 is present in vivo during the resolution phase of inflammation, suggesting that it may function as a feedback regulator of the inflammatory response (7).
Previous studies evaluating PPARγ-dependent inhibition of iNOS expression indicated that 15d-PGJ2 was significantly more effective than synthetic PPARγ ligands, despite binding to PPARγ with lower affinity (4). PGJ2 and its metabolites are characterized by the presence of a cyclopentenone ring system that contains an electrophilic carbon that can react covalently by means of the Michael addition reaction with nucleophiles such as the free sulfhydryls of glutathione and cysteine residues in cellular proteins (1, 8, 9). This reactive center is not present in the synthetic PPARγ ligands and has been proposed to account for some of the receptor-independent biological actions of PGJ2, its metabolites, and the related cyclopentenone prostaglandins PGA2 and PGA1 (8, 9).
The transcription factor NF-κB plays a key role in the activation of inflammatory response genes (10). In resting cells, NF-κB is sequestered in the cytoplasm by association with an inhibitory protein IκB. In response to signaling by inflammatory cytokines, IκB kinase (IKK) is activated and phosphorylates IκB on two serine residues. IκB is then ubiquitinated and degraded by the proteasome, freeing NF-κB to migrate into the nucleus and activate gene expression (10). We show here that 15d-PGJ2 inhibits IKK and also directly inhibits DNA binding of NF-κB. These results provide a mechanistic explanation for the PPARγ-independent repression of NF-κB by 15d-PGJ2. Moreover, the relative importance of the two mechanisms, inhibition of IKK and inhibition of NF-κB DNA binding, differs among different cell types.
Methods
Cell Culture.
HeLa cells were obtained from G. Sato (11), and RAW264.7 cells were obtained from the American Type Culture Collection. Both cell lines were cultured in DMEM supplemented with 10% FBS plus penicillin (100 units/ml) and streptomycin (100 μg/ml). The medium for RAW264.7 cells was supplemented with 0.1 mM nonessential amino acids (GIBCO/BRL). Prostaglandins were obtained from Cayman Chemical (Ann Arbor, MI) or Biomol (Plymouth Meeting, PA). 2-Cyclopenten-1-one was purchased from Aldrich. BRL49653 was obtained from Glaxo Wellcome. LPS (Escherichia coli serotype O127:B8) was from Sigma. Recombinant human TNFα was obtained from R & D Systems.
Transient Transfection.
Transient transfections were carried out as described previously by using Lipofectamine (4) or calcium phosphate (9, 12) to transfect RAW264.7 and HeLa cells, respectively. The PPARγ expression vector pCMX-PPARγ, iNOS promoter-luciferase construct, and NF-κB-dependent reporter construct (3× NF-κB) have been described previously (4). The AP-1-dependent reporter construct (3× AP-1)-TATA-luciferase has been described separately (13). The previously described cyclooxygenase 2 (COX-2) promoter (TIS10) (14) was subcloned into the BNXH luciferase vector (4). Transfections were performed at 1–3 × 105 cells per well in 6-well plates. The cells were allowed to rest for 8 h or overnight in medium containing 0.5% FBS, followed by treatment with the indicated compounds for 18 h. PPARγ-dependent effects were determined by cotransfection of cells with the mouse PPARγ expression construct pCMX-PPARγ (4). Cell extracts were assayed for luciferase and β-galactosidase activity, as described previously (9, 12). Luciferase activity was then normalized to β-galactosidase activity (9, 12). Results were expressed as mean ± SE of three determinations, except where indicated.
Preparation of Nuclear Protein Extracts and Electrophoretic Mobility-Shift Assays (EMSAs).
Nuclear protein extracts were prepared as described (15). EMSAs of nuclear protein extracts were performed as described previously (12), using an oligonucleotide probe containing the high-affinity NF-κB binding site of the mouse Igκ enhancer (Promega). In competition experiments, a 100-fold molar excess of unlabeled oligonucleotide was added to the binding mixture before the addition of nuclear protein extract. For EMSAs of purified p65/p50 heterodimers, binding reactions were performed by using various concentrations of 15d-PGJ2 or cyclopentenone with constant amounts of NF-κB p50/p65 (20 nM). After a 6-h incubation at 20°C, constant amounts of DNA (200 pM) and 0.1 mM dithiothreitol were added to the reactions and incubated at 20°C for 30 min. Protein–DNA complexes were resolved by nondenaturing SDS/PAGE.
Western Blot Analysis.
Western blot analysis was performed by standard methods. All incubations with antibodies were for 1 h at room temperature. Cells were pretreated with 15d-PGJ2, cyclopentenone, or solvent (DMSO) in 0.5% serum media for 1 h before inducing agents were added. An anti-IκBα antibody (SC-371; Santa Cruz Biotechnology) was used for detection of IκB. For detection of PPARγ, a mouse monoclonal antibody (SC 7273, clone E8; Santa Cruz Biotechnology) was used at a 1/300 dilution. For detection of NF-κB p65, rabbit polyclonal anti-p65 (SC 372; Santa Cruz Biotechnology) was used.
