Abstract
Out of the nine male-specific gene families in the human Y chromosome amplicons, we investigate the origin and evolution of seven families for which gametologous and orthologous sequences are available. Proto-X/Y gene pairs in the original mammalian sex chromosomes played major roles in origins and gave rise to five gene families: XKRY, VCY, HSFY, RBMY, and TSPY. The divergence times between gametologous X- and Y-linked copies in these families are well correlated with the former X-chromosomal locations. The CDY and DAZ families originated exceptionally by retroposition and transposition of autosomal copies, respectively, but CDY possesses an X-linked copy of enigmatic origin. We also investigate the evolutionary relatedness among Y-linked copies of a gene family in light of their ampliconic locations (palindromes, inverted repeats, and the TSPY array). Although any pair of copies located at the same arm positions within a palindrome is identical or nearly so by frequent gene conversion, copies located at different arm positions are distinctively different. Since these and other distinct copies in various gene families were amplified almost simultaneously in the stem lineage of Catarrhini, we take these simultaneous amplifications as evidence for the elaborate formation of Y ampliconic structure. Curiously, some copies in a gene family located at different palindromes exhibit high sequence similarity, and in most cases, such similarity greatly extends to repeat units that harbor these copies. It appears that such palindromic repeat units have evolved by and large en bloc, but they have undergone frequent exchanges between palindromes.
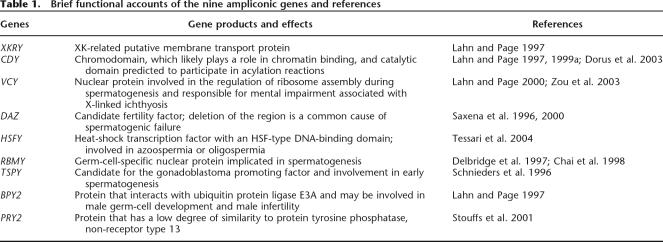
The male-specific region of the human Y chromosome (MSY), also previously called the nonrecombining portion of the Y chromosome (NRY), consists of three different classes of euchromatic sequences: X-transposed, X-degenerate, and ampliconic sequences (Skaletsky et al. 2003; Ross et al. 2005; see also Hughes et al. 2005 and Kuroki et al. 2006 for the available chimpanzee Y sequences of 9.5–12.7 Mb). The X-transposed sequences originated from an X-to-Y transposition 3–5 million years ago (Mya) and encode only two or three genes. The X-degenerate sequences are relics of ancient autosomes that have evolved toward the sex chromosomes (Ohno 1967; for reviews, see Graves 1995, 2002). The sequences encode 16 genes and 13 pseudogenes with their individual X-homologs. The ampliconic segments are composed of eight palindromes (P1–P8), three inverted repeats (IR1–IR3), and two arrays of no long open reading frames (NORF) and TSPY repeats. The segments encode nine gene families, seven of which are implicated in spermatogenesis or sperm production (Lahn and Page 1997; Skaletsky et al. 2003). These gene families are referred to as X Kell blood-related Y (XKRY), chromodomain Y (CDY), variable charge (VCY), deleted in azoospermia (DAZ), heat-shock transcription factor Y (HSFY), RNA-binding motif Y (RBMY), testis-specific Y (TSPY), basic protein Y2 (BPY2), and PTP-BL related Y (PRY) (see Table 1 for their brief accounts of functions and references).
Table 1.
Brief functional accounts of the nine ampliconic genes and references
There are quite a few reports on origins for CDY, DAZ, HSFY, RBMY, and TSPY. Any model for the origin of these genes invokes retroposition of autosomal gene transcripts, duplicated transposition of autosomal loci, or proto-X/Y pairs of genes in the original mammalian sex chromosomes. Lahn and Page (1999a) suggested that autosomal CDYL gave rise to CDY by retroposition in the stem lineage of simian primates and that CDYL and CDY have since undergone functional partitioning of housekeeping and testis-specific functions. Later, by examining CDYL and its early autosomal duplicate CDYL2, Dorus et al. (2003) pushed the origin of CDY back before the Eutherian radiation. Regarding other gene families, it was hypothesized that retroposition resulted in HSFY (Tessari et al. 2004) and transposition resulted in DAZ (Saxena et al. 1996; Lahn and Page 1999a), RBMY (Chai et al. 1998; Elliott et al. 2000), and TSPY (Lahn and Page 1999a). However, another group proposed the proto-X/Y pair origin of RBMY (Delbridge et al. 1998, 1999; Lingenfelter et al. 2001) and TSPY (Delbridge et al. 2004).
The ampliconic gene families are expressed predominantly or exclusively in testes (Skaletsky et al. 2003), and six of them are found only in primates. It is therefore tempting to ask where they came from and how their origin affected the function on the primate Y chromosome. It is also interesting to examine the evolutionary relationships between ampliconic genes and surrounding ampliconic regions per se (Kuroda-Kawaguchi et al. 2001; Rozen et al. 2003; Skaletsky et al. 2003). The purpose of this paper is twofold. First, we study the origin of ampliconic genes based on currently available genome sequence data. We show that XKRY, VCY, and HSFY originated from proto-X/Y gene pairs in addition to RBMY and TSPY. We examine whether the divergence times of these five genes from the X-linked homologs agree with the stepwise differentiation pattern of the Eutherian sex chromosomes, as the evolutionary stratum hypothesis can predict (Lahn and Page 1999b). Second, as a gene family usually scatters multiple copies within and between different ampliconic regions, we study the evolutionary relationships among copies in light of their ampliconic locations. We pay special attention to low sequence similarity among copies that are located in different arm positions within a palindrome as well as high sequence similarity among copies that are located in different palindromes. We extend this analysis to the entire region of the 2.9-Mb palindrome P1 and demonstrate massive sequence transfers with autosomal repeat units and other Y-chromosomal palindromes.
