Abstract
The human Y chromosome, because it is enriched in repetitive DNA, has been very intractable to genetic and molecular analyses. There is no previous evidence for developmental stage- and testis-specific transcription from the male-specific region of the Y (MSY). Here, we present evidence for the first time for a developmental stage- and testis-specific transcription from MSY distal heterochromatic block. We isolated two novel RNAs, which localize to Yq12 in multiple copies, show testis-specific expression, and lack active X-homologs. Experimental evidence shows that one of the above Yq12 noncoding RNAs (ncRNAs) trans-splices with CDC2L2 mRNA from chromosome 1p36.3 locus to generate a testis-specific chimeric β sv13 isoform. This 67-nt 5′UTR provided by the Yq12 transcript contains within it a Y box protein-binding CCAAT motif, indicating translational regulation of the β sv13 isoform in testis. This is also the first report of trans-splicing between a Y chromosomal and an autosomal transcript.
The Y chromosome plays a role in sex determination and spermatogenesis in mammals. The genes responsible for these have been localized to the short and euchromatic long arms of the human Y. Although the human genome has been sequenced, the 40-Mb heterochromatic block on the long arm of human Y chromosome remains recalcitrant with respect to sequence information and gene content. There has been no evidence of transcription from this region. Classically, constitutive heterochromatin has been considered as compact and inert and the degree of decondensation of a chromatin region is considered as a reflection of gene activity (Weintraub and Groudine 1976; Guttenbach et al. 1989). Yq heterochromatin undergoes developmental stage-specific condensation and expansion in testicular Sertoli cells and germ cells, although no evidence for transcription was found in the decondensed signals (Speed et al. 1995). Nevertheless, examples of transcription from heterochromatic sequences are available in literature (Tulin et al. 2002). Transcription from opposite strands of DNA were reported from the long tracts of satellite DNA present on lampbrush chromosome loops within histone gene clusters of the newt Notophthalmus (Diaz et al. 1981). The complementary strands are differentially transcribed from the mouse major γ satellite during development (Rudert et al. 1995) and from the satellite III sequences localizing to human chromosome 9q12 during heat shock (Jolly et al. 2004). Heterogeneously sized transcripts have been reported from the Asty sequences localizing to the mouse Y long arm (Toure et al. 2005). Few genes have been identified in the heterochromatic Y chromosome of Drosophila, suggesting that there could be some genes in the heterochromatic block of the human Y as well (Bachtrog and Charlesworth 2001). However, there is no evidence for transcription from the major distal heterochromatic block (Skaletsky et al. 2003). With increasing evidence on transcription from various repeats, the 40-Mb heterochromatic block, enigmatically poised at the rear end of the long arm of human Y, was pursued for traces of gene activity.
Results
Developmental stage-specific expression
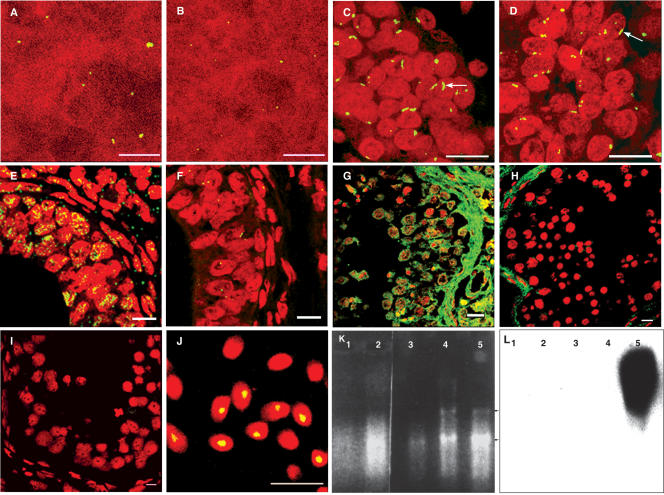
A sex- and species-specific genomic clone 102d(2) from the long arm of human Y chromosome was identified earlier in the lab (Singh et al. 1994). Fluorescence in situ hybridization (FISH) on to normal metaphase plates showed that these sequences localized to the distal heterochromatic block of Yq (Yq12). Sequencing of the genomic clone 102d(2) showed short ORFs within these sequences. This clone was then used as a probe for FISH on human testicular sections at various developmental stages. Testis and skin sections at 20 wk showed a single compacted signal within each nucleus (Fig. 1A,B) indicative of hybridization to corresponding sequences in the genome. Testis sections at 32 wk showed an elongated signal in ∼30% of the nuclei (Fig. 1C), indicating decondensation of this region of the chromosome. RNase treatment, however, showed a marginal reduction in the fluorescence intensity (Fig. 1D). In situ hybridization done on 36-wk testicular sections showed widely spread intense fluorescence signals in the nuclei of most cells (Fig. 1E). RNase-treated, 36-wk testicular sections showed only single dot-like signals in the majority of the cells, corresponding with hybridization to DNA from the locus (Fig. 1F). This indicated that the intense fluorescence signals filling the nuclei correspond to the presence of abundant RNA at 36 wk. Adult testicular sections also showed intense widely spread fluorescence signals in all the cells (Fig. 1G), which were erased by RNase treatment (Fig. 1H). A control experiment using a nontranscribing sequence (50f2) as a probe (Jobling 1994) showed only dot-like signals corresponding to DNA (Fig. 1I) in adult testis sections, further confirming that the fluorescence signals generated using 102d(2) were due to transcripts. FISH on ejaculated sperms showed condensed signals in sperm nuclei containing Y chromosomes (Fig. 1J). Thus, the Yq12 genomic probe reveals a temporal decondensation and subsequent transcription from the locus from late embryogenesis onward. Northern analysis using RNA from 20-wk testis, 32-wk testis, kidney, and spleen, and testis tissues from 36-wk confirmed developmental stage-specific and tissue-specific expression (Fig. 1L). Figure 1K shows the corresponding RNA lanes. Large heterogeneous transcripts were observed only in testicular RNA from 36-wk testis. Heterogeneously sized low-intensity signal was seen at 32 wk after prolonged exposure, indicating the presence of low levels of RNA at this stage (data not shown). Earlier developmental stages and other tissues did not show any signal.