Immunohistochemistry.
For immunolocalization of NF-κB, monolayer cultures growing on glass coverslips in DMEM plus 10% FBS were transferred to DMEM plus 0.5% FBS plus antibiotics for 30 min at 37°C. 15d-PGJ2 or cyclopentenone was then added, and incubation was continued for 60 min. Finally, TNFα (50 ng/ml) or LPS (1 μg/ml) was added for 30 min. Cells were fixed with 4% paraformaldehyde in 0.1 M sodium phosphate (pH 7.5) and permeabilized with ice-cold methanol for 5 min. The fixed cells were sequentially treated with polyclonal anti-p65 antibody (SC 372, 1/70 dilution; Santa Cruz Biotechnology), biotinylated goat anti-rabbit IgG (Vector Laboratories), and fluorescein-avidin (Vector Laboratories). Labeled cells were then photographed with a Nikon fluorescence photomicroscope (HeLa cells) or a Bio-Rad confocal microscope (RAW264.7 cells).
Immunoprecipitations and Kinase Assays.
RAW264.7 cells were pretreated for 1 h with 12 μM 15d-PGJ2 and then stimulated with LPS (1 μg/ml) for the indicated times. The cells were harvested and the pellets were incubated for 30 min with lysis buffer [20 mM Tris⋅HCl (pH 8)/0.15 M NaCl/0.25% Triton X-100/1 mM EDTA/10 mM β-glycerophosphate/10 mM NaF/10 mM 4-nitrophenyl phosphate/300 μM Na3VO4/1 mM benzamidine/0.5 mM PMSF/10 μg/ml aprotinin/1 μg/ml leupeptin/1 mM dithiothreitol] at 4°C. Then, 300 μg of total cell extract was diluted to 500 μl with pull-down buffer [40 mM Tris⋅HCl (pH 8)/0.5 M NaCl/0.1% Nonidet P-40/6 mM EDTA/10 mM β-glycerophosphate/10 mM NaF/10 mM 4-nitrophenyl phosphate/300 μM Na3VO4/1 mM benzamidine/0.5 mM PMSF/10 μg/ml aprotinin/1 μg/ml leupeptin/1 mM dithiothreitol]; 8-10 μl of IKAP antibody (gift of Frank Mercurio, Signal Pharmaceuticals, San Diego) was incubated with samples at 4°C with gentle shaking for 2 h. A total of 20 μl of protein A-Sepharose beads was then added and incubated for an additional 2 h at 4°C. The immunoprecipitates were washed extensively and subjected to a kinase assay as previously described (16). Briefly, immunocomplex kinase assays were performed in kinase buffer [20 mM Hepes (pH 7.7)/2 mM MgCl2/2 mM MnCl2/10 μM ATP/5 μCi of [γ-32P]ATP/10 mM β-glycerophosphate/10 mM NaF/10 mM 4-nitrophenyl phosphate/300 μM Na3VO4/1 mM benzamidine/2 μM PMSF/10 μg/ml aprotinin/1 μg/ml leupeptin/1 μg/ml pepstatin/1 mM dithiothreitol] at 30°C for 30 min in the presence of glutathione S-transferase (GST)-IκBα (1–54). The reaction was stopped by addition of 6× SDS sample buffer, and the products were subjected to SDS/PAGE and visualized by autoradiography. In vitro IKK assays were performed by incubating 0.8 mg/ml GST-IκBα-(1–54) with various concentrations of 15d-PGJ2 from 5 μM to 2 mM. The reaction buffer was 50 mM Tris (pH 8.0)/50 mM NaCl/10 mM MgCl2. Kinase reactions were performed by adding 0.01 mg/ml IKK for 45 min with 0.1 mM dithiothreitol. The reaction products were resolved by SDS/PAGE, and the gel was phosphorimaged the next day. Jun N-terminal kinase activity was determined in RAW264.7 cells by using a SAPK/JNK assay kit (New England Biolabs).
Prostaglandin E2 (PGE2) Analysis.
RAW264.7 cells (0.5 × 106 cells per well) were cultured in 24-well plates and pretreated with various concentrations of 15d-PGJ2, cyclopentenone, or DMSO for 2 h in medium containing 5% FBS. The cells were then induced with LPS (1 μg/ml) for 18 h. PGE2 was assayed by using a monoclonal antibody/enzyme immunoassay kit from Cayman Chemical.
RNA Analysis.
RAW264.7 cells were pretreated with various concentrations of 15d-PGJ2, cyclopentenone, or DMSO for 2 h in 0.5% FBS before stimulation with LPS (30 ng/ml) for 8 h. RNA was isolated, transferred to nitrocellulose membranes, and hybridized with cDNA probes for COX-2 and glyceraldehyde-3-phosphate dehydrogenase as previously described (4).