Results
Gametologs, orthologs, and amplicons
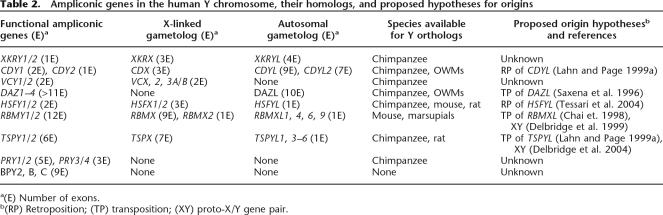
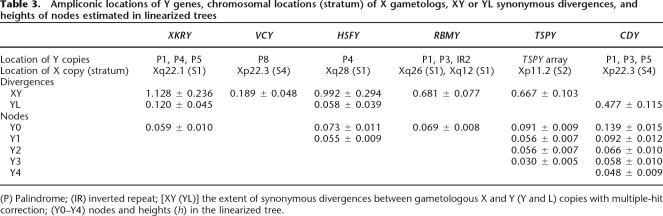
In searching for origins of the ampliconic genes, it is essential to include X-linked and autosomal homologs or more precisely gametologs, if present in the genome, as well as their orthologs in critical phylogenetic positions. Despite the fact, by definition, that the ampliconic genes are located outside the X-degenerate segments, it turns out that each of six gene families has an X-linked gametolog in evolutionary strata 1, 2, or 4. Lahn and Page (1999b) found that during mammalian evolution, the X/Y chromosomal differentiation took place in a stepwise fashion; one stratum at a time. Stratum 1, covering almost the entire long arm of the human X chromosome, is the oldest among four or five strata, and the formation predated the divergence between Monotherians and Metatherians/Eutherians, 210 Mya (Lahn and Page 1999b; Skaletsky et al. 2003; Ross et al. 2005; Waters et al. 2005). Stratum 2 occupies a rather small proximal region of the X short arm and was formed before Metatherians and Eutherians diverged, 180 Mya. On the other hand, stratum 4 was formed independently within individual Eutherian orders (Iwase et al. 2003) and in primates; it was deposited in the stem lineage of simian primates, >50 Mya. Stratum 1 harbors gametologous XKRX of XKRY, HSFX of HSFY, and RBMX of RBMY; stratum 2 harbors TSPX of TSPY; and stratum 4 harbors CDX of CDY and VCX of VCY. In addition to these X-linked copies, each of XKRY, CDY, DAZ, HSFY, RBMY, and TSPY has at least one autosomal copy in the human genome. On the other hand, the availability of orthologs varies among gene families, sometimes reflecting true presence or absence in the genome or in the databases.
Table 2 lists the nine gene families with their gametologs, proposed origin hypotheses, and species for which we found orthologs of human Y ampliconic genes. The number of exons per gene is included to infer molecular mechanisms involved in the origins, although intronless genes such as XKRY1 and XKRY2 did not always result from retroposition, and conversely, intron-containing genes such as CDY1 originated via retroposition and subsequent acquisition of introns (Lahn and Page 1999a). For the BPY2 family (Lahn and Page 1997), there exist three nearly identical genes—BPY2B and BPY2C in palindrome P1 and BPY2 in a proximal region adjacent to palindrome P2. For the PRY family (Stouffs et al. 2001), there exist paired PRY1 and PRY2 and their shorter versions of paired PRY3 and PRY4 in palindrome P3 and P4, respectively. Unfortunately, neither BPY2 nor PRY possesses any detectable X-linked or autosomal copy, and little is known about their orthologs. Under these circumstances of available sequence data, we exclude these two gene families from our phylogenetic analysis.
Table 2.
Ampliconic genes in the human Y chromosome, their homologs, and proposed hypotheses for origins
a(E) Number of exons.
b(RP) Retroposition; (TP) transposition; (XY) proto-X/Y gene pair.
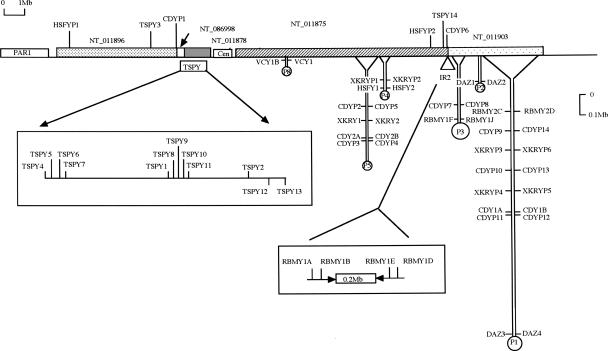
Supplemental Table 1 gives the accession numbers of the sequences of the seven ampliconic gene families and their X-linked and autosomal gametologs, as well as their orthologs when they are used in the phylogenetic analysis. Figure 1 represents ampliconic structure in the human Y chromosome and the chromosomal locations of Y copies we use. Among the eight palindromes, P1 of 1.45-Mb arm length is the largest and harbors two pairs of XKRY, four pairs of CDY, and one pair of each of RBMY and DAZ in our data set. Similarly, the second largest P5 spans nearly 0.50 Mb in its arm length and harbors one pair of XKRY and three pairs of CDY. Inverted repeat 2 (IR2) encodes two pairs of RMBY, and the TSPY array encodes 12 copies.
Figure 1.
Chromosomal locations of human ampliconic genes and pseudogenes based on information about contigs, as well as MapViewer display of BLASTN results (see also Supplemental Table 1). The abscissa represents the ∼24-Mb euchromatic region with the Yp pseudoautosomal region at the left-most end. Palindromes are shown as hairpin-like structures; expanded views of the TSPY array and IR2 are shown in boxes.
