Abstract
Class 2, or DNA transposons, make up ∼3% of the human genome, yet the evolutionary history of these elements has been largely overlooked and remains poorly understood. Here we carried out the first comprehensive analysis of the activity of human DNA transposons over the course of primate evolution using three independent computational methods. First, we conducted an exhaustive search for human DNA transposons nested within L1 and Alu elements known to be primate specific. Second, we assessed the presence/absence of 794 human DNA transposons at orthologous positions in 10 mammalian species using sequence data generated by The ENCODE Project. These two approaches, which do not rely upon sequence divergence, allowed us to classify DNA transposons into three different categories: anthropoid specific (40–63 My), primate specific (64–80 My), and eutherian wide (81–150 My). Finally, we used this data to calculate the substitution rates of DNA transposons for each category and refine the age of each family based on the average percent divergence of individual copies to their consensus. Based on these combined methods, we can confidently estimate that at least 40 human DNA transposon families, representing ∼98,000 elements (∼33 Mb) in the human genome, have been active in the primate lineage. There was a cessation in the transpositional activity of DNA transposons during the later phase of the primate radiation, with no evidence of elements younger than ∼37 My. This data points to intense activity of DNA transposons during the mammalian radiation and early primate evolution, followed, apparently, by their mass extinction in an anthropoid primate ancestor.
Transposable elements (TEs) are mobile repetitive sequences that make up large fractions of mammalian genomes, including at least 45% of the human genome (Lander et al. 2001), 37.5% of the mouse genome (Waterston et al. 2002), and 41% of the dog genome (Lindblad-Toh et al. 2005). TEs may be classified based upon their method of transposition. Class 1 elements transpose via an RNA intermediate using reverse transcriptase and include long and short interspersed nuclear elements (LINEs and SINEs), and long terminal repeat elements. Class 2 elements, or DNA transposons, transpose via a DNA intermediate through a so-called cut-and-paste mechanism (Craig et al. 2002).
Information on human DNA transposons is currently very scarce. This type of element makes up 3% of our genome (Lander et al. 2001), yet only a limited number of studies have focused on DNA transposons in any mammalian genomes (Auge-Gouillou et al. 1995; Morgan 1995; Oosumi et al. 1995; Robertson 1996; Smit and Riggs 1996; Robertson and Martos 1997; Robertson and Zumpano 1997; Demattei et al. 2000). In contrast, the evolutionary history and genomic impact of mammalian retrotransposons has been the subject of intensive investigation (for examples, see Smit et al. 1995; Kapitonov and Jurka 1996; Szak et al. 2002; Xing et al. 2003; Price et al. 2004; Khan et al. 2006; for review, see Deininger et al. 2003). This gap in knowledge can largely be explained by the relatively recent discovery of DNA transposons. Just a decade ago, several groups independently reported the presence of two different families of mariner-like elements in the human genome (Morgan 1995; Oosumi et al. 1995; Smit and Riggs 1996), now called Hsmar1 and Hsmar2. The evolutionary history of these two families was analyzed in detail by Robertson and colleagues using the genomic sequence data available at that time. These studies indicated that Hsmar1 was active during early primate evolution, about 50 million years ago (Mya) (Robertson and Zumpano 1997), while Hsmar2 was older, having propagated at least 80 Mya (Robertson and Martos 1997).
In a seminal study dedicated to human DNA transposons, Smit and Riggs (1996) estimated that over 150,000 nonautonomous miniature inverted-repeat transposable elements (MITEs) from three major evolutionary groups, hAT, pogo and mariner, were integrated in the human genome. In addition to these MITEs, multiple lineages of elements with coding capacity for seemingly full-length but corrupted transposase were identified in the human genome (for examples, see Morgan 1995; Smit and Riggs 1996; Smit 1999). More recently, members of other eukaryotic superfamilies have been identified in the human genome, including the piggyBac, Merlin, and Mutator superfamilies (Smit 1999; Sarkar et al. 2003; Feschotte 2004; C. Feschotte, unpubl.; Repbase Update) as well as single-copy genes derived from transposases of the P-element and PIF/Harbinger superfamilies (Hagemann and Pinsker 2001; Kapitonov and Jurka 2004; Zhang et al. 2004). Overall, seven of nine known eukaryotic superfamilies of DNA transposons are represented in the human genome, and 125 different families are currently listed in Repbase Update (Jurka et al. 2005; www.girinst.org) that have a copy number of ≥100. Only a handful of these families have been subject to a detailed analysis.
The most comprehensive age analysis of human DNA transposons published to date appeared in the initial analysis of the human genome sequence. This study concluded that, “there is no evidence for DNA transposon activity in the past 50 My in the human genome” (Lander et al. 2001). However, this statement should be taken with caution, since this conclusion was drawn solely from the average level of nucleotide divergence of individual copies to their reconstructed family consensus sequence. This approach has proven generally reliable to date the most prominent bursts of TE amplification and for comparing groups of elements that evolve at the same or almost the same rate. However, due to rapid fluctuations in substitution rates during the mammalian and primate radiation (Goodman 1985; Yi et al. 2002) and possible variations in the substitution rate of different types of TEs (for example, see Xing et al. 2004), the results of these analyses should be interpreted carefully. In addition, the method is entirely dependent on the reconstruction of an accurate consensus sequence, a process that is sensitive to the number and genomic distribution of the elements. Since human DNA transposons are anticipated to be of relatively ancient origin and have likely diversified over the entire course of mammalian evolution, sequence divergence should not be the sole method used for accurately dating these types of elements. Furthermore, there has been no published effort to characterize the tempo and mode of accumulation of every DNA transposon family in any mammalian genome. Therefore an accurate and detailed picture of DNA transposon history in humans and other mammals is still lacking.
Here we present the first detailed analysis of the age of the 125 DNA transposon families currently recognizable in the human genome. In particular, we sought to evaluate which human DNA transposon families were actively transposing during primate evolution. To this end, we used a combination of three independent computational methods, two of which do not rely upon sequence divergence. We estimate that at least 40 families of DNA transposons were active during the primate radiation. We conclude that ∼98,000 individual elements were added to the primate genome in the last ∼80 My of evolution. Eleven of these families, or ∼23,000 individual elements, inserted into the primate genome between the split of prosimian primates and new world monkeys (∼40 to 63 Mya). However, we found no evidence that any human DNA transposon family was active within the last ∼37 My of primate evolution. Our results suggest an intriguing history of intense activity of diverse DNA transposons during the first half of the primate radiation, followed by a striking cessation of transposition activity in an anthropoid primate ancestor and no detectable germ-line reinfiltration of the primate lineages leading to humans over the last 37 My.