Figure 1.
Developmental stage-specific decondensation and expression from human Yq12 using the clone 102d(2) (AY598345) from the locus. (A,B) Condensed fluorescence signals from testis and skin sections at 20-wk embryonic stage. (C) Elongated signals (arrow) indicating decondensation of the region at 32 wk of gestation. (E) A testicular section from 36 wk of gestation showing abundant transcripts in nuclei. (G) The expression profile from an adult testicular section, showing the presence of Yq12 complementary RNA in the cytoplasm also. (D,F,H) The corresponding RNase treated sections. (I) The result of hybridization using a nontranscribing 50f2 probe. (J) FISH on to ejaculated sperms with condensed signals in the nuclei. (Bars, 10 μm). (K,L) The developmental stage-specific RNAs and corresponding Northern blot with the Yq12 genomic clone 102d(2) as the probe. (Lanes 1–5) 20-wk testis, 36-wk kidney, 36-wk spleen, 32-wk testis, and 36-wk testis, respectively. (Arrows indicate the positions of 18s and 28s rRNAs). A heterogeneous signal of ∼5–9 kb is obtained only in 36-wk testis, thus indicating a testis-specific and developmental stage-specific expression.
cDNA clones localizing to Yq12 show testis-specific expression
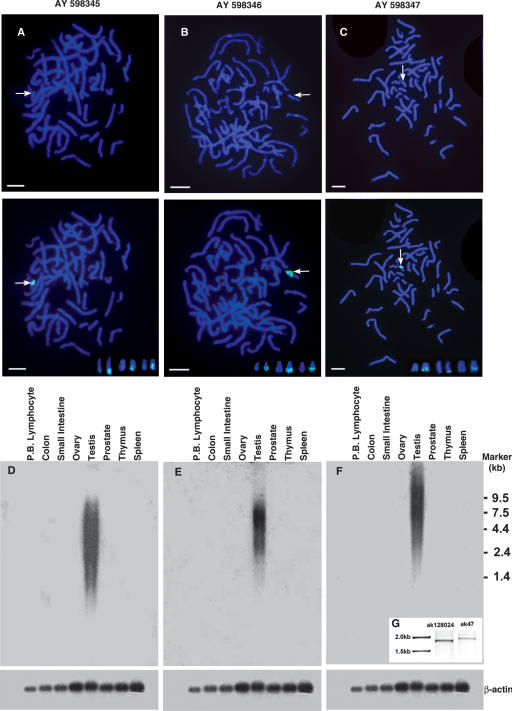
The human testicular cDNA library and multi-tissue Northern blots containing RNA from a panel of human tissues purchased from Clontech were used for further experiments. The localization of the genomic clone to Yq12 was confirmed once again by chromosomal FISH (Fig. 2A). Screening of the cDNA library with 102d(2) identified two male-specific clones localizing to Yq12 on normal metaphase spreads by FISH (Fig. 2B,C). The DNA sequences of genomic clone 102d(2) (2189 bp) and the two cDNA clones (816 and 1441 bp, respectively) have been deposited in GenBank (accession nos. AY598345, AY598346, and AY598347). These cDNA sequences are novel polyadenylated RNAs. When the genomic clone 102d(2) and the cDNA clones localizing to Yq12 were used as probes on multi-tissue poly(A) RNA Northern blots, heterogeneously sized transcripts were observed only in testicular samples (Fig. 2D,E,F). The RNA profiles on the autoradiograms with the two cDNAs (AY598347 and AY598346) differed with respect to molecular weight range, indicating that the RNAs identified by the two cDNAs are different. Poly(A) ends of the two cDNAs AY598346 and AY598347 were obtained by 3′RACE reaction and sequencing more clones from the library, respectively.
Figure 2.
Testis-specific expression of the genomic clone AY598345 and the cDNAs (AY598346, AY598347) and their localization to Yq12. (A,B,C, top) Arrows indicate the brightly fluorescing, distal heterochromatic blocks on the Y. (A,B,C, middle) Localization of the genomic and the cDNA clones to the heterochromatic blocks. (Insets, bottom) Signals on Yq12 from three more metaphases for each of the clones (Bars, 5 μm). (D,E,F) A testis-specific expression of the genomic and the cDNA clones on multi-tissue RNA blots. Large heterogeneously sized transcripts are observed for all of the clones. (D) A smear ranging from ∼9.5 to 1 kb. (E) A smear ranging from about 8 to 1.2 kb, with AY598346; (F) a smear ranging from about 20 to 1.2 kb, with the AY598347 probe. Corresponding β-actin controls are given below. (G) The results of the RT–PCR for the AK128024 (obtained from NCBI database, Primers: Forward- TCCTTTCGAGCCCTTTCAATTTG, Reverse- ATGCCCTTGAATTAAATGGACTG), and AK47, obtained by merger of AK128024 and our cDNA AY598347 (Forward- TCCTTTCGAGCCCTTTCAATTTG, Reverse- TGGAACAGAGAGCAATGGTATAG).