Proto-X/Y origins
XKRY
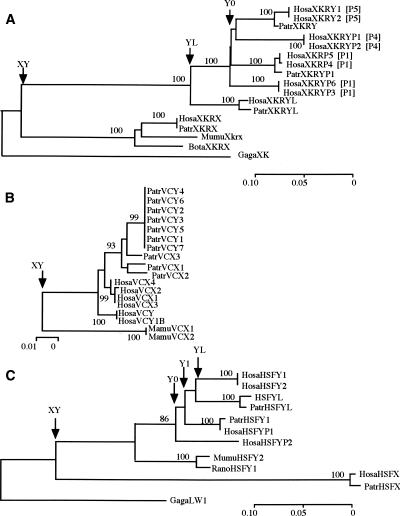
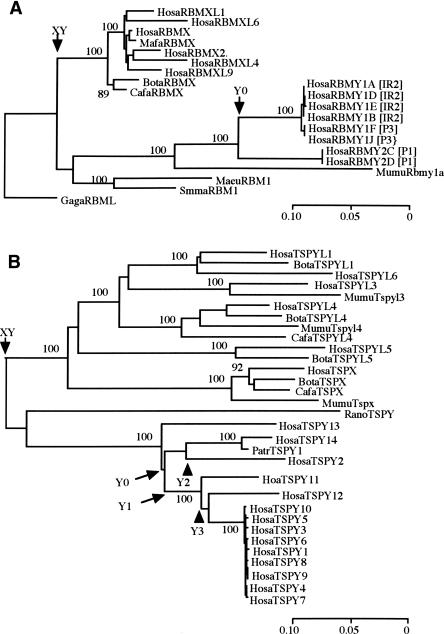
We found eight XKRY copies, one X-linked XKRX, and one autosomal XKRYL in the human genome. The coding sequence of human XKRX shows a high degree of evolutionary conservation with some Eutherian orthologs being >80% similar at the DNA sequence level. The presence of XKRX raises the possibility that XKRY has an origin similar to that of genes in the X-degenerate class. Phylogenetic comparison of XKRY-related gametologous and orthologous sequences identifies three distinct clusters: Eutherian XKRX; a pair of human and chimpanzee XKRYL; and 10 intermingled human and chimpanzee XKRY genes and pseudogenes (Fig. 2A). It is clear that two monophyletic clusters of XKRX and XKRY/XKRYL differentiated from each other well before the Eutherian radiation and that later in simian primate evolution, XKRY and XKRYL began to evolve independently. The estimated extent of synonymous divergences (kXY) between XKRX and XKRY is as large as 1.128 ± 0.236, in agreement with the values in stratum 1 reported by Lahn and Page (1999b) and Skaletsky et al. (2003). The putative coding region of XKRY starts in exon 4 of XKRX/XKRYL. The translated and 3′-untranslated regions of XKRY have sequence similarity to the corresponding regions of XKRX/XKRYL. Moreover, the 5′-flanking region of XKRY shows sequence similarity to exon 1 and part of intron 3 of XKRYL. All of these features support the notion that XKRY did not result from retroposition, but from a proto-X/Y gene pair.
Figure 2.
NJ trees of (A) XKRY, (B) VCY, and (C) HSFY. Whenever chicken homologs are available, they are used for rooting the trees. In A, palindromic locations of human XKRY copies are indicated by bracketed P1, P4, and P5. The numbers next to nodes show the bootstrap values in 1000 replications. Node XY, YL, and Y0 stand for X/Y differentiation, retroposition from Y to autosome or vice versa, and coalescence of all human Y copies, respectively. The accession numbers of the DNA sequences and the sequence alignments are given in Supplemental Table 1 and Supplemental Figure 1, respectively. The gene nomenclature follows a four-letter prefix system to indicate genus and species names: (Hosa) Homo sapiens; (Patr) Pan troglodytes; (Mamu) Macaca mulatta; (Mafa) Macaca fascicularis; (Paha) Papio hamadryas; (Ceap) Cebus apella; (Saoe) Saguinus oedipus; (Sasc) Saimiri sciurea; (Cafa) Canis familiaris; (Susc) Sus scrofa; (Bota) Bos taurus; (Mumu) Mus musculus; (Rano) Rattus norvegicus; (Maeu) Macropus eugenii; (Smma) Sminthopsis macroura; (Gaga) Gallus gallus.
A duplicated copy of XKRY was transposed to create autosomal XKRYL. Based on the extent of synonymous divergences (kYL = 0.120 ± 0.045) between XKRY and XKRYL, we date the transposition event as 41 ± 15 Mya (see Methods). At this point, proto-XKRY still had an exon and intron structure similar to XKRX (Table 2). Yet, there is one base-pair deletion specific to a pair of nearly identical XKRY1/2 copies, and this deletion creates a new initiation codon and new coding frame. It therefore appears that XKRY1/2 are functional, but the remaining six copies are nonfunctional and designated as XKRYP1–6 with suffix P (Figs. 1 and 2A).
VCY
We found two VCY and four X-linked VCX copies in the human genome. The presence of VCX in stratum 4 suggests that VCX and VCY evolved from a proto-X/Y gene pair in the X added region (XAR) (Graves 1995, 2002). The nucleotide differences between X-linked sequences in stratum 4 and the gametologous Y sequences are ∼10% per site on average (Lahn and Page 1999b; Iwase et al. 2003). However, the observed synonymous and nonsynonymous differences between human VCX and VCY are merely 2.2% and 3.3% per site, respectively. These values are too small to be consistent with the expected extent of synonymous divergences for a proto-X/Y gene pair in stratum 4 (but see Skaletsky et al. 2003).
Both VCX and VCY can also be found in chimpanzees, but other sequence data are available only for rhesus monkeys; two VCX genes and one fragmental copy. Despite this limited availability of VCY-related sequences, they show some interesting evolutionary features. First, chimpanzee VCX and VCY are more closely related to each other than they are to their orthologous genes in humans (Fig. 2B), suggesting that chimpanzee VCY was recently converted by the gametolog and then amplified to generate seven nearly identical copies. Second, the average kXY value between rhesus monkey VCX and hominoid VCX and VCY is 0.189 ± 0.048. This extent is too large to be expected from the silent substitution rate (Ebersberger et al. 2002) and the 30-Myr divergence between hominoids and Old World monkeys (OWMs) (Martin 1993; Takahata 2001; but see also Kumar and Hedges 1998). However, the observed kXY value is very similar to that for STS adjacent to VCX in stratum 4 (Skaletsky et al. 2003). It is therefore likely that the differentiation between rhesus monkey VCX and the ancestral lineage of hominoid VCX and VCY predated the species divergence, owing to the earlier differentiation between gametologous VCX and VCY. Consistent with this proto-X/Y origin model, Lahn and Page (2000) and Fukami et al. (2000) detected both VCX and VCY in simian primates by low-stringency hybridization.
HSFY
In addition to two functional HSFY and several pseudogenes, we found one X-linked HSFX and one autosomal (HSFYL) copy in the human genome. Here we used only two pseudogenes in our analysis because inclusion of all of the pseudogenes would have prevented us from comparing HSFY with HSFX and HFSYL, owing to mutually exclusive gaps among these genes. Figure 2C shows that a proto-X/Y gene pair of HSFX and HSFY existed in the original mammalian sex chromosomes and differentiated from each other well before the Eutherian radiation. This is consistent with the location of HSFX in stratum 1 and the kXY value of 0.992 ± 0.294 between HSFX and HSFY.