Results
Census of DNA transposons in the human genome
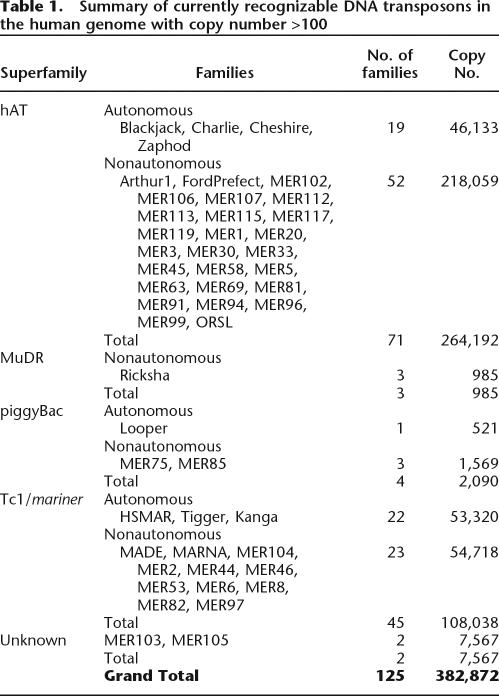
We began our investigation by assessing the diversity and copy number of all DNA transposon families currently recognized in the human genome. Copy numbers were calculated from the RepeatMasker tables of the May 2004 assembly of the human genome, available through the UCSC Genome Browser (http://genome.ucsc.edu). In agreement with previous reports (Smit and Riggs 1996; Smit 1999; Lander et al. 2001), we found that two superfamilies, hAT and Tc1/mariner, are predominant in the human DNA transposon population (Table 1). hAT elements account for approximately two-thirds of the census and more than half of the 125 families. Human Tc1/mariner elements account for one-third of the population and can be divided into three evolutionary distinct lineages: pogo-like, mariner-like, and Tc2-like (Smit and Riggs 1996, Robertson 2002). The former is the most abundant and diversified lineage, and includes eight families of transposase-encoding Tigger elements and 22 related MITE families. The prevalence of nonautonomous MITEs (74% of the total number of DNA TEs) over transposase-encoding elements (26%) is particularly striking in the human genome, and this phenomenon affects all superfamilies (Table 1). It is also a characteristic of the DNA transposon population of plants and nematodes (Feschotte et al. 2002).
Table 1.
Summary of currently recognizable DNA transposons in the human genome with copy number >100
Analysis of DNA transposons nested into other elements
In order to obtain a first assessment of human DNA transposon families that were active during the primate radiation, we took advantage of the fine-scale evolutionary histories of L1 and Alu elements in the primate lineage produced by others. We used these primate-specific families as historical markers for dating DNA transposons. We reasoned that any DNA transposon inserted or nested within a primate-specific L1 element (Khan et al. 2006) or a primate-specific dimeric Alu element (Kapitonov and Jurka 1996) should itself be primate specific. Using a Perl script and data from the UCSC Genome Browser RepeatMasker tables, we conducted an exhaustive search of human L1 and Alu retroposons that had suffered a nested insertion of a DNA transposon. A DNA transposon was considered to be nested within one of these retroposons if: (1) the upstream and downstream retroposon fragments were within 50 bp of the 5′ and 3′ ends of the DNA transposon, (2) the orientation of the upstream and downstream retroposon fragments were the same, and (3) the 3′ end of the upstream retroposon fragment and the 5′ end of the downstream retroposon fragment were within 20 bp of each other according to matching positions in their consensus nucleotide sequence.
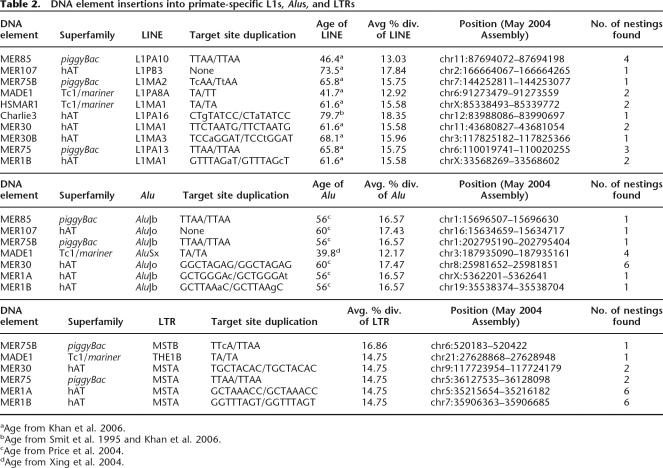
This analysis revealed the presence of elements representing 10 distinct DNA element families that were inserted into primate-specific L1s (Table 2). Each of these insertions were validated by visual inspection of expected target-site duplications (TSDs) flanking the DNA transposon insertion based on alignment to the family consensus sequence (one example per family is shown in Table 2). The only exception was MER107 elements, whose insertion into L1 created a short deletion (from 11 to 19 bp) at the integration site accompanied by the coinsertion of unrelated “filler” DNA (11–27 bp), instead of the 8-bp TSD canonical of hAT elements (see Supplemental Fig. 1). The youngest L1 elements that suffered a DNA transposon insertion belong to the L1PA8A family, estimated to be between 42 and 50 My old (Khan et al. 2006; Supplemental Table 1). L1PA8A elements suffered insertions from two separate MADE1 elements previously proposed to be among the youngest DNA element families (Lander et al. 2001; Table 2).
Table 2.
DNA element insertions into primate-specific L1s, Alus, and LTRs
aAge from Khan et al. 2006.
bAge from Smit et al. 1995 and Khan et al. 2006.
cAge from Price et al. 2004.
dAge from Xing et al. 2004.
In each case, we found that the length of the nested DNA transposons was similar to the length of its respective consensus sequence and that the nucleotide divergence of the L1 copy to its consensus sequence was congruent with the age of the corresponding L1 subfamily. For example, four distinct MER85 elements were found inserted into L1PA10 elements (one example is reported in Table 2). A TSD of the tetranucleotide sequence TTAA, a hallmark of piggyBac transposons, could be associated with each insertion. The length of the nested MER85 elements ranged from 126 to 130 bp, which is in good agreement with the length of the family consensus sequence (140 bp). The nested MER85 copies differ by 6.3% to 9.3% from their consensus sequence, while the average divergence for the family is 7.3% (as calculated from the May 2004 Repeatmasker files from the UCSC Genome Browser, see Methods). The L1PA10 elements that suffered insertions by MER85 elements were 8.5% to 23.9% divergent from their consensus (with three of the four being 15.3% diverged or less), which is consistent with the average 14.7% pairwise divergence of the L1PA10 subfamily (Khan et al. 2006). These data indicate that both nested DNA transposon and disrupted L1 elements are fully representative of their respective families.
We found that six of the 10 DNA element families that had copies inserted into primate-specific L1 elements also comprise copies nested into dimeric Alu elements (Table 2), all of which are known to be primate specific (Kapitonov and Jurka 1996). One family, MER1A, was found to be nested within a dimeric Alu (AluJb) but was not found to be nested within a primate-specific L1. Here again, each DNA transposon insertion was validated as a bona fide transposition event by the presence of expected TSD (Table 2), except for MER107, which once again created a short deletion upon insertion rather than a typical TSD (Supplemental Fig. 1). The youngest Alu element that suffered a DNA element insertion was a member of the AluSx subfamily, which amplified ∼44 Mya (Xing et al. 2004). Four unambiguous instances were found of an AluSx element that had suffered an insertion of a MADE1 transposon (one example is shown in Table 2). Each MADE1 was integrated at a different position within each AluSx element, indicating that they all resulted from independent MADE1 transposition events rather than from propagation of a composite element.
Though the evolutionary history of primate LTR elements has not been analyzed as fully as those of L1 and Alu elements, we drew upon the available literature to provide further validation for the nested insertion analysis. Smit (1993) demonstrated that five families of LTR retrotransposons (THE1A, THE1B, THE1C, MSTA, and MSTB) were primate specific. We found that six of the 11 DNA elements that had nested within the primate-specific L1 and Alus had nested within these LTR elements (Table 2). Each insertion was validated by the presence of the expected TSD. The youngest primate-specific LTR to suffer an insertion from a DNA element was THE1B.