DYZ1 repeats are transcribed
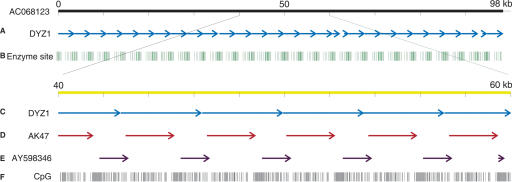
A search of the nucleotide database with all three of our clones picked up two Y chromosomal sequences, AC068123, a 98-kb BAC clone, and X06228, described as DYZ1 with zero e-values. Comparison of the two against each other showed that the BAC clone was comprised entirely of repeats of the 3.5-kb DYZ1 clone. There were 27 tandem copies of DYZ1 within the 98-kb clone (Fig. 3A). Analysis of restriction enzyme sites on the 98-kb BAC clone showed a clustering of 27 restriction enzyme sites (Fig. 3B). Beside the genomic clones AC068123 and X06228, BLASTN analysis using 102d(2) and our cDNAs identified a human testicular cDNA AK128024 (1986 bp). Rigorous pairwise alignment of our cDNAs with AK128024, using matcher from the EMBOSS suite of programs showed that AK128024 aligned with AY598347 end-to-end with 92% identity over a stretch of 600 bp. The two sequences were merged using the merger program from EMBOSS to form a larger fragment of 2151 bp, which we named AK47. Using total RNA from testicular tissue purchased from Clontech, expression of AK128024 and AK47 were experimentally confirmed by RT–PCR (Fig. 2G). The two partial cDNAs, AK47 and AY598346, spanned the complete BAC clone, alternating with each other repeatedly (Fig. 3D,E). Thus, expression analysis as well as computational screening indicate multiple transcripts from within the Yq12 region. As the BAC clone was comprised entirely of DYZ1 sequences, what we observe here is the transcription from DYZ1 repeats. We also observed a clustering and 2.8 times the frequency of CpG dinucleotides within the region (Fig. 3F).
Figure 3.
Mapping of DYZ1 repeats and cDNA clones on to the 98-kb BAC clone. (A) Twenty-seven copies of DYZ1 on the BAC. (B) The clustering of restriction enzyme sites (one inch represents 10 kb). (Bottom) A zoom in into a 20-kb region of the BAC. (C) The head to tail tandem repetition of DYZ1 on the BAC. (D,E) The arrangement of the cDNAs AK47 and AY598346 with respect to one another and the DYZ1 repeat. (F) The distribution of CpG dinucleotides (one inch represents 2 kb).
Testis-specific chimeric transcript between Yq12 noncoding RNA and CDC2L2 from chromosome 1p36.3
In order to find any known transcripts that may correspond to the Yq12 region, the UTR database (http://bighost.area.ba.cnr.it/BIG/ UTRHome/) was searched. This revealed that a 67-nt region of AY598346 is present within the 5′UTR of testicular β sv13 isoform of CDC2L2 mRNA (AF067525) (Fig. 4A) with 98.5% identity. There was a single nucleotide change from T in the noncoding RNA to C in the UTR of CDC2L2 mRNA at position 57. The coding sequence begins from 68 nt onward in this isoform. BLAST analysis using the 67-nt UTR alone against the NCBI nucleotide database and human genome resources identified three sequences with significant e-values. They were the CDC2L2 β sv13 isoform mRNA and the genomic clones for DYZ1 and the 98-kb BAC clone (AC068123) localizing to Yq12. The CDC2L2 genomic clone, however, was not identified in the database when the 67-nt UTR was used as the query, showing that this segment is not present in the cognate genomic locus at chromosome 1p36.3. This clearly shows that 67-nt UTR is present only on the Y chromosome and on the β sv13 isoform mRNA of CDC2L2.
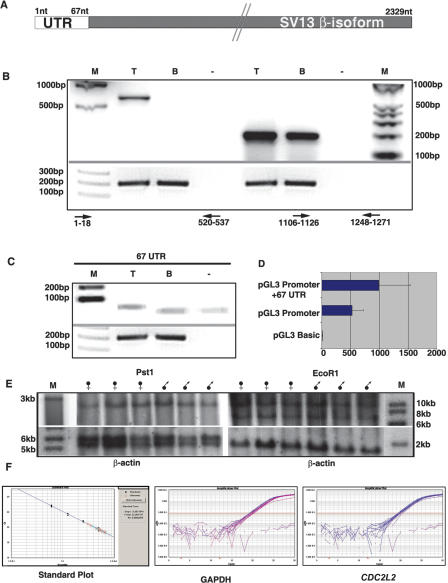
Figure 4.
RT–PCRs showing the presence of chimeric transcript. (A) A diagram of the 2329-nt-long CDC2L2 β sv 13 isoform with the 67-nt UTR at the 5′ end and the downstream coding region. (B) The presence of a chimeric transcript: a product of the expected size is seen by RT–PCR in testis (T) but not in the brain (B); (Primers: Forward- GAATTCCATTCCATTCCA, Reverse GTTCGGTGATGTGCAGAC). (Lanes 5,6) Amplification of CDC2L2 in both testis and brain using primers to exons common to both the isoforms (Forward- CCCAAGTACCTGCCGGCCCTG, Reverse- AGCCCTCCTTCTCCTTCTCCATCT). (–) Negative control. (C) Amplification of the 67-nt UTR from testis (T) but not from brain tissue (B) (Primers: Forward- GAATTCCATTCCATTCCA, Reverse- GCAGTAGAAGAGAATAGA). (M) The marker lanes. RT–PCR using GAPDH primers as loading controls is given below the corresponding lanes. (D) The results of the reporter assay. pGL3 promoter containing the 67-nt UTR shows a ∼1.88-fold enhancement of luciferase activity when compared with the promoter vector alone. Standard deviations are indicated by the bars on the histograms. (E) The Southern blots of normal male and female DNAs digested with Pst1 and EcoR1 and probed with CDC2L2. Comparison with the β-actin control shows that there is no copy number difference between the male and female DNAs, thereby confirming the presence of the gene on an autosome. (F) The standard plot and amplification plots of CDC2L2 [blue] (Primers: Forward- CCCAAGTACCTGCCGGCCCTG, Reverse- AGCCCTCCTTCTCCTTCTC CATCT) and GAPDH [purple] (Primers: Forward- ACGGGAAGCTCACTGGCATGG, Reverse- CAACAGC GACACCCACTCCTC) using DNA from XO, XX, XY, and XYY individuals. These clearly show that there is no copy number difference with increasing copies of Y chromosome in CDC2L2 and GAPDH. Therefore, there is no copy of CDC2L2 on the Y chromosome.