GST Assays.
GST assays were performed at 25°C by using whole cell extracts and 1-choloro-2,4-dinitrobenzene as substrate. GST specific activity is expressed as nmol of substrate converted per min per mg of cellular protein.
Results and Discussion
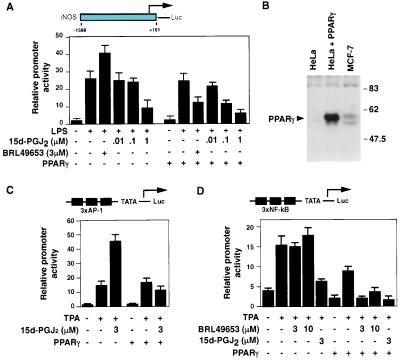
Previous studies evaluating PPARγ-dependent inhibition of iNOS expression indicated that 15d-PGJ2 was significantly more effective than synthetic PPARγ ligands, despite binding to PPARγ with lower affinity (4). Transfection experiments performed in RAW264.7 macrophages that lack PPARγ (4) indicated that 15d-PGJ2 could inhibit the induction of the iNOS promoter in response to LPS when used at concentrations higher than required for PPARγ-dependent inhibition (Fig. 1A). Transcriptional activation of iNOS in response to LPS is mediated by combinatorial actions of AP-1 and NF-κB transcription factors (17, 18). To assess effects of 15d-PGJ2 on these factors, experiments were performed in PPARγ-negative HeLa cells (Fig. 1B) using AP-1 and NF-κB-specific reporter constructs. PPARγ-dependent effects of 15d-PGJ2 were assessed by cotransfection of a PPARγ-expression plasmid. As previously reported (4), 15d-PGJ2 inhibited AP-1-dependent activity in a PPARγ-dependent manner (Fig. 1C). In the absence of PPARγ, 15d-PGJ2 actually potentiated the stimulation of AP-1 by phorbol 12-tetradecanoate 13-acetate (TPA), an effect that may be attributable to the ability of 15d-PGJ2 to stimulate Jun N-terminal kinase (see below). In contrast, 15d-PGJ2 inhibited NF-κB activity in the absence of PPARγ, with expression of PPARγ reducing the concentration of 15d-PGJ2 required for half-maximal inhibition (Fig. 1D). The synthetic PPARγ agonist BRL49653 inhibited NF-κB only in the presence of PPARγ and was less effective than 15d-PGJ2, as previously described (4). Western blotting experiments indicated similar levels of PPARγ in cells cotransfected with a PPARγ expression plasmid for each of the treatment conditions (data not shown). These experiments established PPARγ-dependent and PPARγ-independent mechanisms of NF-κB inhibition by 15d-PGJ2.
Figure 1.
15d-PGJ2 inhibits NF-κB by PPARγ-dependent and PPARγ-independent mechanisms. (A) 15d-PGJ2 inhibits LPS induction of the iNOS promoter in PPARγ-negative RAW264.7 cells. Inhibition occurs at a lower concentration of 15d-PGJ2 in the presence of coexpressed PPARγ. (B) HeLa cells lack endogenous PPARγ. Whole cell extracts were obtained from HeLa cells, from HeLa cells transfected with CMV-PPARγ expression plasmid, and from MCF-7 cells that contain endogenous PPARγ. PPARγ was detected by Western blotting with a PPARγ-specific antibody (Santa Cruz Biotechnology). (C) Inhibition of AP-1 activity by 15d-PGJ2 is PPARγ-dependent. HeLa cells were transfected with the (AP-1)3-TATA-Luc reporter and pCMV-β-gal (1 μg each), with or without 1 μg of pCMX-PPARγ. Transfected cells were treated with TPA (100 nM) with or without 15d-PGJ2 (3 μM). (D) Effect of 15d-PGJ2 on NF-κB activity in the presence or absence of PPARγ. HeLa cells were transfected with the (NF-κB)3-TATA-Luc reporter and β-actin-β-gal (1 μg each), with or without pCMX-PPARγ (1 μg). Transfected cells were treated with TPA (100 nM) and BRL49653 or 15d-PGJ2, as indicated. In D, results represent the mean of duplicate determinations, with the error bars representing the range for the duplicates.