Cross-species genomic analysis of orthologous insertions
To refine the age of the DNA transposon families, we turned to another method that did not rely on sequence divergence. It is possible to ascertain when individual TE insertions occurred by investigating their presence/absence at orthologous genomic regions in multiple species whose phylogenetic relationships are confidently established. If an element is present and fixed within the population in one species, but is absent at the orthologous position in another species, then the element must have been transposing some time after the split of the two species. Note that for DNA transposons, which transpose through a cut-and-paste mechanism, this pattern could be caused by either the insertion of the element or by its excision in one of the two species examined. In either case, nonetheless, the absence of the element in one of the two species can be interpreted as a manifestation of transposon activity after the divergence time of the two species. Conversely, if an element is present at orthologous positions in two different species, it is almost certain that this insertion predates the divergence of the two species. This is because the probability of two elements of the same family inserting at the exact same position independently in two different lineages is extremely small, especially for DNA elements, as these occur in relatively low copy-number families in mammalian genomes.
Taking advantage of the ongoing ENCODE Project and of other genomic resources accessible through the UCSC Genome Browser, we assessed the presence/absence of 794 human DNA transposon copies at orthologous positions in at least eight (and up to 10) other mammalian species, including three primate species (see Methods). These elements represent 111 of 125 families known in the human genome.
We found members of 11 DNA element families that were present at orthologous positions in human, Rhesus macaque (Macaca mulatta), and marmoset (Callithrix jacchus), but absent in the galago (Otolemur garnettii), a prosimian primate. We were able to identify “empty sites” in the respective orthologous galago loci for members of each DNA element family (one example per family is shown in Table 3) except for MER75B, since there is only one copy of MER75B within an ENCODE region and there is a large deletion within the galago lineage at that locus. These were clear “empty sites,” with only one copy of the TSD and no additional sequences indicative of transposon excision. DNA transposon excisions are typically imprecise in that they leave behind one of the two TSD and/or a few terminal nucleotides of the transposon at the excision site (for example, see Plasterk 1991; for review, see Craig et al. 2002). This data suggests that these elements were inserted in the anthropoid lineage, rather than excised in the galago lineage. We assume that these individual copies are representative of their respective families, because their individual percent divergences are all within the 95% confidence interval for average divergence of the family (average divergence ± 1.96 SD). As such, these elements do not represent statistical outliers. These data demonstrate that at least 11 families of human DNA transposons were transpositionally active after the split of prosimians and anthropoid primates, or during the last ∼63 My (Goodman 1999). Seven of these 11 families also include copies nested within L1 or Alu elements known to be primate specific (see above; Table 2). Assuming that these individual copies are representative of their respective families and that their activity is contemporary to the activity of their entire family, these 11 families make up a total of 23,570 transposons in our genome. Therefore, these data imply that many thousands of DNA transposons were inserted in the lineage of anthropoid primates, that is, within the last ∼63 My.
Table 3.
Preintegration empty sites in galago
Members of the remaining 100 human DNA transposon families represented in the ENCODE regions were found at orthologous positions in human, marmoset, and galago. To investigate which of these families was primate specific, each of the 316 copies, along with at least 100 bp flanking both the 5′ and 3′ ends, was visually inspected in the UCSC Genome Browser for their presence/absence at orthologous positions in at least five nonprimate mammals using the ENCODE Comparative Genomics tracks. Phylogenetically, these five species form two separate outgroups to the primates, with the mouse/rat/rabbit lineage being closer to the primates than the cow/dog lineage (Murphy et al. 2004). Hence, a transposon present in primates but absent in the five other mammals most likely resulted from an insertion in the primate lineage. A less parsimonious explanation for this pattern would be that the element inserted in a eutherian (i.e., placental) ancestor and subsequently excised twice, independently, in the mouse/rat/rabbit lineage and in the dog–cow lineage. We found 163 elements from 23 families that were clearly present in humans as well as three of the five other primate genomes (chimp, baboon, rhesus, marmoset, and galago), but absent in all other mammals examined. Thus, these 23 families were classified as primate specific.
The remaining 77 families were represented by elements present at orthologous sites in all the primates and at least one of the other eutherian mammals; thus, these families were classified as eutherian-wide. It should be noted, however, that several families (such as Charlie1 and Charlie1a) included some copies that were apparently primate specific, as well as copies present at orthologous positions in primates and at least one of the nonprimate mammals. It could well be that the activity of these families initiated prior to the emergence of the primates, but continued in a primate ancestor, generating primate-specific insertions. Alternatively, lineage-specific sorting of ancient alleles with or without the insertions could also account for these patterns. In the absence of further evidence to distinguish between these possibilities, we adopted a conservative classification of such families as eutherian-wide.
To determine whether the other 14 DNA element families not represented within the ENCODE regions were primate specific, we searched the finished mouse, rat, and dog genomes for the presence or absence of TEs at orthologous positions to the human DNA elements (see Methods). We found that six families of elements (MER6C, Ricksha, Ricksha_b, Ricksha_c, Tigger5a, and Tigger5b) were present in primates, but clearly absent in the mouse, rat, and dog. These families were additionally classified as primate specific. The other eight families were found to be present in the primates and in the mouse, rat, or dog genomes. These families were classified as eutherian-wide. Together, the cross-species analysis of orthologous insertions suggests that a minimum of 40 distinct DNA transposon families, which accounts for 98,300 DNA elements currently fixed in the human genome, were active in the primate lineage, i.e., within the last ∼80 My.
Age of DNA transposons based on sequence divergence
The results above allowed us to distinguish among DNA transposon families that have been active in an anthropoid (40–63 Mya), primate (63–80 Mya), or eutherian ancestor (80–150 Mya) (Goodman 1999; Murphy et al. 2004). In order to obtain a better resolution of the age of each family, we used the median of these three evolutionary periods (51.5, 72, and 115.5 My, respectively) to determine the median substitution rate per million years of all DNA transposons within each age class (anthropoid, primate, or eutherian). We also calculated a low- and high-substitution rate using the upper and lower bounds of the age class. This was done by calculating the corrected number of nucleotide substitutions per site in all individual copies to their respective family consensus sequence (as given by Repeatmasker), and excluding consensus CpG positions, using the REV model (Tavare 1986) (see Methods). The REV, or general reversible model, calculates five different rate parameters based upon nucleotide composition, thereby correcting for differing nucleotide compositions between DNA element families. The same analysis was performed separately with the L1s and Alus falling into the anthropoid- or primate-specific class ages (plus an additional class of <40 My) and four eutherian-wide L1 elements (L1PA17, PB4, and MA4-5), following the most recently published dating of L1 (Khan et al. 2006) and Alu subfamilies (Price et al. 2004; Xing et al. 2004).