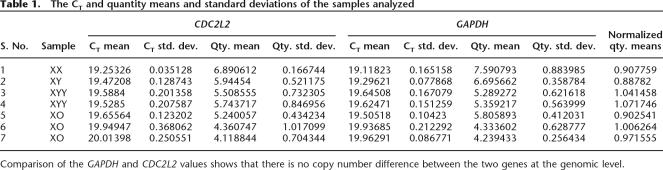
RT–PCR using testicular RNA procured from Clontech with primers spanning the 67-nt and at ∼540 nt of CDC2L2 β SV 13 isoform gave a product in testis, but not in brain (Fig. 4B), confirming that a chimeric RNA is indeed present in testis. PCR analysis of CDC2L2 internal exons present within both the testis and brain isoforms showed the transcription of CDC2L2 in both the tissues (Fig. 4B). Primers to the 67-nt UTR alone gave a product in testis but not in brain, further showing testis specificity of the UTR (Fig. 4C). The 67-bp 5′UTR region was cloned into the pGL3 promoter vector to assay for its effect on the translation of the reporter gene. Analysis of the luciferase activity in the transfected HeLa cells showed that the activity of the gene is enhanced by ∼1.88-fold in the presence of the 67-bp fragment as compared with the pGL3 promoter vector (Fig. 4D). Southern blot analysis using DNA from three normal male and three female individuals after digestion with two different restriction enzymes, EcoR1 and Pst1, and on probing with CDC2L2 gave comparable intensities of signals (Fig. 4E). This showed that there is no difference in copy number of CDC2L2 between males and females, thereby ruling out the possibility of the presence of this gene on the Y chromosome. RT–PCR analysis showed that CT values of GAPDH and CDC2L2 in DNA from XO, XX, XY, and XYY individuals containing varying numbers of Y chromosomes do not differ significantly (Fig. 4F; Table 1). This further confirmed that there are no copies of CDC2L2 on the Y chromosome.
Table 1.
The CT and quantity means and standard deviations of the samples analyzed
Comparison of the GAPDH and CDC2L2 values shows that there is no copy number difference between the two genes at the genomic level.
Discussion
Testis-specific transcription from the heterochromatic block Yq12
Since the inception and the coining of the term “heterochromatin” by Emil Heitz in 1928 (Heitz 1928), heterochromatic regions have been accepted as highly condensed loci devoid of genes and, hence, functionally inert. In fact, the extent of compaction or expansion of chromatin structure is considered as a reflection of gene activity from the corresponding region (Weintraub and Groudine 1976; Guttenbach et al. 1989). We have observed decondensation and subsequent expression from the distal heterochromatic block of the long arm of human Y chromosome, as we not only observe testis-specific expression on multi-tissue Northerns, but have also localized cDNAs to Yq12. Indeed, what we see on tissue in situ hybridizations is a decondensation of the region at 32 wk of gestation and expression from 36 wk onward. Decondensation and condensation of satellite sequences were reported from within the testicular tissues of human and mouse embryos, but transcription was found only in mouse (Rudert et al. 1995) and not in human (Speed et al. 1995). Here we show definitive evidence for transcription from the heterochromatic Yq12 block. There is growing evidence that repeated sequences of the eukaryotic genome are transcribed (Diaz et al. 1981; Fornace Jr. and Mitchell 1986; Kimura et al. 2001; Tulin et al. 2002; Jolly et al. 2004). Although the function of most of the satellite DNA remained questionable a while ago (Csink and Henikoff 1998), more recently there are reports of functional noncoding RNAs, which are transcribed from repeats within the genome (Mattick and Makunin 2006).
DYZ1 repeats are transcribed
The only sequence information available regarding the Yq12 region was the localization of the DYZ1 and DYZ2 repeats in ∼4000 and 2000 copies, respectively (Manz et al. 1992). Sequence comparison of our clones with the 98-kb BAC showed that this clone is a repeat of DYZ1, containing 27 copies of the same within it. Our cDNAs also localize to the clone in a repetitive fashion; therefore, it is the transcription of the DYZ1 repeat that is observed here. DYZ1 RNAs are transcribed during embryonic stages, similar to some of the other testicular transcripts, and these transcripts are confined to the nuclei at this stage. In the adult testicular tissue, the transcripts localize mainly in the cytoplasm. Testicular RNAs are known to be transcribed earlier in development and sequestered as translationally repressed messenger Ribonucleoprotein Particles (mRNPs) until required (Schafer et al. 1995; Kleene 2003, and references therein). 3′RACE and sequencing confirmed that our cDNAs are polyadenylated. Some testicular transcripts have long poly(A) tails, which are reduced to the size of other potentially translated poly(A) RNAs found in cytoplasmic fractions (Kleene 1989). Transcripts from Yq12 are large and heterogeneous, as seen from Northern blots. It is not clear at the moment whether this is due to multiple start sites, variable poly(A) shortening, or read-through transcription. As the DYZ1 cDNAs (AY598346, AY598347) showed regions of identity to the 98-kb BAC clone in an alternating manner (see Fig. 3), and as the two mRNAs did not have any significant identity with each other, there are at least two different RNAs from the region. The above observations confirm transcription from the Yq12 region. Thus, the DYZ1 transcripts localizing in multiple copies to this heterochromatic region provide insight into the block that was not known to be transcribed and classically described as “junk.”
Class 2 genes at Yq12?
Sequence analysis of the genomic clone (AY598345) and the cDNA clones AY598346 and AY598347 by the Tandem Repeats Finder (TRF) program (Benson 1999) revealed a consensus sequence GGAAT. Fickett’s TestCode (Fickett 1982), GeneScan (Tiwari et al. 1997), and GenScan (http://genes.mit.edu/GENSCAN.html) did not recognize the cDNAs AY598346, AY598347, and AK47 as protein-coding genes or predict any genes on the BAC clone AC068123. Translated BLAST search using the cDNAs did not pick up any known protein from the database. These cDNAs have semblance to mRNA like noncoding RNAs (ncRNAs) in the sense that these are large, polyadenylated, tissue-specific transcripts, which do not have long ORFs. GenScan predicted short ORFs within the 98-kb BAC that have no correspondence to the experimental cDNAs, which is a characteristic feature of mRNA like noncoding RNAs. Apparently, no single-copy gene is buried within the span of the 98-kb heterochromatin region analyzed. Similar to transcripts described from the euchromatic Yq (Foote et al. 1992), these Yq heterochromatic cDNAs are also present in multiple copies. These cDNA sequences, which localize to distal MSY heterochromatic block in multiple copies, show testis-specific expression, and lack active X homologs conform to the class 2 genes of the human Y (Lahn et al. 2001).