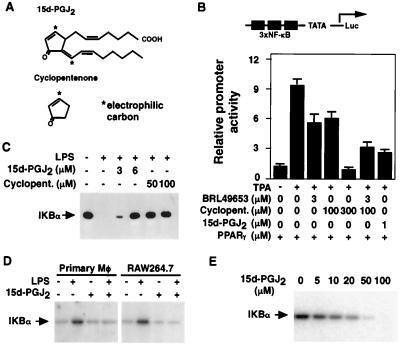
PGJ2 and its metabolites are characterized by the presence of a cyclopentenone ring system containing an electrophilic carbon (Fig. 2A). This ring system can react covalently by means of the Michael addition reaction with nucleophiles such as the free sulfhydryls of glutathione and cysteine residues in cellular proteins (1, 8, 9). We next considered whether this reactive ring system might account for the PPARγ-independent actions of 15d-PGJ2. Consistent with this hypothesis, cyclopentenone (2-cyclopenten-1-one) itself inhibited NF-κB activity, although concentrations approximately 100-fold higher than 15d-PGJ2 were required for similar levels of inhibition (Fig. 2B). In contrast, the related compounds cyclopentanone and cyclopentene, which do not contain a reactive center, did not inhibit NF-κB activity (data not shown). In the presence of coexpressed PPARγ, the effects of BRL49653 and cyclopentenone were additive at a concentration of 3 μM BRL49653, which is saturating for repression by this PPARγ ligand (Fig. 2B). This result is consistent with the hypothesis that the potent repressive activity of 15d-PGJ2 is partly attributable to its ability to act as a PPARγ ligand and partly to the presence of the reactive cyclopentenone ring in this compound, which acts in a receptor-independent manner.
Figure 2.
Effects of 15d-PGJ2 on activity of IKK and IκB degradation. (A) Structures of 15d-PGJ2 (11-oxoprosta-5Z,9,12E,14Z-tetraen-1-oic acid) and cyclopentenone. The positions of chemically reactive, electrophilic carbons are indicated by asterisks. (B) Inhibition of NF-κB by cyclopentenone and the PPARγ-specific ligand BRL49653 is additive in the presence of PPARγ. HeLa cells were transfected with the (NF-κB)3-TATA-Luc reporter and β-actin-β-gal (1 μg each), with pCMX-PPARγ (200 ng). Transfected cells were treated for 16 h with TPA (100 nM) and other additions as indicated. (C) 15d-PGJ2 and cyclopentenone inhibit degradation of IκB in LPS-stimulated RAW264.7 cells. RAW cells were treated with 15d-PGJ2 or cyclopentenone for 1 h. Cells were stimulated with LPS (1 μg/ml) for 30 min, and whole cell extracts were assayed for IκBα by Western blotting. (D) 15d-PGJ2 inhibits IKK activity. PPARγ-negative resident murine peritoneal macrophages and RAW264.7 cells were incubated with 15d-PGJ2 (6 μM) as indicated for 1 h and then stimulated with LPS (1 μg/ml). Whole cell extracts were prepared 10 min later, and IKK activity was assayed by using GST-IKBα (1–54) as a substrate. (E) 15d-PGJ2 inhibits kinase activity of purified IKK. IκBα (1–54) was incubated with purified IKK and [γ-32P]ATP in the presence of the indicated concentrations of 15d-PGJ2 and 0.1 mM dithiothreitol. Incorporation of 32P was determined by SDS/PAGE and phosphorimaging.
The predominant form of NF-κB consists of p50 and p65 subunits that are sequestered in the cytoplasm of unstimulated cells by the inhibitory proteins IκBα and IκBβ (10). Signal-dependent activation of IKK results in phosphorylation and rapid degradation of IκBα and IκBβ, allowing p50/p65 heterodimers to translocate to the nucleus and bind to target genes. To investigate PPARγ-independent mechanisms responsible for inhibition of NF-κB functional activity by 15d-PGJ2, IκBα levels were assessed in RAW264.7 cells stimulated with LPS in the presence or absence of 15d-PGJ2 or cyclopentenone. 15d-PGJ2 and cyclopentenone significantly inhibited degradation of IκBα after treatment with LPS (Fig. 2C). These results are consistent with a previous report that the related cyclopentenone prostaglandin PGA1 inhibits IκBα degradation (19) and suggested an inhibitory effect of these compounds on IKK. PPARγ-negative resting peritoneal macrophages (4) and RAW264.7 cells were therefore stimulated with LPS in the presence or absence of 15d-PGJ2, and whole cell extracts were tested for IKK activity. 15d-PGJ2 inhibited IKK activity in both cell types (Fig. 2D). In contrast, 15d-PGJ2 weakly stimulated the activity of Jun N-terminal kinase and had no effect on protein kinase A activity (data not shown), indicating that its inhibitory effects were kinase-specific. 15d-PGJ2 also inhibited the activity of highly purified IKK in vitro (Fig. 2E). The ability of 15d-PGJ2 to inhibit IKK activity in vitro was exquisitely sensitive to the presence of dithiothreitol, which would be expected to quench the reactive ring system of 15d-PGJ2 (data not shown).