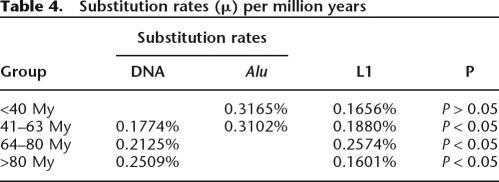
This approach allowed us to calculate a substitution rate per million years that takes into account possible fluctuations in evolution rates during the different time periods and among the three types of elements (see Methods; Supplemental Table 2). The results reveal that DNA transposons, L1s, and Alus each have different average substitution rates in the different evolutionary periods (Table 4). There are statistically significant differences among the three types of TEs within the same period. For example, within the anthropoid-specific lineage (41–63 My), which was the only period for which mutation rates could be estimated for all three types of TEs, the differences were statistically significant (P < 0.05, ANOVA). These differences may be attributed to several factors, such as biased genomic distributions (e.g., Alus preferentially accumulate in GC-rich regions), compositional biases, replication mechanisms (reverse transcriptase for RNA elements, DNA polymerase for DNA transposons), and amplification dynamics (subfamily structure). Regardless, this data provides a rationale for treating DNA transposons separately from the other types of TEs in the human genome in these calculations as opposed to using substitution rates estimated for other type of elements or for other neutrally evolving sequences such as pseudogenes (for example, see Robertson and Martos 1997).
Table 4.
Substitution rates (μ) per million years
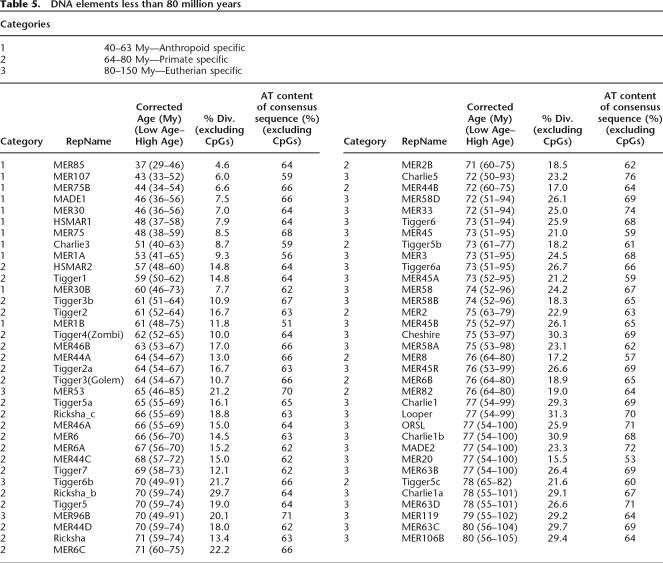
We next applied the substitution rate of each type of element for each period to estimate the age of individual families based on their average nucleotide divergence to the respective consensus sequence, excluding CpG sites (see Methods). This method differed slightly from other datings of TEs in which a single, constant substitution rate for the entire span of primate evolution was used (for examples, see Price et al. 2004; Khan et al. 2006). Our estimates for the age of L1 and Alu subfamilies using these period-specific substitution rates differed somewhat from recently published datings (Fig. 1; Price et al. 2004; Xing et al. 2004; Khan et al. 2006). This is to be expected since all three published datings used the Kimura 2-parameter correction not the REV model (Kimura 1980). Overall, L1 elements appeared to be slightly older, due to their higher consensus AT content, and Alu elements appeared to be slightly younger, due to their lower consensus AT content, using the REV model rather than using the Kimura model. The resulting estimated ages of human DNA transposon families less than or equal to 80 My is given in Table 5, and the dating for all 125 families is available in the Supplemental Table 1. Table 5 gives the median age for the family and the upper and lower boundaries of the age.
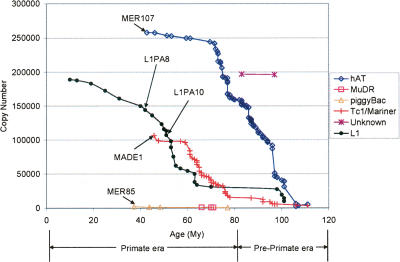
Figure 1.
Cumulative copy numbers for DNA element superfamilies and L1 elements according to age. The age of each family is plotted against the cumulative copy number of the superfamily. See Results and Methods sections for details on the calculation of the age for each family. (1) hAT superfamily elements were more intensively active during the pre-prosimian era (>63 My), decreasing in proliferation during the subsequent primate radiation (<63 My). (2) MuDR and piggyBac (e.g., MER85) superfamily elements were strictly active during the primate radiation. (3) Tc1/mariner (e.g., MADE1, Tigger1) superfamily elements were active before the primate radiation, but experienced a marked burst of activity during the primate radiation.
Table 5.
DNA elements less than 80 million years
This data was used to generate a plot of the age of DNA transposons and L1s as a function of their copy number (Fig. 1). This representation shows that our dating of DNA transposons is in good agreement with the nested insertion analysis, but provides a better resolution of the age of an individual family. For example, MER85 and MADE1 are among the youngest DNA elements, with estimated ages of ∼37 and ∼46 Mya, respectively, and both families included members nested within L1PA10 elements (Table 2), a subfamily that we dated as ∼51-My old (in agreement with Khan et al. 2006). Conversely, there are no DNA transposon families significantly younger than L1PA8, as indeed we found no instances of any DNA element nested within L1PA8 or younger. However, we could detect several L1PA8 elements nested within MER85 and MADE1 transposons (data not shown).
Our dating of individual DNA transposon families based on sequence divergence and calculated age is also largely congruent with the cross-species analysis. All elements classified as eutherian-wide from this analysis were found to be older than 65 My based on sequence divergence, with all but one family (MER53) being older than 70 My. MER53 is an outlier due to its unusually high 70% AT content (Table 5). All DNA transposon families classified as primate specific by the cross-species analysis were estimated to be between 57 and 78 My based on sequence divergence.
Figure 1 reveals that there were two bursts of DNA transposon activity in the time period between the mammalian radiation and the split of New World Monkeys from the primate ancestor. The first peak is the most pronounced and involves primarily members of the hAT superfamily and spanned a period of ∼40 My from a pre-primate era to early primate evolution (∼70 Mya). The second subsequent peak is strictly primate specific (from 80 to 63 Mya) and mostly implicated Tc1/mariner elements, although the greatest diversity of DNA transposons was active during this time, including hAT, MuDR, and PiggyBac elements (Fig. 1). The relatively more recent activity of Tc1/mariner elements compared with the most abundant hAT elements has been previously noticed (Lander et al. 2001). Our data indicates that the burst of Tc1/mariner activity reached a plateau that seems to coincide with the emergence of anthropoid primates, at ∼63 Mya. Three superfamilies (hAT, Tc1/mariner, PiggyBac) continued to be active in the anthropoid lineage, but there seems to be a sudden loss of activity of all DNA transposons shortly after the emergence of the new world monkeys ∼40 Mya.
Discussion
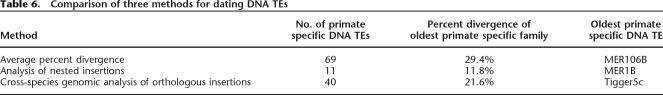
While the evolutionary history of human Alu and L1 retrotransposons has been studied intensively, the history of DNA transposons has largely been overlooked. In this study we have combined three different approaches to determine the average age of all 125 DNA transposon families known in the human genome. The results of the three approaches converge to reveal that a substantial fraction of human DNA transposon families (at least 40 and up to 69 families, see Tables 5, 6), representing at least ∼98,000 elements in our genome, were transpositionally active in the primate lineage. Below we first discuss the value of combining various methods for estimating the age of TEs, then turn to the specific implications of our findings for primate genome evolution and for understanding the forces underlying the amplification dynamics of TEs in mammalian genomes.
Table 6.