The above-described highly conserved pentamer GGAAT (Grady et al. 1992) in the genomic clone 102d(2) and the cDNA clones AY598346, AY598347, and AK47 is also shared with repeat sequences on all human chromosomes, including the human satellite III on pericentromeric regions of chromosomes 9 and 15. The satellite III sequences are also transcribed following heat shock and form the stress granules (Jolly et al. 2004). It is significant in this context that 102d(2) and the two cDNA clones hybridized only with the Yq12 region. This suggests that the organization of the GGAAT pentamer is different at different chromosome regions, and thus the transcripts detected by in situ and Northern analysis in the present study are Yq12 specific.
Generation of testicular isoform of CDC2L2 by trans-splicing with Y noncoding RNA
Gururajan et al. (1998) reported a testis-specific isoform of CDC2L2, differing from others in its 5′UTR. The 67-nt 5′UTR reported in their study is 98.5% identical to the Yq12-derived 67-nt sequence. As the 67-nt long 5′ terminal exon is not present at the cognate CDC2L2 genomic locus and since there is no copy of CDC2L2 on the Y, it is reasonable to assume that the testis-specific β sv13 isoform arises by trans-splicing of the CDC2L2 mRNA and the noncoding RNA from Yq12. Splicing of precursor RNAs is necessary for maturation of many mRNAs in higher organisms. Compared with the numerous examples of cis-splicing, a few recent reports of trans-splicing between two distinct mRNAs generating chimeric RNAs are known (Kikumori et al. 2002; Takahara et al. 2002; Peatson et al. 2004). The present study provides evidence for generation of mRNA for a testis-specific CDC2L2 cell division kinase by trans-splicing between a noncoding RNA from Yq12, which provides the 5′UTR. This appears to be a novel regulatory role for the Y heterochromatin in testis.
When a database of all reported snRNA sequences was created and the 67-nt UTR was queried against it, a few of these had short regions of identity (11–12 nt) within the loops of the 67-nt UTR. This may indicate a role for this transcript in RNA splicing. There is a single Y-box consensus sequence 5′CCAAT3′ within the 67-nt UTR from position 15–19. YBX1 is reported to enhance splicing, control mRNA translation, and protect mRNA against degradation (Kohno et al. 2003). Reporter assays of a construct with the 67-nt UTR between the promoter and the lucifrerase reporter in HeLa cells showed a slight enhancement of luciferase activity. The trans-splicing event could be involved in regulating the expression of CDC2L2 in testis.
The gain and retention of genes that specifically benefit male fecundity and promote spermatogenesis in particular seem to be the global theme in Y evolution. The present study demonstrates that the heterochromatic block of human Y chromosome consisting of the consensus sequence of GGAAT, thus far described as transcriptionally inactive, is transcribed specifically in the testis. This noncoding RNA probably modulates translation of sv13 isoform of CDC2L2 within testis. As some CDC2L2 isoforms are associated with cyclins beside being substrates for caspases, a fine tuning of the cell division-related and apoptotic activities of CDC2L2 in testis is perhaps achieved in-house via the noncoding transcripts from Yq12 heterochromatin. It is worth recalling that McClintock (1951) predicted that the study of heterochromatin would lead to the understanding of regulation of euchromatic gene expression. Trans-splicing of a part of noncoding transcript from a Y chromosome with an autosome-derived coding mRNA (CDC2L2) to generate a testis-specific isoform may be one such example of regulation of euchromatic gene expression by heterochromatin.
Methods
Procurement of tissues and probes
A male-specific clone 102d(2) localizing to the long arm of human Y chromosome, first isolated by Singh et al. (1994) was used as the probe for studying the distal Yq heterochromatic block. The cDNAs used as probes for Southern, Northern, and FISH studies were obtained by screening of a testis cDNA library with 102d(2). The human embryonic tissues were obtained from Professor C.M. Habibullah’s laboratory, Owaisi Hospital and Research Center, Kanchanbagh, Hyderabad, India, where the liver tissues of the fetus are routinely used for liver transplantation studies. They have clearance from the Ethical Committee of the Indian Council of Medical Research, New Delhi, for the use of human tissues. These samples have clearance from our Internal Review Board. The testes were collected from 20-, 32-, and 36-wk-old abortuses. A piece of skin collected from a 20-wk-old embryo was used as a control. An adult testicular sample was collected from a fresh accident case brought to the Osmania General Hospital mortuary, Hyderabad, India. Part of the tissue was snap-frozen in liquid nitrogen for isolation of RNA and a part fixed in 4% formaldehyde in 1× PBS. The fixed tissues were processed for paraffin sections using a routine procedure. Eight to10-μm sections were obtained on gelatin-coated slides (2% gelatin in 0.2% chrome alum). Sections were firmly adhered on to the slides by baking them at 45°C for 3 d. The slides were then processed for tissue FISH as described below.