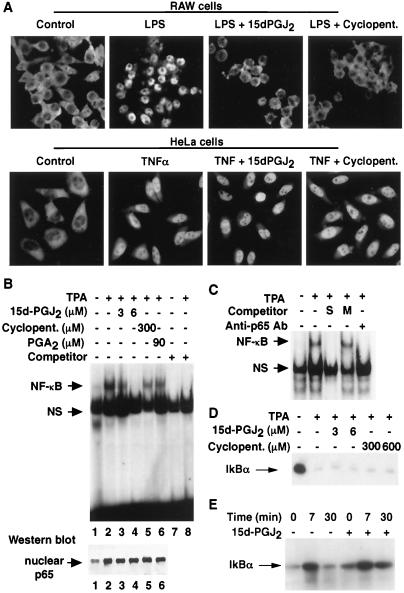
Immunohistochemistry experiments indicated that 15d-PGJ2 and cyclopentenone inhibited nuclear entry of p65 in LPS-treated RAW264.7 cells, consistent with their effects on IKK activity and IκBα degradation (Fig. 3A, Upper). Intriguingly, the degree to which these compounds inhibited nuclear entry of NF-κB varied among different cell types. In particular, immunohistochemistry experiments performed with HeLa cells indicated that nuclear import of p65 was not impaired at concentrations of 15d-PGJ2 and cyclopentenone that abolished nuclear entry of NF-κB in RAW264.7 cells (Fig. 3A, Lower). These results raised the interesting question of how 15d-PGJ2 inhibited NF-κB functional activity in HeLa cells (as illustrated in Figs. 1D and 2B). Gel shift experiments indicated that treatment of HeLa cells with 15d-PGJ2 resulted in inhibition of TPA-stimulated NF-κB DNA binding activity (Fig. 3B, Upper). NF-κB DNA binding activity was diminished in cells treated with 3 μM 15d-PGJ2 and abolished in cells treated with 6 μM 15d-PGJ2. The inhibited band was verified as NF-κB because it was eliminated by incubation with anti-p65 antibody or a competing κB oligonucleotide, but not by an oligonucleotide containing a mutated κB recognition site (Fig. 3C). Although less potent than 15d-PGJ2, cyclopentenone and PGA2 also inhibited the DNA-binding activity of NF-κB (Fig. 3B, Upper). Western blot analysis confirmed that inhibition of DNA binding activity occurred even though nuclear entry of NF-κB was not impaired (Fig. 3B, Lower). Finally, consistent with the observation that nuclear entry of NF-κB was not impaired by 15d-PGJ2 in HeLa cells, 15d-PGJ2 did not inhibit IκB degradation or IKK activity in these cells (Fig. 3 D and E). These findings indicated that 15d-PGJ2 strongly inhibited the DNA binding activity of nuclear NF-κB, even in cells in which IKK activity and nuclear entry of NF-κB were not inhibited.
Figure 3.
15d-PGJ2 inhibits NF-κB DNA binding in HeLa cells without inhibiting nuclear entry of NF-κB. (A) (Upper) 15d-PGJ2 (6 μM) and cyclopentenone (300 μM) prevent nuclear entry of p65 in LPS-treated (1 μg/ml) RAW264.7 cells. (Lower) Significant nuclear entry of NF-κB occurs in TNFα-stimulated (50 ng/ml) HeLa cells treated with 6 μM 15d-PGJ2 or cyclopentenone (300 μM). (B) (Upper) 15d-PGJ2, PGA2 and cyclopentenone inhibit the DNA binding activity of NF-κB. HeLa cells were treated with TPA (100 nM) and the indicated additions, and nuclear extracts were assessed for DNA-binding activity by EMSA. (Lower) Western blot of p65 in nuclear extracts used in EMSA (Upper), lanes 1–6. Each lane was loaded with 10 μg of protein. (C) Specific NF-κB oligonucleotide (S) and anti-p65 antibody, but not mutant NF-κB oligonucleotide (M), abolish formation of the NF-κB/DNA complex. (D) Effect of 15d-PGJ2 on IκB degradation in HeLa cells. Cells were treated with TNFα with or without 15d-PGJ2, and IκB was quantified by Western blotting. (E) Effect of 15d-PGJ2 on IKK activity in HeLa cells. Cells were treated with TNFα with or without 15d-PGJ2, and IKK activity was assayed as described for Fig. 2D.