Tessari et al. (2004) proposed that HSFY arose via retroposition of HSFYL, whereas we demonstrate that the reverse happened. That is, intronless HSFYL arose via retroposition of HSFY with two exons. Conversely, if HSFY arose via retroposition of HSFYL, it becomes difficult to account for the presence of HSFY in mice and rats. Actually, the kYL value of 0.058 ± 0.039 between HSFY1/2 and intronless HSFYL indicates that the retroposition of HSFY occurred as recently as 20 ± 13 Mya. Prior to this retroposition, HSFY began to amplify, producing at least three distinct pseudogene lineages in humans. In the linearized tree (Takezaki et al. 1995), the height (h) of nodes Y0 and Y1 leading to the three distinct HSFY lineages is estimated as 0.073 ± 0.011 and 0.055 ± 0.009, respectively (see Methods). Thus, these gene lineages diverged 34–46 Mya in the stem lineage of Catarrhini (hominoids and OWMs).
RBMY and TSPY
As mentioned earlier, RBMY and TSPY possess their X-linked gametologs and multiple Y-linked copies in ampliconic segments. Here we confirm their proto-X/Y origins and examine the kXY value between X and Y copies. In the next section, we examine these gene families with respect to their Y-chromosomal locations in P1, P3, and IR2 as well as in the TSPY array.
The RBMY family consists of six subfamilies (RMBY1–6) of ∼30 genes and pseudogenes on both arms of the Y chromosome (Skaletsky et al. 2003) and exhibits sequence similarity with X-linked functional RBMX and intronless RBMX2 as well as autosomal intronless RBMXL. Sequence similarity between RBMY subfamilies is as low as 60% even at the amino acid level, and some sequences in different subfamilies are difficult to align. For this reason, we used only RBMY1 and RBMY2 subfamilies. The phylogenetic analysis indicates an early dichotomous occurrence of mammalian RBMX and RBMY (Fig. 3A). It is also clear that the X and Y differentiation predated the emergence of Metatherians (Delbridge et al. 1999) such that RBMY in Macropus eugenii and Sminthopsis macroura is orthologous to Eutherian RBMY. The kXY value between RBMX and RBMY is 0.681 ± 0.077. It is smaller than the value given in Skaletsky et al. (2003). However, the previous report included an autosomal sequence (accession no. Z23064). Without this autosomal sequence, the estimated value becomes somewhat small, compared with that for XKRY and HSFY in stratum 1.
Figure 3.
NJ trees of (A) RBMY and (B) TSPY drawn in the same way and with the same gene nomenclature system as in Figure 2. Ampliconic locations of human RBMY copies are indicated by bracketed P1, P3, and IR2.
TSPY is the first gene that was isolated from any Y chromosome (Arnemann et al. 1987) and was subsequently found in primates (Manz et al. 1993), artiodactyls (Jakubiczka et al. 1993), and rodents (Mazeyrat and Mitchell 1998). There are ∼35 copies in the TSPY array (Skaletsky et al. 2003), several autosomal copies (TSPYL), and one X-linked copy (TSPX previously designated as TSPYL2) under X inactivation (Delbridge et al. 2004). We examined 14 TSPY copies from humans together with TSPY, TSPX, and TSPYL orthologs from Eutherians. Although there is no obvious outgroup sequence in this data set, there exist two distinct clusters of TSPY and TSPX/TSPYL, and this dichotomy is consistent with the ancient origin hypothesis of X and Y copies (Fig. 3B). The kXY value between TSPX and TSPY is 0.667 ± 0.103 (Table 3), as expected in stratum 2. In the stem lineage of Eutherians, intronless TSPYL was retroposed and duplicated such that various TSPYL genes exhibit multiple orthologous relationships among Eutherians. It is TSPX rather than TSPY that was retroposed so as to generate autosomal TSPYL1, 3, 4, 5, and 6 (Fig. 3B).
Table 3.
Ampliconic locations of Y genes, chromosomal locations (stratum) of X gametologs, XY or YL synonymous divergences, and heights of nodes estimated in linearized trees
(P) Palindrome; (IR) inverted repeat; [XY (YL)] the extent of synonymous divergences between gametologous X and Y (Y and L) copies with multiple-hit correction; (Y0–Y4) nodes and heights (h) in the linearized tree.
Ages of amplified copies and ampliconic structure
For a given gene family, there is a striking pattern in sequence similarity among copies within and between palindromes. All pairs of copies at symmetric arm positions within a palindrome are identical or nearly so (Skaletsky et al. 2003). However, not only may copies located between different palindromes be significantly different, but also different pairs within a palindrome may differ greatly. Two pairs of XKRYP3/P6 and XKRYP4/P5 within P1 differ from each other by nearly the same amount that they differ from XKRYP1/P2 in P4 and XKRY1/2 in P5 (Figs. 1 and 2A). The height of node Y0 (0.059 ± 0.010) in the linearized tree (Table 3) suggests that XKRYP1/P2 in P4, XKRY1/2 in P5, and two distinct pairs of XKRYP3/P6 and XKRYP4/P6 in P1 all diverged from one another 37 ± 6 Mya.
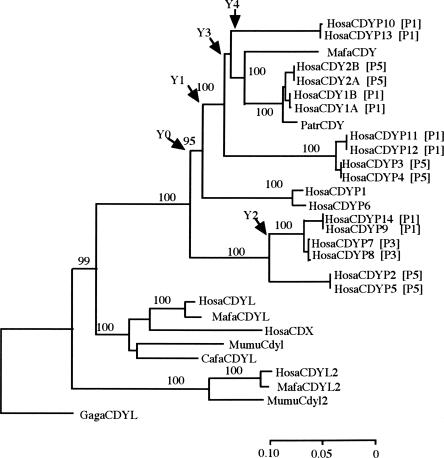
More informative and perplexing than the XKRY family is the CDY family. The CDY copies are scattered over different palindromes similar to XKRY, although more densely (Fig. 1). Consistent with the early retroposition of CDY (Dorus et al. 2003), we date the differentiation from CDYL as 159 ± 13 Mya (Fig. 4; Table 3). Subsequently in the primate lineage, the family expanded mostly in P1, P3, and P5. Four pairs of CDY1A/1B, CDYP9/P14, CDYP10/P13, and CDYP11/P12 in P1 are substantially different from each other, and so are three pairs of CDY2A/2B, CDYP2/P5, and CDYP3/P4 in P5. In terms of node heights in the linearized tree, CDY1A/1B and CDYP10/P13 differ from each other by 0.048 ± 0.009, these four differ from CDYP11/P12 by 0.058 ± 0.010, and these six differ from the most distinct CDYP9/P14 by 0.139 ± 0.015. We can find almost the same extent of sequence divergences among CDY copies in P5. Thus, the earliest divergence of CDY copies within P1 or P5 goes back to >80 Mya, and the latest is as old as 30 Myr.