FISH
Sections were deparaffinized in xylene for 40 min, with four changes of 10 min each, followed by a brief fixation (1 min) in methanol:acetic acid (3:1) and air-dried. After Proteinase K (1 μg/μL) treatment for 2–3 min at room temperature, sections were washed two times in 1× PBS for 3 and 5 min, respectively, at room temperature and air-dried. The genomic DNA clone, 102d(2), labeled by multipriming using the Stratagene kit (Prime-It Fluor Fluorescence labeling kit) was used for tissue FISH. A total of 100 ng of the probe was applied to the sections in hybridization mix (dextran sulphate [10%], heparin sulphate [500 μg/mL], 4× SET, sheared Escherichia coli genomic DNA [20 μg/mL]; 4× SET: 3 M NaCl, 40 mM EDTA, 0.6 M Tris [pH 7.4]). The probe and targets were denatured simultaneously on a 90°C hotplate as follows: 6 min on the plate at 90°C, 2 min at room temperature, 5 min at 90°C, 1 min at room temperature, and 4 min at 90°C, followed by incubation for 16–20 h at 37°C in a moist chamber. The slides were washed in 2× SSC at room temperature for 10 min and subsequently in 2× SSC, 50% formamide at 45°C three times for 10 min each. The sections were then processed through 1× SSC for 10 min at 45°C. They were then left overnight at 4°C in 2× SSC. The sections were counterstained with Vectashield containing propidium iodide (Vector Labs) and observed under a Confocal microscope. RNase-treated sections served as controls.
The genomic clone 102d(2) and male-specific cDNAs were used as probes on normal metaphase spreads obtained from peripheral blood lymphocytes. After RNase treatment (50 μg/mL for 1 h at 37°C), slides were rinsed in four changes of 2× SSC at room temperature for 2 min each, dehydrated through alcohol series (2 min each), and denatured in 70% formamide, 2× SSC at 70°C/slide for 2 min. An increment of 1°C was given for every extra slide. These slides were then taken through a chilled alcohol series and air-dried. Denatured probes were applied (100 ng/slide) in the hybridization mix, sealed, and incubated in a humid chamber at 37°C for 16–20 h. The slides were rinsed in 2× SSC at room temperature for 10 min and washed under the same conditions as that for tissue FISH. Chromosomes were counterstained with Vectashield containing DAPI. Signals were captured under a fluorescence microscope.
Identification of male-specific cDNAs and their testis-specific expression
A human testis cDNA library was obtained from Clontech (cat.# HL1010B). Then, 5 × 103 PFU of the library were plated on 150-mm plates, filters lifted, and screened with 102d(2) according to standard protocol at a stringency of 0.5× SSC/0.2% SDS at 70°C. Male-specific cDNA clones were identified using the above clones as probes on Southern blots containing human male and female DNA at a final stringency of 0.1× SSC/0.2% SDS at 65°C. The cDNA clones were sequenced using λgt11 forward and reverse primers (GGTGGCGACGACTCCTGGAGCCCG, TTGACACC AGACCAACTGGTAATG). Sequencing of AY598347 was completed using the internal primer TGGACTGGAAAGCAATGG ACTG. Multi-tissue Northern blots containing poly(A) RNA were obtained from Clontech (cat # 636805). These blots contained RNA from peripheral blood lymphocytes, colon, small intestine, ovary, testis, prostate, thymus, and spleen. Genomic clone and the cDNA clones were labeled with [α32P]dATP (NEN/BRIT, India), by multipriming and used as probes following the manufacturer’s protocol. Blots were washed after 16–20 h of hybridization at 37°C to a stringency of 0.1× SSC at 65°C for 15–20 min. Signals were captured by PhosphorImaging and by exposure to X-ray sheets.
Real-time quantitative PCR
Real-Time quantitative PCR was performed by adding 5 μL of 2× SYBR Green PCR Master Mix (Applied Biosystems) to each well containing 1 μL of 10 ng template and 0.5 μL of a primer mix with 5 pM of each primer in a 10-μL reaction volume. The (CT) threshold cycle value was determined on the linear phase of PCRs using the Sequence Detector Software version 2.1 (Applied Biosystems). A standard curve was generated using serial dilutions of the target genomic DNA. A complete four-stage reaction of the initial stage I at 50°C for 50 sec, stage II at 95°C for 10 min, followed by a 30-cycle stage III PCR cycle profile of 95°C for 15 sec, 67°C for 30 sec, and 68°C for 30 sec, and a stage IV dissociation was followed. The final result with its respective Standard plot with a slope of −3.357, Amplification plot with the automated threshold value, and the Dissociation constant was thoroughly analyzed to come to a final conclusion.
Analysis of the regulatory activity of the 5′UTR
The 67-bp UTR of the CDC2L2_SV13 RNA isoform was amplified and cloned into the pGL3 promoter vector (Promega, Inc) at the HindIII restriction enzyme site. The fragment was placed between the SV40 promoter and the luciferase reporter gene. The orientation and sequence were confirmed by sequencing. The positive clone was then transfected into HeLa cells for analysis of reporter activity. Transfection experiments were carried out in 24-well plates using liposome-mediated methods; 105 cells were seeded per well and the transfection was done 24 h later. The transfection mix contained 0.5 μg of the reporter construct and 0.125 μg of pCMV-β gal vector complexed with 1 μL of LipofectAMINE reagent (GIBCO-BRL) and 4 μL of Plus reagent in 200 μL of serum-free medium. The cells were washed twice with 1× PBS and overlaid with the transfection mix for 5 h, after which complete medium was added to the wells. The cells were grown for a period of 48 h and harvested. The whole-cell extracts were prepared using the Reporter Lysis Buffer (RLB) (Promega, Inc.). The transfected cells were washed twice with 1× PBS, overlaid with 1× RLB, and kept on a shaker for 10 min. The cells were then scraped using a rubber policeman, vortexed for 15 sec, and quick frozen at −70°C. The cells were then thawed at room temperature and centrifuged at 13,000 rpm for 5 min at 4°C. The extract was then assayed for luciferase activity with the substrate luciferin in a luminometer. The luciferase activity was normalized with respect to their corresponding β galactosidase activities. The fold differences in the reporter activity were calculated with respect to the values obtained with the basic vector.