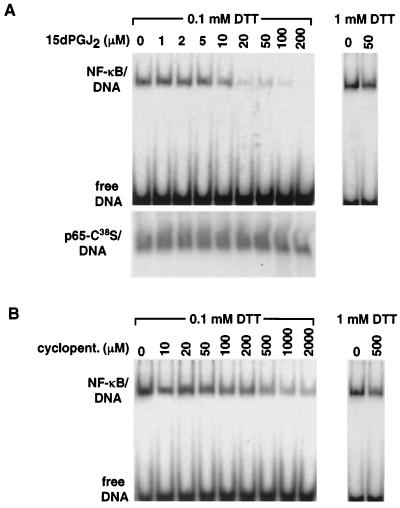
In all Rel proteins, including p65 and p50, there is a conserved cysteine residue located in the DNA-binding domain (Cys62 in p50, Cys38 in p65) (20). Because alkylation of this cysteine impairs DNA binding (21, 22), it was plausible that the cyclopentenone prostaglandins might inhibit NF-κB DNA binding by alkylation of p50/Cys62 and/or p65/Cys38. Consistent with this possibility, 15d-PGJ2 abolished the DNA-binding activity of highly purified p65/p50 DNA-binding domain heterodimers in vitro (Fig. 4A). Three lines of evidence suggest that this effect involved direct modification of NF-κB by the cyclopentenone ring system of 15d-PGJ2. First, similar effects were observed when cyclopentenone was used (Fig. 4B), although 200–600 μM concentrations were required, consistent with the different potencies of 15d-PGJ2 and cyclopentenone observed in cell-based experiments. Second, the effects of 15d-PGJ2 and cyclopentenone on NF-κB binding were eliminated by 1 mM dithiothreitol (Fig. 4 A and B). Third, DNA binding of p65 homodimers containing a Cys38 to Ser mutation was resistant to inhibition by 15d-PGJ2 (Fig. 4A, Lower). These observations raise the possibility that adduct formation by means of the Michael addition also accounts for the inhibitory effects of 15d-PGJ2 on IKK, as there are several cysteine residues near the active site that are potentially subject to covalent modification. The reactivity of 15d-PGJ2 with the DNA-binding domain of p65 appears to be relatively specific. 15d-PGJ2 did not inhibit DNA binding of PPARγ, which contains eight essential cysteine residues within its DNA-binding domain, and binds as an obligate heterodimer with the retinoid X receptor, which also contains eight essential cysteines (data not shown).
Figure 4.
15d-PGJ2 and cyclopentenone directly inhibit the DNA-binding activity of NF-κB. (A) Inhibition of NF-κB DNA binding by 15d-PGJ2 involves the conserved Cys38 in p65. (Upper) The indicated concentrations of 15d-PGJ2 were incubated with 20 nM NF-κB p50/p65 and 200 pM labeled DNA, and protein–DNA complexes were assessed by EMSA. (Lower) Identical experimental setup as above using NF-κB p65 homodimers in which Cys38 was mutated to Ser. (B) Inhibition of NF-κB DNA binding by cyclopentenone. The indicated concentrations of cyclopentenone were incubated with 20 nM of NF-κB p50/p65 and 200 pM labeled DNA.
As illustrated by data presented in Figs. 2 and 3, there was a dramatic difference in the ability of 15d-PGJ2 to inhibit IKK and nuclear entry of NF-κB in RAW264.7 as compared with HeLa cells. Cytoplasmic GST is the major enzyme responsible for metabolic elimination of cyclopentenone prostaglandins by conjugation to glutathione (23). Based on the exquisite in vitro sensitivity of 15d-PGJ2 to dithiothreitol, we examined whether differences in GST activity correlated with the degree to which 15d-PGJ2 inhibited IKK activity and nuclear localization of NF-κB in cells. The GST specific activity was 9.3 ± 5.4 nmol/min per mg of cellular protein in the RAW264.7 cells and 65.6 ± 5.1 nmol/min per mg in the HeLa cells. These observations suggested that the rate of elimination of 15d-PGJ2 influences the overall sensitivity of the NF-κB pathway to 15d-PGJ2 and the relative importance of inhibition of IKK activity. Consistent with this possibility, 15d-PGJ2 inhibited IKK in another HeLa subline expressing lower GST (23.7 + 1.8 nmol/min per mg) (data not shown).
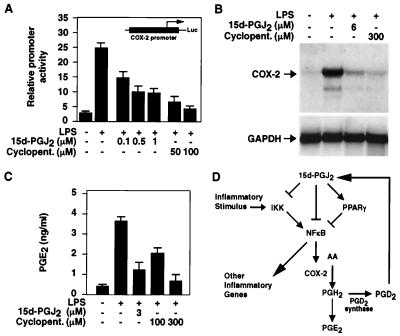
The rate-limiting step of prostaglandin biosynthesis in response to inflammatory stimuli is catalyzed by COX-2 (24, 25). Because COX-2 is under transcriptional control of NF-κB (26, 27), these findings suggested a mechanism for negative feedback regulation by cyclopentenone prostaglandins. Consistent with this, 15d-PGJ2 and cyclopentenone inhibited LPS induction of the COX-2 promoter in RAW264.7 macrophages, and 15d-PGJ2 prevented induction of COX-2 mRNA and blocked production of PGE2 in RAW264.7 macrophages (Fig. 5 A–C). These findings suggest that 15d-PGJ2 may function as a component of a negative feedback loop that regulates prostaglandin biosynthesis (Fig. 5D).
Figure 5.