Figure 4.
NJ tree of CDY drawn in the same way and with the same gene nomenclature system as in Figure 2. Palindromic locations of human CDY copies are indicated by bracketed P1, P3, and P5.
RBMY and TSPY also provide useful information about the age of IR2 and the TSPY array, respectively (Fig. 1). The height of node Y0 between RBMY2C/2D in P1 and six nearly identical RBMY1 copies in P3 and IR2 is 0.069 ± 0.008 (Table 3). The two subfamilies thus diverged 43 ± 5 Mya. TSPY underwent massive duplication independently in individual Eutherians, and some of 14 human TSPY copies exhibit ancient duplication. There exist distinct TSPY2 and TSPY13 within the TSPY array, as well as TSPY14 located between P3 and IR2. The height of node Y0 leading to the most distinct TSPY13 is 0.091 ± 0.009 (57 ± 6 Myr), and that of node Y1 and Y2 is 0.056 ± 0.007 (35 ± 4 Myr).
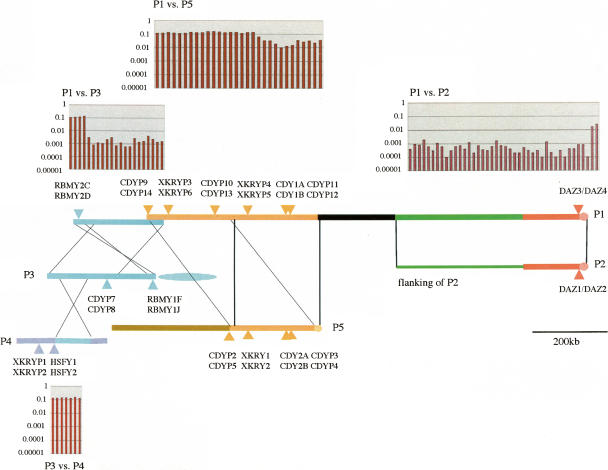
In short, major expansions of the gene families mentioned above occurred in the stem lineage of Catarrhini, 30–50 Mya. However, different palindromes and inverted repeats contain nearly identical copies as well. Like four DAZ genes in P1 and P2, mutually distinct CDY1A/1B and CDYP11/P12 pairs in P1 are closely related to mutually distinct CDY2A/2B and CDYP3/P4 pairs in P5, respectively (Figs. 1 and 4). Also, CDYP7/P8 in P3 show high sequence similarity to CDYP9/P14 in P1, and RBMY1F/1J in P3 are nearly identical to four other RBMY1 copies in IR2. If such high sequence similarity is restricted to genetic loci, transposition or retroposition of genes is likely involved. A dot-plot analysis between P3 and IR2 indicates that sequence similarity is restricted to RBMY1 loci (data not shown). Since these RBMY1 copies show the same exon–intron structure, they are apparently related via recent duplicated transposition of genes. However, this is not the case for DAZ and CDY. Indeed, Kuroda-Kawaguchi et al. (2001) demonstrated that P1 shares distinct amplicons (massive repeat units) with P2 and P3, and proposed a model of Alu-mediated tandem duplication and subsequent inversion for the P1 mosaicism.
After carrying out a dot-plot analysis, we aligned homologous sequences among palindromes, to show more precisely how and to what extent P1 is related to P2, P3, and P5, as well as P3 to P4 (Fig. 5). Several features are noteworthy. Above all, four amplicons in P1, which are nicknamed as red, green, yellow, and blue in Kuroda-Kawaguchi et al. (2001), are evolutionarily related to those in other palindromes. The red and green P1 amplicons are the same as the entire P2 region and a proximal, nonpalindromic region adjacent to P2, respectively (Kuroda-Kawaguchi et al. (2001). Using the aligned sequences over the red and green amplicons, we measured the per-site nucleotide differences (P) in non-overlapping windows of size 10 kb each. Except for a small DAZ encoding region in which P = 1% ∼ 3%, the value is as low as ∼0.1%. We can also find such a low P-value between a 170-kb blue P1 ampliconic unit and the corresponding region in P3. The blue P1 amplicon contains another 40-kb inverted region of P3 with P = 13% ∼ 15%. Moreover, P3 and P4 share an ampliconic unit with P = 13% ∼ 15%. The relationship of the yellow P1 amplicon to P5 is more complicated than that of the blue P1 amplicon. For convenience, the yellow P1 amplicon is divided into three units. Of these, one 200-kb unit (a thick black line in Fig. 5) has 92%–94% similarity to a 60-kb region of Chromosome 15 and ∼80% similarity to 140-kb regions in some other chromosomes, but it does not have any homology with P5. Another 100-kb unit shows high similarity (P = 1% ∼ 3%), and the remaining 460-kb unit is a tandem duplication of a 280-kb region of P5 (P = 13% ∼ 15%). Thus, sequence similarity among ampliconic genes is a reflection of the P1 mosaicism with respect to P2, P3, P4, and P5.
Figure 5.
Sequence similarity of P1 against P2, P3, P4, and P5. The yellow amplicon in Kuroda-Kawaguchi et al. (2001) is divided into two: black, which has homology with autosomes, and yellow, which contains tandem duplication of a P5 ampliconic unit. Each thick line represents both arms in a palindrome, and a thin green line adjacent to P2 is nonpalindromic. Three nearly identical BYP2 copies are located in the green ampliconic unit, and two distinct pairs of PRY copies are located in a blue ampliconic unit shared by P3 and P4. In non-overlapping windows of each size 10 kb, the nucleotide differences per site (P) between P1 and other palindromic amplicons are given above P1 and below P4. Note that the P-value (ordinate) is represented in common logarithms.