Acknowledgments
We thank Dr. B. Sesikeren, National Institute of Nutrition, Hyderabad, for help with paraffin sections, Nandini Rangarajan for the confocal microscopy, Koushik Chatterjee for part of the bioinformatics analysis, and Professor S.C Lakhotia and Dr. V. Radha for reading the manuscript and providing useful inputs. Professor Habibullah, Deccan College of Medical Sciences, Hyderabad, provided the human tissue samples. The DNA samples for Real Time PCR analysis were kindly provided by Ms. Lakshmi Rao. We were supported by funds from the Department of Biotechnology, India (R.A.J.) and the Council for Scientific and Industrial Research, India (Y.R.A.).
Footnotes
[The sequence data from this study have been submitted to GenBank under accession nos. AY598345, AY598346, and AY598347.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5155706
References
- Bachtrog D., Charlesworth B., Charlesworth B. Towards a complete sequence of the human Y-chromosome. Genome Biol. 2001;2:10161–10165. doi: 10.1186/gb-2001-2-5-reviews1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csink A.K., Henikoff S., Henikoff S. Something from nothing: The evolution and utility of satellite repeats. Trends Genet. 1998;14:200–204. doi: 10.1016/s0168-9525(98)01444-9. [DOI] [PubMed] [Google Scholar]
- Diaz M.O., Barsacchi-Pilone G., Mahon K.A., Gall J.G., Barsacchi-Pilone G., Mahon K.A., Gall J.G., Mahon K.A., Gall J.G., Gall J.G. Transcripts from both strands of a satellite DNA occur on lampbrush chromosome loops of the newt Notophthalmus. Cell. 1981;24:649–659. doi: 10.1016/0092-8674(81)90091-x. [DOI] [PubMed] [Google Scholar]
- Fickett J.W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982;10:5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foote S., Vollrath D., Hilton A., Page D.C., Vollrath D., Hilton A., Page D.C., Hilton A., Page D.C., Page D.C. The human Y chromosome: Overlapping DNA clones spanning the euchromatic region. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- Fornace A.J., Jr., Mitchell J.B., Mitchell J.B. Induction of B2 RNA polymerase III transcription by heat shock: Enrichment for heat shock induced sequences in rodent cells by hybridization subtraction. Nucleic Acids Res. 1986;14:5793–5811. doi: 10.1093/nar/14.14.5793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady D.L., Ratlif R.L., Robinson D.L., McCanlies E.C., Meyene J., Moyzis R.K., Ratlif R.L., Robinson D.L., McCanlies E.C., Meyene J., Moyzis R.K., Robinson D.L., McCanlies E.C., Meyene J., Moyzis R.K., McCanlies E.C., Meyene J., Moyzis R.K., Meyene J., Moyzis R.K., Moyzis R.K. Highly conserved repetitive DNA sequences are present at human centromeres. Proc. Natl. Acad. Sci. 1992;89:1695–1699. doi: 10.1073/pnas.89.5.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururajan R., Lahti M., Grenet J., Easton J., Gruber I., Ambros P.F., Kidd V.J., Lahti M., Grenet J., Easton J., Gruber I., Ambros P.F., Kidd V.J., Grenet J., Easton J., Gruber I., Ambros P.F., Kidd V.J., Easton J., Gruber I., Ambros P.F., Kidd V.J., Gruber I., Ambros P.F., Kidd V.J., Ambros P.F., Kidd V.J., Kidd V.J. Duplication of genomic region containing the Cdc2L1-2 and MMP21-22 genes on human chromosome 1p36.3 and their linkage to D1Z2. Genome Res. 1998;8:929–939. doi: 10.1101/gr.8.9.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttenbach M., Schmid M., Jausch A., Vogt P., Schmid M., Jausch A., Vogt P., Jausch A., Vogt P., Vogt P. The Y chromosome of the mouse is decondensed in Sertoli cells. Chromosoma. 1989;97:429–433. doi: 10.1007/BF00295026. [DOI] [PubMed] [Google Scholar]
- Heitz E. Das Heterochromatin der Moose. Jahrb. Wiss. Botanik. 1928;69:762–818. [Google Scholar]
- Jobling M.A. A survey of long-range DNA polymorphisms on the human Y chromosome. Hum. Mol. Genet. 1994;3:107–114. doi: 10.1093/hmg/3.1.107. [DOI] [PubMed] [Google Scholar]
- Jolly C., Metz A., Govin J., Vigneron M., Turner B.M., Khochbin S., Vourih C., Metz A., Govin J., Vigneron M., Turner B.M., Khochbin S., Vourih C., Govin J., Vigneron M., Turner B.M., Khochbin S., Vourih C., Vigneron M., Turner B.M., Khochbin S., Vourih C., Turner B.M., Khochbin S., Vourih C., Khochbin S., Vourih C., Vourih C. Stress induced transcription of satellite III repeats. J. Cell Biol. 2004;164:25–33. doi: 10.1083/jcb.200306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikumori T., Gilbert C.J., Gagel R.F., Gilbert C.J., Gagel R.F., Gagel R.F. Naturally occurring heterologous trans-splicing cellular transcripts during infection. FEBS Lett. 2002;522:41–46. doi: 10.1016/s0014-5793(02)02878-8. [DOI] [PubMed] [Google Scholar]
- Kimura R.H., Choudary P.V., Stone K.K., Schmid C.W., Choudary P.V., Stone K.K., Schmid C.W., Stone K.K., Schmid C.W., Schmid C.W. Stress induction of Bm1 RNA in silkworm larvae: SINEs, an unusual class of stress genes. Cell Stress Chaperones. 2001;6:263–272. doi: 10.1379/1466-1268(2001)006<0263:siobri>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene K.C. Poly(A) shortening accompanies the activation of translation of five mRNAs during spermatogenesis in the mouse. Development. 1989;106:367–373. doi: 10.1242/dev.106.2.367. [DOI] [PubMed] [Google Scholar]
- Kleene K.C. A possible meiotic function of the peculiar patterns of gene expression in mammalian spermatogenic cells. Mech. Dev. 2003;106:3–23. doi: 10.1016/s0925-4773(01)00413-0. [DOI] [PubMed] [Google Scholar]