Comparison of three methods for dating DNA TEs
Combined methods provide a detailed estimate of the age of human DNA transposons
As new genome sequences are released, different methods for dating transposable elements are being developed that allow greater accuracy in estimating the age of TEs (Price et al. 2004; Salem et al. 2005; Caspi and Pachter 2006). This is a crucial aspect of genome research because TEs not only provide a rich fossil record to determine the pace and mode of molecular evolution (for example, see Waterston et al. 2002), but they are also major players in the structural evolution and regulation of genes and genomes (Feschotte et al. 2002; Deininger et al. 2003; Kazazian 2004; Cordaux et al. 2006). To our knowledge, our study is the first that uses a combination of three independent methods to evaluate the age of a broad population of TEs on a genome-wide scale.
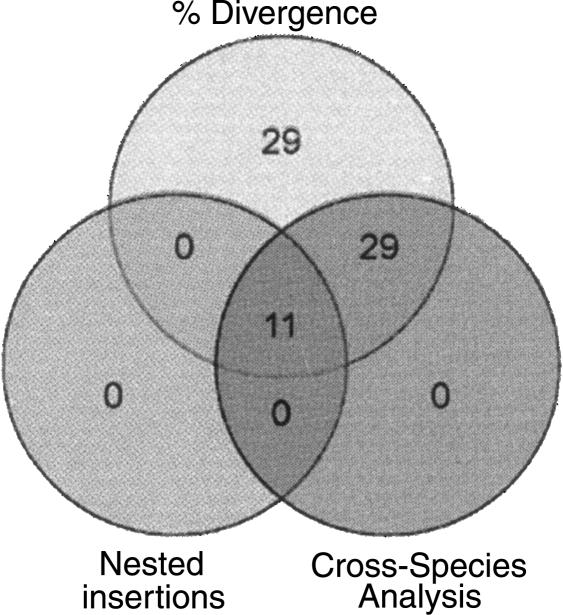
The first method, nested insertion analysis, capitalizes on the well-characterized history of L1 and Alu elements and provides an estimation of the relative age of TE families. This method does not rely on the molecular clock and can be performed even in the absence of genomic sequences for other closely related species. The shortcoming of this approach is that not every TE family member will necessarily suffer an insertion from every other TE family, thereby leading to gaps in the data, especially for TE families with low copy numbers. The second method (cross-species analysis of orthologous insertions) takes advantage of the large amount of sequence data recently generated for several mammalian species and is also independent of the molecular clock. These first two methods deliver a rough, yet robust evaluation of the time periods when the elements were active. This, in turn, provided us the means to calibrate the molecular clock at three different evolutionary time points, which allowed the refinement of the age of each human DNA transposon family based on nucleotide divergence of individual copies to their ancestral consensus sequence. There is only partial overlap between the results gathered by the three methods (Fig. 2), which emphasizes the value of combining the three methods to derive a reliable history of the entire population of human DNA transposons.
Figure 2.
Comparison of three independent methods for dating DNA transposons. Eleven distinct families of DNA transposons were shown to be primate specific by all three methods. Twenty-nine additional DNA element families were found to be primate specific by both the percent divergence and cross-species genomic analysis methods. Twenty-nine families predicted to be primate specific based solely on percent divergence and age were determined to be eutherian-wide by cross-species analysis.
Of the 125 DNA element families currently recognizable in the human genome, a total of 11 families could be classified as primate specific by all three methods (Fig. 2). Since nested insertion analysis does not allow us to examine all DNA transposon families, but only a subset of them, one cannot expect a complete overlap between the results produced by the three different methods. Sixty-nine families of DNA transposons were predicted to be primate specific based on sequence divergence data and thus, calculated age, alone (Table 5), and 40 of these families were confirmed to be primate specific by at least one of the two alternative methods. Hence, we believe that this set of 40 families provides a reliable, yet conservative estimate of DNA transposons that were integrated during primate evolution. The corresponding families range in age from MER85 (37 My) to Tigger5c (78 My) and have contributed 98,300 elements (totaling ∼33 Mb of sequence) to the human genome (Fig. 2). Furthermore, the results of divergence and cross-species and/or nested insertion analysis confirm that nearly one-fourth of these transposons (23,462 elements, ∼5 Mb) have likely been inserted since the split of anthropoid primates from prosimian primates, or within the last ∼63 My (according to Goodman 1999).
Thirty families were predicted to be primate specific based on their sequence divergence and calculated age but were shown to be eutherian-wide by cross-species analysis. However, for some of these families, such as MER53, Charlie5, and MER33 (65, 72, 72 My, respectively), we could detect copies present at orthologous positions in all eutherian species examined, which strongly indicates that at least a subset of family members inserted prior to the divergence of placental mammals. Thus, sequence divergence alone may not always be an accurate measure of the age of TEs. It could be that, for various reasons, members of these families evolve more slowly overall than other families. A non-mutually exclusive explanation is that these families include a subfamily of primate-specific elements as well as older elements. Further analyses are required to distinguish between these possibilities.
A general extinction of DNA transposon activity in the anthropoid lineage
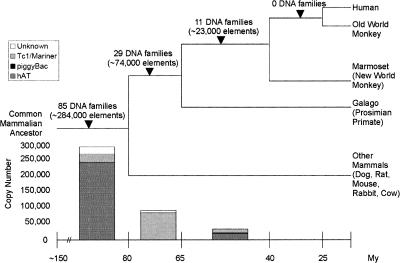
An interesting phenomenon is observed when looking at the overall history of DNA transposons in the mammalian and primate lineages (Fig. 3). Eighty-five families, or ∼291,000 DNA transposons, are shared between primates and other mammals. In contrast, 29 families, or ∼74,000 elements, were active specifically in primates prior to the split of emergence of anthropoids, and 11 families, or ∼23,000 elements, were integrated in anthropoid species. Thus, there was a steady decline in the activity of DNA transposons during primate evolution (see Figs. 1, 3). According to our combined age calculations, we found no evidence for DNA transposon families significantly younger than the divergence of new world monkeys, that is ∼40 Mya (Fig. 1; Table 5). Furthermore, we conducted a systematic survey for the presence/absence of human DNA transposons at orthologous positions in the nearly complete genome of the Rhesus macaque (an old world monkey) and could not uncover a single conclusive instance of a DNA transposon copy present in human, but missing in the macaque (data not shown). Thus, there is no evidence for the activity of any DNA transposons after the emergence of old world monkeys. The last active DNA transposon families represented in the human genome seem to have all become extinct in the relatively short evolutionary window (∼23 My) that separated prosimians and new world monkeys (Fig. 1). Yet, our study predicts that at the dawn of this extinction, some ∼40–55 Mya, there were at least 11 families from three different superfamilies active in the anthropoid genome (Fig. 1; Table 5). The majority of these elements were from the hAT superfamily (six families), while two Tc1/mariner and three piggyBac families comprised the rest of this group (Figs. 1, 3). This suggests that at least three distinct sources of transposases, with some of them represented by hundreds of seemingly intact copies (e.g., Hsmar1) (Robertson and Zumpano 1997; Cordaux et al. 2006) were shut down around the same evolutionary period.
Figure 3.
Summary of the activity of DNA transposons through primate evolution. The bar graph at the bottom of the figure represents the number of DNA elements active during each major lineage, broken down per superfamily. Note that no DNA elements were found to be active after the emergence of new world monkeys.