Discussion
Out of seven families in human Y amplicons, we showed that five were derived from proto-X/Y gene pairs. The extent of synonymous divergences of these ampliconic genes from their X-linked gametologs differs greatly, depending on the latter chromosomal locations (Table 3). When an X-linked gametolog is located in stratum 1 (XKRX, HSFX, and RBMX), stratum 2 (TSPX), and stratum 4 (VCX), the differentiation between X and Y copies occurred more than 210, 180, and 50 Mya, respectively. Thus, the proto-X/Y-derived genes in the ampliconic class of euchromatic sequences began to differentiate at the same time as those in the X-degenerate class, and both classes are relics of ancient autosomes (Skaletsky et al. 2003). However, these two classes of sequences and genes encoded therein were subjected to different fates. For instance, AMELY (Iwase et al. 2003) and VCY derived in XAR exemplify the difference: Whereas AMELY remains a single, housekeeping copy, VCY was duplicated so as to produce an identical pair in P8 and a testis-specific function. Incidentally, the presence of proto-X/Y-derived VCY in P8 implies per Ockham’s razor that this palindrome cannot be older than the age of stratum 4, >50 Mya.
Transposition and retroposition of autosomal genes were once regarded as major mechanisms for generating ampliconic genes, but they actually contributed to generation of the DAZ and CDY gene families only. As discussed by Saxena et al. (1996, 2000), Agulnik et al. (1998), and Gromoll et al. (1999), a complete copy of autosomal DAZL was transposed on the Y chromosome in the stem lineage of Catarrhini, and the whole intron-containing transcript unit of this newly transposed gene (DAZ) was tetraplicated in P1 and P2. High sequence similarity between DAZ1/2 in P2 and DAZ3/4 in P1 (Fig. 5) indicate the hominoid or hominid-specific duplication or conversion between P1 and P2. Likewise, the retroposition origin is unique to CDY (Lahn and Page 1997, 1999a; Dorus et al. 2003). However, the presence of intron-containing CDX, annotated as a pseudogene in NCBI (Build 36.1), is enigmatic. Sequence similarity between the 5′ portion of CDX and mature CDYL transcripts suggests that CDX arose via duplicated transposition of CDYL (Fig. 4). If the transposition occurred after the formation of stratum 4, CDX is likely to be specific to the X chromosome. In the opposite situation, the transposition could generate an intron-containing CDY copy, as the sex chromosomal differentiation proceeded in stratum 4. In either case, retroposition and transposition of autosomal genes played relatively minor roles in originating ampliconic genes. However, the reverse process occurred more frequently. Transposition of XKRY produced XKRYL, and retroposition of HSFY, RBMX, and TSPX produced HSFYL, RBMXL, and TSPYL (more logically designated as TSPXL), respectively. Since the latter three genes are highly expressed in germ-cell lines, transcripts have ample opportunity to be retroposed and integrated into the genome (Zhang and Gerstein 2004; Cheng et al. 2005).
Unlike the X-degenerate segments, the ampliconic segments underwent massive amplifications during primate evolution. We showed that most distinct copies in XKRY, HSFY, RBMY, TSPY, and CDY families were established in the stem lineage of Catarrhini. There are two notable features in the pattern and degree of sequence identity among those copies. First, within a palindrome, we can always find two nearly identical copies of a given gene family at symmetric arm positions (Skaletsky et al. 2003), yet copies at different arm positions are substantially different from each other. Palindrome P1 harbors two such paired XKRY and four such paired CDY copies in our data set. The estimated ages of these paired copies are similar and older than 30 Myr irrespective of gene families. Palindrome P5 harbors three such paired CDY. Again, these paired copies in P5 are as distinct as those in P1. Second, there are paired copies that are identical or nearly so even when they are located in different palindromes P1, P2, P3, P5, and/or IR2 (Figs. 3 and 4). Such high sequence similarity among copies compelled us to extend our analyses. As mentioned, the previous dot-plot analysis for P1, P2, and P3 indicated the elaborate mosaicism of P1 (Kuroda-Kawaguchi et al. 2001). We further found that (1) P1 and P3 are also related to P5 and P4, respectively; (2) some P1 amplicons are reticulately related to other palindromes via small shared ampliconic units; and (3) sequence similarity among gene copies is well correlated with that among those ampliconic units (Fig. 5).
The mosaicism of P1 is so extensive that >80% of P1 is related directly to P2, P3, and P5 and indirectly to P4. The observed P-values (per-site nucleotide differences) indicate three distinct levels of evolutionary relatedness of ampliconic units between different palindromes; the lowest level of P < 0.1% in the red, green, and blue amplicons, the middle level of P = 1% ∼ 3% in the red and yellow amplicons, and the highest level of P = 13% ∼ 15% in the yellow and blue amplicons. The yellow amplicon in P1 also contains a black ampliconic unit (Fig. 5) that is totally unrelated to any other palindrome in the Y chromosome. Rather, a subregion of the black ampliconic unit encoding transcription units CSPG4LY and GOLGA2LY (Kuroda-Kawaguchi et al. 2001; Skaletsky et al. 2003) is closely related to a region in chromosome 15 (P < 5%), whereas the remaining subregions show the P-values of ∼20% with various chromosomes. It is conceivable that P1 originally consisted of a small region and then repeatedly acquired other ampliconic units in P2, P3, P5, and autosomes or that P1 simply exchanged ampliconic sequences without substantial expansion in its size. In either case, the extent of sequence differences in Figure 5 dates the earliest sequence acquisition or exchange as ∼50 Mya in the stem lineage of Catarrhini, the second as 3–9 Mya in the hominid or hominoid lineage, and the latest as <0.3 Mya in modern humans. Rozen et al. (2003) reported the presence of P1 in chimpanzees and bonobos. However, since they examined two inner and outer boundaries of a palindrome, the internal palindromic structure is not known in these and other apes. It is extremely unlikely that these relatives possess the same red and green P1 ampliconic structure as humans do. Also, it is questionable if the yellow and blue P1 amplicons in chimpanzees are the same as those in humans.
In conclusion, owing to extensive sequence transfers of ampliconic units, the age of a palindrome differs from region to region. Nonetheless, the age of an ampliconic unit is well correlated with the age of genes encoded therein. Undoubtedly, like genes, major ampliconic units must have been established in the stem lineage of Catarrhini. While these repeat units have since evolved en bloc, palindromes have been extensively modified by acquisition and/or exchange of repeat units in other palindromes. It would be surprising if such modifications in ampliconic gene contents and structures had nothing to do with changes in spematogenesis or sperm production in the lineage leading from the Catarrhini ancestor to modern humans. Indeed, the AZFc (azoospermia factor c) region, whose deletion is known as the most common cause of spermatogenetic failure in humans, largely consists of P1 and P2 (Kuroda-Kawaguchi et al. 2001). Our finding that P2 is quantitatively similar to and qualitatively redundant to P1 may imply that evolution of P1 has played critical roles in developing human-specific spermatogenesis.