15d-PGJ2 inhibits production of PGE2 and expression of COX-2 in macrophages. (A) 15d-PGJ2 and cyclopentenone inhibit COX-2 promoter activity. RAW264.7 macrophages were transfected with the indicated COX-2 reporter construct (14) and stimulated with LPS (10 μg/ml) for 18 h. (B) RAW264.7 macrophages were treated with the indicated concentrations of 15d-PGJ2 or cyclopentenone for 2 h and stimulated with LPS (30 ng/ml). The cells were harvested 8 h later, and RNA was analyzed by Northern blotting. (C) RAW264.7 macrophages were treated with the indicated concentrations of 15d-PGJ2 or cyclopentenone, stimulated with LPS (1 μg/ml), and assayed for media content of PGE2 18 h later. (D) Model for mechanisms by which 15d-PGJ2 exerts negative feedback on prostaglandin biosynthesis. COX-2-dependent synthesis of prostaglandin H provides substrate for production of PGD2 in macrophages that express PGD2 synthase. PGD2 is further metabolized to PGJ2 and 15d-PGJ2. 15d-PGJ2 inhibits NF-κB target genes, including COX-2, by inhibition of IKK, direct inhibition of DNA binding, and by PPARγ-dependent transrepression.
15d-PGJ2 has potent effects on cell growth and the expression of inflammatory response genes (4, 5, 28, 29). Although 15d-PGJ2 can be derived from PGD2, which is the most abundant prostaglandin in normal tissues (30), its relevance as a physiologic mediator has been uncertain. Recent studies demonstrating the presence of 15d-PGJ2 in an acute lung inflammation model also provided evidence that it plays an important role in resolution of the inflammatory response (7). The concentrations of exogenously added 15d-PGJ2 required to inhibit NF-κB activity in the present studies were significantly higher than concentrations measured in the extracellular fluid of inflammatory exudates (7). However, 15d-PGJ2 has a very short half-life in cells because of metabolism by GST, limiting its extracellular accumulation. Because its intracellular production would provide direct access to intracellular IKK and NF-κB, it is likely that 15d-PGJ2 functions primarily as an autocrine factor. The relative importance of IKK inhibition, direct inhibition of DNA binding, and PPARγ-dependent transrepression of NF-κB appears to vary from cell type to cell type, but these mechanisms can clearly operate in a combinatorial manner. These studies should stimulate further efforts to understand mechanisms regulating the production and biological roles of 15d-PGJ2 in vivo. Differences in the relative potencies of 15d-PGJ2 and model compounds with respect to inhibition of IKK activity and inhibition of NF-κB DNA binding suggest additional approaches to the development of antiinflammatory drugs.
Acknowledgments
We thank Dr. H. R. Herschman for providing the COX-2 reporter construct and cDNA, A. Morgan for assistance with the EMSAs, N. McDaniel, C. Lytle, and D. DeMason for helpful advice on the immunohistochemistry, and Devin Drew, Yi Chen, and Christopher Phelps for assistance with studies of purified NF-κB and IKK. We thank Frank Mercurio (Signal Pharmaceuticals) for providing IKAP antibodies for precipitation of IKK complexes. These studies were supported by a University of California Multicampus Research Initiative award (to D.S.S. and C.K.G.), a grant from the University of California Cancer Research Coordinating Committee (to D.S.), and grants from the National Institutes of Health (to C.K.G.). M.R. was supported by a Postdoctoral Fellowship from the American Heart Association, and C.K.G. is an Established Investigator of the American Heart Association.
Abbreviations
- PGJ2
prostaglandin J2
- 15d-PGJ2
15-deoxy-Δ12,14-PGJ2
- PGD2
prostaglandin D2
- PGE2
prostaglandin E2
- PPARγ
peroxisome proliferator-activated receptor γ
- iNOS
inducible NO synthase
- TNFα
tumor necrosis factor α
- IKK
IκB kinase
- COX-2
cyclooxygenase 2
- EMSA
electrophoretic mobility-shift assay
- GST
glutathione S-transferase
- TPA
phorbol 12-tetradecanoate 13-acetate
Note Added in Proof
While this paper was under review, another report appeared that also demonstrated inhibition of IKK by the cyclopentenone prostaglandins (31).