- Kohno K., Izumi H., Uchiumi T., Ashizuka M., Kuwano M., Izumi H., Uchiumi T., Ashizuka M., Kuwano M., Uchiumi T., Ashizuka M., Kuwano M., Ashizuka M., Kuwano M., Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. Bioessays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Pearson N.M., Jegalian K., Pearson N.M., Jegalian K., Jegalian K. The human Y chromosome, in the light of evolution. Nat. Rev. Genet. 2001;2:207–215. doi: 10.1038/35056058. [DOI] [PubMed] [Google Scholar]
- Manz E., Alkan M., Buhler E., Schmidtke J., Alkan M., Buhler E., Schmidtke J., Buhler E., Schmidtke J., Schmidtke J. Arrangement of DYZ1 and DYZ2 repeats on the human Y-chromosome: A case with presence of DYZ1 and absence of DYZ2. Mol. Cell. Probes. 1992;6:257–259. doi: 10.1016/0890-8508(92)90025-s. [DOI] [PubMed] [Google Scholar]
- Mattick J.S., Makunin I.V., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- McClintock B. Chromosome organization and genic expression. Cold Spring Harb. Symp. Quant. Biol. 1951;16:13–47. doi: 10.1101/sqb.1951.016.01.004. [DOI] [PubMed] [Google Scholar]
- Peatson A.E., Eviskov A.V., Graber J.H., de Vries W.N., Hollbrook A.E., Solter D., Knowles B.B., Eviskov A.V., Graber J.H., de Vries W.N., Hollbrook A.E., Solter D., Knowles B.B., Graber J.H., de Vries W.N., Hollbrook A.E., Solter D., Knowles B.B., de Vries W.N., Hollbrook A.E., Solter D., Knowles B.B., Hollbrook A.E., Solter D., Knowles B.B., Solter D., Knowles B.B., Knowles B.B. Retrotransposons regulate host genes in mouse oocyte and pre-implantation embryos. Dev. Cell. 2004;7:597–606. doi: 10.1016/j.devcel.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Rudert F., Bronner S., Garnier J.M., Dolle P., Bronner S., Garnier J.M., Dolle P., Garnier J.M., Dolle P., Dolle P. Transcripts from opposite strands of γ satellite DNA are differentially expressed during mouse development. Mamm. Genome. 1995;6:76–83. doi: 10.1007/BF00303248. [DOI] [PubMed] [Google Scholar]
- Schafer M., Nayernia K., Engel W., Schafer U., Nayernia K., Engel W., Schafer U., Engel W., Schafer U., Schafer U. Translational control in spermatogenesis. Dev. Biol. 1995;172:344–352. doi: 10.1006/dbio.1995.8049. [DOI] [PubMed] [Google Scholar]
- Singh L., Panicker S.G., Nagaraj R., Majumdar K.C., Panicker S.G., Nagaraj R., Majumdar K.C., Nagaraj R., Majumdar K.C., Majumdar K.C. Proc. Indian Natn. Sci. Acad. B60. 1994. Functional significance of molecular organization of sex chromosomes; pp. 455–470. [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Repping S., Pyntikova T., Ali J., Bieri T., Pyntikova T., Ali J., Bieri T., Ali J., Bieri T., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Speed R.M., Vogt P., Kohler M.R., Hargreave T.B., Chandley A.C., Vogt P., Kohler M.R., Hargreave T.B., Chandley A.C., Kohler M.R., Hargreave T.B., Chandley A.C., Hargreave T.B., Chandley A.C., Chandley A.C. Chromatin condensation behaviour of the Y chromosome in the human testis. 1. Evidence for decondensation of distal Yq in germ cells prior to puberty with a switch to Sertoli cells in adults. Chromosoma. 1995;102:421–427. doi: 10.1007/BF00360407. [DOI] [PubMed] [Google Scholar]
- Takahara T., Kasahara D., Mori D., Yanagisawa S., Akanuma H., Kasahara D., Mori D., Yanagisawa S., Akanuma H., Mori D., Yanagisawa S., Akanuma H., Yanagisawa S., Akanuma H., Akanuma H. The trans-spliced variants of Sp1 mRNA in rat. Biochem. Biophys. Res. Commun. 2002;298:156–162. doi: 10.1016/s0006-291x(02)02419-1. [DOI] [PubMed] [Google Scholar]
- Tiwari S., Ramachandran S., Bhattacharya A., Bhattacharya S., Ramaswamy R., Ramachandran S., Bhattacharya A., Bhattacharya S., Ramaswamy R., Bhattacharya A., Bhattacharya S., Ramaswamy R., Bhattacharya S., Ramaswamy R., Ramaswamy R. Prediction of probable genes by Fourier analysis of genomic sequences. Comput. Appl. Biosci. 1997;13:263–270. doi: 10.1093/bioinformatics/13.3.263. [DOI] [PubMed] [Google Scholar]
- Toure A., Clemente E.J., Ellis P., Mahadevaiah S.K., Ojarikre O.J., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Clemente E.J., Ellis P., Mahadevaiah S.K., Ojarikre O.J., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Ellis P., Mahadevaiah S.K., Ojarikre O.J., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Mahadevaiah S.K., Ojarikre O.J., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Ojarikre O.J., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Ball P.A.F., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Reynard L., Loveland K.L., Burgoyne P.S., Affara N.A., Loveland K.L., Burgoyne P.S., Affara N.A., Burgoyne P.S., Affara N.A., Affara N.A. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol. 2005;6:R102. doi: 10.1186/gb-2005-6-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulin A., Stewart D., Spradling A.C., Stewart D., Spradling A.C., Spradling A.C. The Drosophila heterochromatic gene encoding poly (ADP-ribose) polymerase (PARP) is required to modulate chromatin structure during development. Genes & Dev. 2002;16:2108–2119. doi: 10.1101/gad.1003902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Groudine M., Groudine M. Chromosomal subunits in active genes have an altered conformation. Science. 1976;193:848–856. doi: 10.1126/science.948749. [DOI] [PubMed] [Google Scholar]