What could have provoked the extinction of DNA transposons that would not have affected the propagation of L1, which continued to thrive even after the emergence of new world monkeys (see Fig. 1; Khan et al. 2006)? A distinctive feature of the life cycle of DNA transposons is their apparent propensity for horizontal transmission (Silva and Kidwell 2000; Robertson 2002). Theory predicts that horizontal transfer is indeed critical for the maintenance of DNA transposons (Hickey 1982; Hartl et al. 1997). In contrast, horizontal transfer of LINEs occurs rarely, if ever (Eickbush and Malik 2002) and it is probably not an essential component to their maintenance (Wei et al. 2001; Kulpa and Moran 2006). It is tempting to speculate that the extinction of the DNA transposon population in the anthropoid lineage was linked to a sudden incapacity of these elements to undergo horizontal transmission. This could be due to the emergence of a host barrier aimed against the cellular entrance of TEs and other forms of invasive DNA. This would also explain the parallel regression of endogenous retroviruses during the same period of primate evolution (Lander et al. 2001). Interestingly, several defense mechanisms have been recently characterized that restrict retroviral activity in primates (Emerman 2006; Zennou and Bieniasz 2006). It could be that similar mechanisms have also compromised the propagation of DNA transposons in anthropoid primates.
Contribution of DNA transposons to primate genome evolution
One of the most striking findings of our study is that at least ∼74,000 of the DNA transposons now fixed in the human genome (∼33 Mb of DNA) were integrated during a period of <17 My, prior to the emergence of prosimian primates (∼63 Mya) but after the divergence of a primate ancestor from the closest mammalian clades represented in our data set (rat, mouse, and rabbit; ∼75–85 Mya, see Fig. 1). This is almost twice the number of L1 elements inserted during the same period and now fixed in the human genome (∼43,000 elements from the subfamilies L1PA15–16, L1PB3, L1MA2–3). Clearly, early primate evolution was a period of intense activity for DNA transposons. During the next phase of the primate radiation (63–40 Mya), i.e., after the split of prosimians, but prior to the emergence of new world monkeys, we estimated that ∼23,000 DNA elements were inserted and fixed in the human genome, adding at least ∼5 Mb to an ancestral anthropoid genome (Figs. 1, 3). Hence, the quantitative contribution of DNA-mediated transposition to the primate genome is far from negligible.
Prior to this study, the history of human DNA transposons has been largely neglected relative to those of retroelements (Alus and L1s). One reason for this is the common belief that active DNA transposon families have long been extinct and that they are currently only represented by very ancient molecular “fossils” immobilized in the genome. Indeed, unlike Alus and L1s (Deininger et al. 2003), there are no known cases of de novo insertion of any human DNA transposon. Our results support the conclusion that there has been little, if any, activity of DNA transposons in the ape lineage. On the other hand, our study demonstrates that many thousands of DNA elements have integrated and become fixed during the first half of primate evolution and that several high copy number families with >90% nucleotide identity among copies remain in the human genome (see average sequence divergence in Table 5). It is tempting to speculate that these primate-specific bursts of DNA transposition have had a strong impact on the structural evolution of primate genomes. DNA transposons have been frequently implicated in chromosomal rearrangements in plant and animal species, including deletions, inversions, duplications, translocations, and chromosome breakage mediated by interelement recombination or aberrant transposition events (for examples, see Lim and Simmons 1994; Caceres et al. 1999; Gray 2000; Zhang and Peterson 2004). Given the medical and evolutionary importance of chromosomal rearrangements in humans (Inoue and Lupski 2002; Eichler and Sankoff 2003; Feuk et al. 2006), the possible role of DNA transposons in shaping primate genomes warrants further investigation.
Methods
Calculation of average percent divergence from RepeatMasker output
The average percent divergence of each transposable element family was calculated using the RepeatMasker rmsk files from the UCSC Genome Browser for the May 2004 assembly. The percent divergence (milliDiv) of each distinct element within a transposable element family was weighted by the length of the element. The average percent divergence was weighted to account for the vast difference in sizes between currently recognized elements of the same family. To calculate the average percent divergence for each family, the percent divergence calculated by RepeatMasker was multiplied by the length of the element and the sum of all elements in each family was divided by the sum of the total length of all elements in the family.
Nested insertions
To find nested insertions, the RepeatMasker files from the UCSC Genome Browser and a Perl script (see Supplemental Material) were used to automate the analysis. Each Alu, L1, and DNA element in the RepeatMasker files was evaluated to see whether there was a repeat within 50 bp upstream and a repeat with 50 bp downstream. If so, the upstream and downstream repeats were compared to see whether the repName and Strand fields were the same. Next, the repeat positions of the ends of the upstream and downstream TEs were examined. If the upstream and downstream TEs both began within position 1–20 or both ended within 20 bp of the consensus length, the case was discarded, suggesting that they might represent a cluster of two elements of the same family independently inserted in close proximity and surrounding another element, rather than a single element disrupted by a nested element. Next, the coordinates of the two flanking repeats were checked to verify that the end of the first repeat was within ±20 bp of the start of the second repeat according to the family consensus sequence. If all of these conditions were met, the Alu, L1, or DNA element was classified as nested within each other.
Cross-species genomic analysis of orthologous insertions
To search for the presence or absence of DNA elements in marmoset and galago, sequences for each human DNA element present in an ENCODE region (http://www.nisc.nih.gov) were retrieved from the UCSC Genome Browser in September 2005. BAC sequences for each of the orthologous ENCODE regions in the marmoset and galago were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) using the accession ID given on the ENCODE website. From these sequences, custom BLAST databases were built for both the marmoset and galago using BLAST Version 2.2.11 (Altschul et al. 1990) and the sequences from the GenBank accessions. Each human DNA transposon, along with 100 bp of flanking sequences, were used as queries in BLASTN searches of the marmoset and galago custom databases. A repeat was classified as present at the orthologous position if at least one of the two flanking regions and at least 50% of the element from the human sequence were found in the marmoset or galago.
Calculation of substitution rates and dating according to sequence divergence
The May 2004 human genome sequence (hg17) was downloaded from the UCSC Genome Browser. TEs were masked locally using RepeatMasker version 3.1.5 with the March 14, 2006 library from RepBase Update. A Perl script (see Supplemental Material) was used to parse the RepeatMasker align files and generate a single, concatenated sequence for each different chromosomal repeat along with the corresponding consensus sequence. The concatenated sequences had all CG dinucleotides (for + strand) and GC dinucleotides (for – strand), as well as non-ATGC characters, removed. These chromosomal repeat and consensus sequences were then combined and analyzed using PAML version 3.15 (Yang 1997). Each file was analyzed using the REV model with the clock = 1 option. The corrected number of substitutions per site was calculated as one-half of the branch length, since the consensus sequence does not evolve.
To calculate the substitution rate, the corrected substitutions per site was divided by the median age for the class (anthropoid, primate, or eutherian specific). The rate for each TE within the age class was weighted by the percentage of the total number of bases that TE comprised of the total base length for the entire class. These weighted rates were then summed, giving a corrected substitution rate for the entire class. The age of the family was calculated by multiplying the corrected substitution rate by the corrected percent divergence for the family.