Methods
Our analysis is based on DNA sequence data in the NCBI genome database as of July 31, 2006. We retrieved DNA sequences of the nine ampliconic genes and related pseudogenes from the human genome database. At the same time, we located them in the Y chromosome based on information about locus positions in contigs as well as results of DOTTER and Map Viewer after BLASTN. We also used DOTTER to examine sequence similarity between different amplicons. The current chromosomal locations of the ampliconic genes in the NCBI Master Map appear slightly different from the initial proposal by Skaletsky et al. (2003). Using BLASTN and GENE, we searched for X-linked and autosomal copies for a given gene family. Some genes homologous to human ampliconic genes can be found even in fish. However, we did not use distantly related orthologs in our analysis because the origin of the primate Y chromosome surely postdated the divergence between mammals and birds.
To designate an ortholog in figures, we use a four-letter prefix to identify the genus and species names. For example, XKRY in humans (Homo sapiens) and chimpanzees (Pan troglodytes) is designated as HosaXKRY and PatrXKRY, respectively. Autosomal genes and pseudogenes are indicated by addition of the suffix L or P, respectively. When there are multiple human Y-linked pseudogenes in a given gene family, those without official names are numbered from the short arm end of the Y (Fig. 1; see also Kuroda-Kawaguchi et al. 2001). The sequence alignments are given in Supplemental Figure 1.
For phylogenetic analyses, we use the coding regions only to minimize misalignments among distantly related and rapidly evolving Y-linked sequences. However, some genes, such as DAZ, contain repeated exons or tandem repeats that underwent extensive expansions and contractions. We exclude such repeats, as our main purpose is to comprehend the evolutionary relationships among Y-linked, X-linked, and autosomal copies when they are present. We use the NJ method by Saitou and Nei (1987) to determine the topology as well as the method by Takezaki et al. (1995) to determine the height (h) of a specified node in the linearized tree. However, regarding node XY at which X and Y gametologs differentiate, we cannot use the linearized tree method because these gametologs apparently evolved with different rates. We apply the same caution to node YL at which Y-linked and autosomal copies differentiate via transposition or retroposition. We simply compute the sequence divergences per synonymous site (kXY or kYL) after multiple-hit corrections (Jukes and Cantor 1969; Kimura 1980). To date, for XY, YL, or other nodes in Figures 2 and 3, we use the silent nucleotide substitution rate per site per year of rL = 1.0 × 10−9 for the autosomes, rX = 0.83 × 10−9 for the X chromosome, and rY = 1.6 × 10−9 for the Y chromosome (Ebersberger et al. 2002). These are estimated by the 1.2%, 1.0%, and 1.9% sequence divergences between the human and chimpanzee chromosomes, respectively, and the assumption of the 6-Myr species divergence time (Haile-Selassie 2001). Applying these rates to kXY, we date node XY as
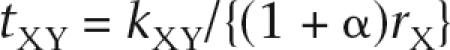 |
where α is the ratio of rY to rX. If we assume α = 2 (Ebersberger et al. 2002), the tXY value is given by kXY/(3rX) rather than kXY/(2rX) under the assumption of rY = rX. Similarly, we date node YL by
Regarding the height (h) of a node leading to a group of Y-linked copies, we date the node simply by
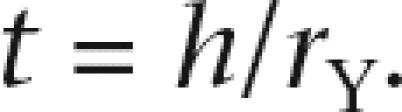 |
Acknowledgments
This work was supported in part by the Japan Society for Promotion of Science Grants 12304046 to N.T., and MEXT scholarship assistance to B.K.B.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5734907
References
- Agulnik A.I., Zharkikh A., Boettger-Tong H., Bourgeron T., McEireavey K., Bishop C.E., Zharkikh A., Boettger-Tong H., Bourgeron T., McEireavey K., Bishop C.E., Boettger-Tong H., Bourgeron T., McEireavey K., Bishop C.E., Bourgeron T., McEireavey K., Bishop C.E., McEireavey K., Bishop C.E., Bishop C.E. Evolution of the DAZ gene family suggests that Y-linked DAZ plays little, or a limited, role in spermatogenesis but underlines a recent African origin for human populations. Hum. Mol. Genet. 1998;7:1371–1377. doi: 10.1093/hmg/7.9.1371. [DOI] [PubMed] [Google Scholar]
- Arnemann J., Epplen J.T., Cooke H.J., Sauermann U., Engel W., Schmidtke J., Epplen J.T., Cooke H.J., Sauermann U., Engel W., Schmidtke J., Cooke H.J., Sauermann U., Engel W., Schmidtke J., Sauermann U., Engel W., Schmidtke J., Engel W., Schmidtke J., Schmidtke J. A human Y-chromosomal DNA sequence expressed in testicular tissue. Nucleic Acids Res. 1987;15:8713–8724. doi: 10.1093/nar/15.21.8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai N.-N., Zhou H., Hernandez J., Najmabadi H., Bhasin S., Yen P.H., Zhou H., Hernandez J., Najmabadi H., Bhasin S., Yen P.H., Hernandez J., Najmabadi H., Bhasin S., Yen P.H., Najmabadi H., Bhasin S., Yen P.H., Bhasin S., Yen P.H., Yen P.H. Structure and organization of the RBMY genes on the human Y chromosome: Transposition and amplification of an ancestral autosomal hnRNPG gene. Genomics. 1998;49:283–289. doi: 10.1006/geno.1998.5255. [DOI] [PubMed] [Google Scholar]