References
- 1.Fukushima M. Prostaglandins Leukotrienes Essent Fatty Acids. 1992;47:1–12. doi: 10.1016/0952-3278(92)90178-l. [DOI] [PubMed] [Google Scholar]
- 2.Forman B M, Tontonoz P, Chen J, Brun R P, Spiegelman B M, Evans R M. Cell. 1995;83:803–812. doi: 10.1016/0092-8674(95)90193-0. [DOI] [PubMed] [Google Scholar]
- 3.Kliewer S A, Lenhard J M, Willson T M, Patel I, Morris D C, Lehmann J M. Cell. 1995;83:813–819. doi: 10.1016/0092-8674(95)90194-9. [DOI] [PubMed] [Google Scholar]
- 4.Ricote M, Li A C, Willson T M, Kelly C J, Glass C K. Nature (London) 1998;391:79–82. doi: 10.1038/34178. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, Ting A T, Seed B. Nature (London) 1998;391:82–86. doi: 10.1038/34184. [DOI] [PubMed] [Google Scholar]
- 6.Marx N, Scheonbeck U, Lazar M A, Libby P, Plutzky J. Circ Res. 1998;83:1097–1103. doi: 10.1161/01.res.83.11.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gilroy D W, Colville-Nash P R, Willis D, Chivers J, Paul-Clark M J, Willoughby D A. Nat Med. 1999;5:698–701. doi: 10.1038/9550. [DOI] [PubMed] [Google Scholar]
- 8.Rossi A, Elia G, Santoro M G. J Biol Chem. 1996;271:32192–32196. doi: 10.1074/jbc.271.50.32192. [DOI] [PubMed] [Google Scholar]
- 9.Bui T, Straus D S. Biochim Biophys Acta. 1998;1397:31–42. doi: 10.1016/s0167-4781(97)00214-5. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh S, May M J, Kopp E B. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- 11.Hutchings S E, Sato G H. Proc Natl Acad Sci USA. 1978;75:901–904. doi: 10.1073/pnas.75.2.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hsiang C-H, Marten N W, Straus D S. Biochem J. 1999;338:241–249. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Lin J H C, Liao H L, Friedli O, Verna L, Marten N W, Straus D S, Stemerman M B. Arterioscler Thromb Vasc Biol. 1998;18:473–480. doi: 10.1161/01.atv.18.3.473. [DOI] [PubMed] [Google Scholar]
- 14.Xie W, Fletcher B S, Andersen R D, Herschman H R. Mol Cell Biol. 1994;14:6531–6539. doi: 10.1128/mcb.14.10.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang G, Nepomuceno L, Hopkins K, Sladek F M. Mol Cell Biol. 1995;15:5131–5143. doi: 10.1128/mcb.15.9.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mercurio F, Murray B W, Shevchenko A, Bennett B L, Young D B, Li J W, Pascual G, Motiwala A, Zhu H, Mann M, Manning A M. Mol Cell Biol. 1999;19:1526–1538. doi: 10.1128/mcb.19.2.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Q W, Kashiwabara Y, Nathan C. J Biol Chem. 1994;269:4705–4708. [PubMed] [Google Scholar]
- 18.Lorsbach R B, Murphy W J, Lowenstein C J, Snyder S H, Russell S W. J Biol Chem. 1993;268:1908–1913. [PubMed] [Google Scholar]
- 19.Rossi A, Elia G, Santoro M G. Proc Natl Acad Sci USA. 1997;94:746–750. doi: 10.1073/pnas.94.2.746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen F E, Huang D-B, Chen Y-Q, Ghosh G. Nature (London) 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Rabson A B, Gélinas C. Mol Cell Biol. 1992;12:3094–3106. doi: 10.1128/mcb.12.7.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toledano M B, Ghosh D, Trinh F, Leonard W J. Mol Cell Biol. 1993;13:852–860. doi: 10.1128/mcb.13.2.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Atsmon J, Freeman M L, Meredith M J, Sweetman B J, Roberts L J, II. Cancer Res. 1990;50:1879–1885. [PubMed] [Google Scholar]
- 24.Vane J R, Bakhle Y S, Botting R M. Annu Rev Pharmacol Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 25.Smith W L, Garavito R M, DeWitt D L. J Biol Chem. 1996;271:33157–33160. doi: 10.1074/jbc.271.52.33157. [DOI] [PubMed] [Google Scholar]
- 26.Yamamoto K, Arakawa T, Ueda N, Yamamoto S. J Biol Chem. 1995;270:31315–31320. doi: 10.1074/jbc.270.52.31315. [DOI] [PubMed] [Google Scholar]
- 27.Schmedtje J F, Jr, Ji Y-S, Liu W-L, DuBoi R N, Runge M S. J Biol Chem. 1997;272:601–608. doi: 10.1074/jbc.272.1.601. [DOI] [PubMed] [Google Scholar]
- 28.Petrova T V, Akama K T, Van Eldik L J. Proc Natl Acad Sci USA. 1999;96:4668–4673. doi: 10.1073/pnas.96.8.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su C G, Wen X M, Bailey S T, Jiang W, Rangwala S M, Keilbaugh S A, Flanigan A, Murthy S, Lazar M A, Wu G D. J Clin Invest. 1999;104:383–389. doi: 10.1172/JCI7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ujihara M, Urade Y, Eguchi N, Hayashi H, Ikai K, Hayaishi O. Arch Biochem Biophys. 1988;260:521–531. doi: 10.1016/0003-9861(88)90477-8. [DOI] [PubMed] [Google Scholar]
- 31.Rossi A, Kapahi P, Natoli G, Takahashi T, Chen Y, Karin M, Santoro M G. Nature (London) 2000;403:103–108. doi: 10.1038/47520. [DOI] [PubMed] [Google Scholar]