Acknowledgments
We thank Ellen Pritham for critical reading of the manuscript, Don Hucks for support with PAML, and members of the Genome Biology Group at UTA for stimulating discussions. This work was supported by funds from the University of Texas at Arlington.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5826307
References
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Auge-Gouillou C., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Pollet N., Hamelin M.H., Meunier-Rotival M., Periquet G., Hamelin M.H., Meunier-Rotival M., Periquet G., Meunier-Rotival M., Periquet G., Periquet G. Human and other mammalian genomes contain transposons of the mariner family. FEBS Lett. 1995;368:541–546. doi: 10.1016/0014-5793(95)00735-r. [DOI] [PubMed] [Google Scholar]
- Caceres M., Ranz J.M., Barbadilla A., Long M., Ruiz A., Ranz J.M., Barbadilla A., Long M., Ruiz A., Barbadilla A., Long M., Ruiz A., Long M., Ruiz A., Ruiz A. Generation of a widespread Drosophila inversion by a transposable element. Science. 1999;285:415–418. doi: 10.1126/science.285.5426.415. [DOI] [PubMed] [Google Scholar]
- Caspi A., Pachter L., Pachter L. Identification of transposable elements using multiple alignments of related genomes. Genome Res. 2006;16:260–270. doi: 10.1101/gr.4361206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R., Udit S., Batzer M.A., Feschotte C., Udit S., Batzer M.A., Feschotte C., Batzer M.A., Feschotte C., Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl. Acad. Sci. 2006;103:8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N.L., Craigie R., Gellert M., Lambowitz A.M., Craigie R., Gellert M., Lambowitz A.M., Gellert M., Lambowitz A.M., Lambowitz A.M. Mobile DNA II. American Society for Microbiology Press; Washington, D.C.: 2002. [Google Scholar]
- Deininger P.L., Moran J.V., Batzer M.A., Kazazian H.H., Jr., Moran J.V., Batzer M.A., Kazazian H.H., Jr., Batzer M.A., Kazazian H.H., Jr., Kazazian H.H., Jr. Mobile elements and mammalian genome evolution. Curr. Opin. Genet. Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Demattei M.V., Auge-Gouillou C., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Auge-Gouillou C., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Pollet N., Hamelin M.H., Meunier-Rotival M., Bigot Y., Hamelin M.H., Meunier-Rotival M., Bigot Y., Meunier-Rotival M., Bigot Y., Bigot Y. Features of the mammal mar1 transposons in the human, sheep, cow, and mouse genomes and implications for their evolution. Mamm. Genome. 2000;11:1111–1116. doi: 10.1007/s003350010204. [DOI] [PubMed] [Google Scholar]
- Eichler E.E., Sankoff D., Sankoff D. Structural dynamics of eukaryotic chromosome evolution. Science. 2003;301:793–797. doi: 10.1126/science.1086132. [DOI] [PubMed] [Google Scholar]
- Eickbush T.H., Malik H.S., Malik H.S. Origins and evolution of retrotransposons. In: Craig N.L., et al., editors. Mobile DNA II. ASM Press; Washington, D.C.: 2002. pp. 1111–1144. [Google Scholar]
- Emerman M. How TRIM5α defends against retroviral invasions. Proc. Natl. Acad. Sci. 2006;103:5249–5250. doi: 10.1073/pnas.0601373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C. Merlin, a new superfamily of DNA transposons identified in diverse animal genomes and related to bacterial IS1016 insertion sequences. Mol. Biol. Evol. 2004;21:1769–1780. doi: 10.1093/molbev/msh188. [DOI] [PubMed] [Google Scholar]
- Feschotte C., Zhang X., Wessler S.R., Zhang X., Wessler S.R., Wessler S.R. Miniature inverted-repeat transposable elements and their relationship to established DNA transposons. In: Craig N.L., et al., editors. Mobile DNA II. ASM Press; Washington, D.C: 2002. pp. 1147–1158. [Google Scholar]
- Feuk L., Carson A.R., Scherer S.W., Carson A.R., Scherer S.W., Scherer S.W. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- Goodman M. Rates of molecular evolution: The hominoid slowdown. Bioessays. 1985;3:9–14. doi: 10.1002/bies.950030104. [DOI] [PubMed] [Google Scholar]
- Goodman M. The genomic record of humankind’s evolutionary roots. Am. J. Hum. Genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray Y.H. It takes two transposons to tango: Transposable-element-mediated chromosomal rearrangements. Trends Genet. 2000;16:461–468. doi: 10.1016/s0168-9525(00)02104-1. [DOI] [PubMed] [Google Scholar]
- Hagemann S., Pinsker W., Pinsker W. Drosophila P transposons in the human genome? Mol. Biol. Evol. 2001;18:1979–1982. doi: 10.1093/oxfordjournals.molbev.a003739. [DOI] [PubMed] [Google Scholar]
- Hartl D.L., Lohe A.R., Lozovskaya E.R., Lohe A.R., Lozovskaya E.R., Lozovskaya E.R. Modern thoughts on an ancyent marinere: Function, evolution, regulation. Annu. Rev. Genet. 1997;31:337–358. doi: 10.1146/annurev.genet.31.1.337. [DOI] [PubMed] [Google Scholar]
- Hickey D.A. Selfish DNA: A sexually-transmitted nuclear parasite. Genetics. 1982;101:519–531. doi: 10.1093/genetics/101.3-4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K., Lupski J.R., Lupski J.R. Molecular mechanisms for genomic disorders. Annu. Rev. Genomics Hum. Genet. 2002;3:199–242. doi: 10.1146/annurev.genom.3.032802.120023. [DOI] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J., Klonowski P., Kohany O., Walichiewicz J., Kohany O., Walichiewicz J., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kapitonov V., Jurka J., Jurka J. The age of Alu subfamilies. J. Mol. Evol. 1996;42:59–65. doi: 10.1007/BF00163212. [DOI] [PubMed] [Google Scholar]
- Kapitonov V.V., Jurka J., Jurka J. Harbinger transposons and an ancient HARBI1 gene derived from a transposase. DNA Cell Biol. 2004;23:311–324. doi: 10.1089/104454904323090949. [DOI] [PubMed] [Google Scholar]
- Kazazian H.H., Jr. Mobile elements: Drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- Khan H., Smit A., Boissinot S., Smit A., Boissinot S., Boissinot S. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 2006;16:78–87. doi: 10.1101/gr.4001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kulpa D.A., Moran J.V., Moran J.V. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat. Struct. Mol. Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lim J.K., Simmons M.J., Simmons M.J. Gross chromosome rearrangements mediated by transposable elements in Drosophila melanogaster. Bioessays. 1994;16:269–275. doi: 10.1002/bies.950160410. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Wade C.M., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Mikkelsen T.S., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Karlsson E.K., Jaffe D.B., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Jaffe D.B., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Kamal M., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Clamp M., Change J.L., Kulbokas E.J., III, Zody M.C., Change J.L., Kulbokas E.J., III, Zody M.C., Kulbokas E.J., III, Zody M.C., Zody M.C., et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Morgan G.T. Identification in the human genome of mobile elements spread by DNA-mediated transposition. J. Mol. Biol. 1995;254:1–5. doi: 10.1006/jmbi.1995.0593. [DOI] [PubMed] [Google Scholar]
- Murphy W.J., Pevzner P.A., O’Brien S.J., Pevzner P.A., O’Brien S.J., O’Brien S.J. Mammalian phylogenomics comes of age. Trends Genet. 2004;20:631–639. doi: 10.1016/j.tig.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Oosumi T., Belknap W.R., Garlick B., Belknap W.R., Garlick B., Garlick B. Mariner transposons in humans. Nature. 1995;378:672. doi: 10.1038/378672a0. [DOI] [PubMed] [Google Scholar]
- Plasterk R.H. The origin of footprints of the Tc1 transposon of Caenorhabditis elegans. EMBO J. 1991;10:1919–1925. doi: 10.1002/j.1460-2075.1991.tb07718.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Eskin E., Pevzner P.A., Eskin E., Pevzner P.A., Pevzner P.A. Whole-genome analysis of Ali repeat elements reveals complex evolutionary history. Genome Res. 2004;14:2245–2252. doi: 10.1101/gr.2693004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H.M. Members of the pogo superfamily of DNA-mediated transposons in the human genome. Mol. Gen. Genet. 1996;252:761–766. doi: 10.1007/BF02173985. [DOI] [PubMed] [Google Scholar]
- Robertson H.M. Evolution of DNA transposons in eukaryotes. In: Craig N.L., et al., editors. Mobile DNA II. ASM Press; Washington, D.C: 2002. pp. 1093–1110. [Google Scholar]
- Robertson H.M., Martos R., Martos R. Molecular evolution of the second ancient human mariner transposon, Hsmar2, illustrates patterns of neutral evolution in the human genome lineage. Gene. 1997;205:219–228. doi: 10.1016/s0378-1119(97)00471-x. [DOI] [PubMed] [Google Scholar]
- Robertson H.M., Zumpano K.L., Zumpano K.L. Molecular evolution of an ancient mariner transposon, Hsmar1, in the human genome. Gene. 1997;205:203–217. doi: 10.1016/s0378-1119(97)00472-1. [DOI] [PubMed] [Google Scholar]
- Salem A., Ray D.A., Hedges D.J., Jurka J., Batzer M.A., Ray D.A., Hedges D.J., Jurka J., Batzer M.A., Hedges D.J., Jurka J., Batzer M.A., Jurka J., Batzer M.A., Batzer M.A. Analysis of the human Alu Ye lineage. BMC Evol. Biol. 2005;5:18–27. doi: 10.1186/1471-2148-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.C., Kidwell M.G., Kidwell M.G. Horizontal transfer and selection in the evolution of P elements. Mol. Biol. Evol. 2000;17:1542–1557. doi: 10.1093/oxfordjournals.molbev.a026253. [DOI] [PubMed] [Google Scholar]
- Sarkar A., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Sim C., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Hong Y.S., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Hogan J.R., Fraser M.J., Robertson H.M., Collins F.H., Fraser M.J., Robertson H.M., Collins F.H., Robertson H.M., Collins F.H., Collins F.H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genomics. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- Smit A.F.A. Identification of a new, abundant superfamily of mammalian LTR-transposons. Nucleic Acids Res. 1993;21:1863–1872. doi: 10.1093/nar/21.8.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F. Interspersed repeats and other mementos of transposable elements in mammalian genomes. Curr. Opin. Genet. Dev. 1999;9:657–663. doi: 10.1016/s0959-437x(99)00031-3. [DOI] [PubMed] [Google Scholar]
- Smit A.F.A., Riggs A.D., Riggs A.D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit A.F.A., Toth G., Riggs A.D., Jurka J., Toth G., Riggs A.D., Jurka J., Riggs A.D., Jurka J., Jurka J. Ancestral, mammalian-wide subfamilies of LINE-1 repetitive sequences. J. Mol. Biol. 1995;246:401–417. doi: 10.1006/jmbi.1994.0095. [DOI] [PubMed] [Google Scholar]
- Szak S.T., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Pickeral O.K., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Makalowski W., Boguski M.S., Landsman D., Boeke J.D., Boguski M.S., Landsman D., Boeke J.D., Landsman D., Boeke J.D., Boeke J.D. Molecular archeology of L1 insertions in the human genome. Genome Biol. 2002;3:1–18. doi: 10.1186/gb-2002-3-10-research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavare S. Lectures in mathematics in the life sciences. Vol 17. American Mathematical Society; Providence, RI: 1986. Some probabilistic and statistical problems on the analysis of DNA sequences; pp. 57–86. [Google Scholar]
- Waterston R.H., Lindblad-Toh K., Bimey E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Lindblad-Toh K., Bimey E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Bimey E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Abril J.F., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwal P., Agarwala R., Ainscough R., Alexandersson M., An P., Agarwala R., Ainscough R., Alexandersson M., An P., Ainscough R., Alexandersson M., An P., Alexandersson M., An P., An P., et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Wei W., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Gilbert N., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ooi S.L., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Lawler J.F., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Ostertag E.M., Kazazian H.H., Boeke J.D., Moran J.V., Kazazian H.H., Boeke J.D., Moran J.V., Boeke J.D., Moran J.V., Moran J.V. Human L1 retrotransposition: cis preference versus trans complementation. Mol. Cell. Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J., Salem A., Hedges D.J., Kilroy G.E., Watkins W.S., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Salem A., Hedges D.J., Kilroy G.E., Watkins W.S., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Hedges D.J., Kilroy G.E., Watkins W.S., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Kilroy G.E., Watkins W.S., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Watkins W.S., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Schienman J.E., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Stewart C., Jurka J., Jorde L.B., Batzer M.A., Jurka J., Jorde L.B., Batzer M.A., Jorde L.B., Batzer M.A., Batzer M.A. Comprehensive analysis of two Alu Yd subfamilies. J. Mol. Evol. 2003;57:S76–S89. doi: 10.1007/s00239-003-0009-0. [DOI] [PubMed] [Google Scholar]
- Xing J., Hedges D.J., Han K., Wang H., Cordaux R., Batzer M.A., Hedges D.J., Han K., Wang H., Cordaux R., Batzer M.A., Han K., Wang H., Cordaux R., Batzer M.A., Wang H., Cordaux R., Batzer M.A., Cordaux R., Batzer M.A., Batzer M.A. Alu element mutation spectra: Molecular clocks and the effect of DNA methylation. J. Mol. Biol. 2004;344:675–682. doi: 10.1016/j.jmb.2004.09.058. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML: A program package for phylogenetic analysis by maximum likelihood. Comput. Appl. Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yi S., Ellsworth D.L., Li W.H., Ellsworth D.L., Li W.H., Li W.H. Slow molecular clocks in Old World monkeys, apes, and humans. Mol. Biol. Evol. 2002;19:2191–2198. doi: 10.1093/oxfordjournals.molbev.a004043. [DOI] [PubMed] [Google Scholar]
- Zennou V., Bieniasz P.D., Bieniasz P.D. Comparative analysis of the antiretroviral activity of APOBEC3G and APOBEC3F from primates. Virology. 2006;349:31–40. doi: 10.1016/j.virol.2005.12.035. [DOI] [PubMed] [Google Scholar]
- Zhang J., Peterson T., Peterson T. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics. 2004;167:1929–1937. doi: 10.1534/genetics.103.026229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Jiang N., Feschotte C., Wessler S.R., Jiang N., Feschotte C., Wessler S.R., Feschotte C., Wessler S.R., Wessler S.R. Distribution and evolution of PIF- and Pong-like transposons and their relationships with Tourist-like MITEs. Genetics. 2004;166:971–986. doi: 10.1093/genetics/166.2.971. [DOI] [PMC free article] [PubMed] [Google Scholar]