- Cheng J., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Kapranov P., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Drenkow J., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Dike S., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Brubaker S., Patel S., Long J., Stern D., Tammana H., Helt G., Patel S., Long J., Stern D., Tammana H., Helt G., Long J., Stern D., Tammana H., Helt G., Stern D., Tammana H., Helt G., Tammana H., Helt G., Helt G., et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- Delbridge M.L., Harry J.L., Toder R., O’Neill R.J., Ma K., Chandley A.C., Graves J.A.M., Harry J.L., Toder R., O’Neill R.J., Ma K., Chandley A.C., Graves J.A.M., Toder R., O’Neill R.J., Ma K., Chandley A.C., Graves J.A.M., O’Neill R.J., Ma K., Chandley A.C., Graves J.A.M., Ma K., Chandley A.C., Graves J.A.M., Chandley A.C., Graves J.A.M., Graves J.A.M. A human candidate spermatogenesis gene, RBM1, is conserved and amplified on the marsupial Y chromosome. Nat. Genet. 1997;15:131–136. doi: 10.1038/ng0297-131. [DOI] [PubMed] [Google Scholar]
- Delbridge M.L., Ma K., Subbarao M.N., Cooke H.J., Bhasin S., Graves J.A.M., Ma K., Subbarao M.N., Cooke H.J., Bhasin S., Graves J.A.M., Subbarao M.N., Cooke H.J., Bhasin S., Graves J.A.M., Cooke H.J., Bhasin S., Graves J.A.M., Bhasin S., Graves J.A.M., Graves J.A.M. Evolution of mammalian HNRPG and its relationship with the putative azoospermia factor RBM. Mamm. Genome. 1998;9:168–170. doi: 10.1007/s003359900712. [DOI] [PubMed] [Google Scholar]
- Delbridge M.L., Lingenfelter P.A., Disteche C.M., Graves J.A.M., Lingenfelter P.A., Disteche C.M., Graves J.A.M., Disteche C.M., Graves J.A.M., Graves J.A.M. The candidate spermatogenesis gene RBMY has a homolog on the human X chromosome. Nat. Genet. 1999;22:223–224. doi: 10.1038/10279. [DOI] [PubMed] [Google Scholar]
- Delbridge M.L., Longepied G., Depetris D., Mattei M.-G., Disteche C., Graves J.A.M., Michell M.J., Longepied G., Depetris D., Mattei M.-G., Disteche C., Graves J.A.M., Michell M.J., Depetris D., Mattei M.-G., Disteche C., Graves J.A.M., Michell M.J., Mattei M.-G., Disteche C., Graves J.A.M., Michell M.J., Disteche C., Graves J.A.M., Michell M.J., Graves J.A.M., Michell M.J., Michell M.J. TSPY, the candidate gonadoblastoma gene on the human Y chromosome, has a widely expressed homologue on the X—Implications for Y chromosome evolution. Chromosome Res. 2004;12:345–356. doi: 10.1023/B:CHRO.0000034134.91243.1c. [DOI] [PubMed] [Google Scholar]
- Dorus S., Gilbert S.L., Foster M.L., Foster M.L., Barndt R.J., Lahn B., Gilbert S.L., Foster M.L., Foster M.L., Barndt R.J., Lahn B., Foster M.L., Foster M.L., Barndt R.J., Lahn B., Foster M.L., Barndt R.J., Lahn B., Barndt R.J., Lahn B., Lahn B. The CDY-related gene family: Coordinated evolution in copy number, expression profile and protein sequence. Hum. Mol. Genet. 2003;12:1643–1650. doi: 10.1093/hmg/ddg185. [DOI] [PubMed] [Google Scholar]
- Ebersberger I., Metzler D., Schwarz C., Pääbo S., Metzler D., Schwarz C., Pääbo S., Schwarz C., Pääbo S., Pääbo S. Genomewide comparison of DNA sequences between humans and chimpanzees. Am. J. Hum. Genet. 2002;70:1490–1497. doi: 10.1086/340787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott D.J., Venables J.P., Newton C.S., Lawson D., Boyle S., Eperon I.C., Cooke H.J., Venables J.P., Newton C.S., Lawson D., Boyle S., Eperon I.C., Cooke H.J., Newton C.S., Lawson D., Boyle S., Eperon I.C., Cooke H.J., Lawson D., Boyle S., Eperon I.C., Cooke H.J., Boyle S., Eperon I.C., Cooke H.J., Eperon I.C., Cooke H.J., Cooke H.J. An evolutionarily conserved germ cell-specific hnRNP is encoded by a retrotransposed gene. Hum. Mol. Genet. 2000;9:2117–2124. doi: 10.1093/hmg/9.14.2117. [DOI] [PubMed] [Google Scholar]
- Fukami M., Kirsch S., Schiller S., Richter A., Benes V., Franco B., Muroya K., Rao E., Merker S., Niesler B., Kirsch S., Schiller S., Richter A., Benes V., Franco B., Muroya K., Rao E., Merker S., Niesler B., Schiller S., Richter A., Benes V., Franco B., Muroya K., Rao E., Merker S., Niesler B., Richter A., Benes V., Franco B., Muroya K., Rao E., Merker S., Niesler B., Benes V., Franco B., Muroya K., Rao E., Merker S., Niesler B., Franco B., Muroya K., Rao E., Merker S., Niesler B., Muroya K., Rao E., Merker S., Niesler B., Rao E., Merker S., Niesler B., Merker S., Niesler B., Niesler B., et al. A member of a gene family on Xp23.3, VCX-A, is deleted in patients with X-linked nonspecific mental retardation. Am. J. Hum. Genet. 2000;67:563–573. doi: 10.1086/303047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves J.A.M. The origin and function of the mammalian Y chromosome and Y-borne genes—An evolving understanding. Bioessays. 1995;17:311–320. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves J.A.M. The rise and fall of SRY. Trends Genet. 2002;18:259–264. doi: 10.1016/s0168-9525(02)02666-5. [DOI] [PubMed] [Google Scholar]
- Gromoll J., Weinbauer G.F., Skaletsky H., Schlatt S., Rocchietti-March M., Page D.C., Niesclag E., Weinbauer G.F., Skaletsky H., Schlatt S., Rocchietti-March M., Page D.C., Niesclag E., Skaletsky H., Schlatt S., Rocchietti-March M., Page D.C., Niesclag E., Schlatt S., Rocchietti-March M., Page D.C., Niesclag E., Rocchietti-March M., Page D.C., Niesclag E., Page D.C., Niesclag E., Niesclag E. The Old World monkey DAZ (Deleted in Azoospermia) gene yields insights into the evolution of the DAZ gene cluster on the human Y chromosome. Hum. Mol. Genet. 1999;8:2017–2024. doi: 10.1093/hmg/8.11.2017. [DOI] [PubMed] [Google Scholar]
- Haile-Selassie Y. Late Miocene hominids from the middle Awash, Ethiopia. Nature. 2001;412:178–181. doi: 10.1038/35084063. [DOI] [PubMed] [Google Scholar]
- Hughes J.F., Skaletsky H., Pyntikova T., Minx P.J., Graves T., Rozen S., Wilson R.K., Page D.C., Skaletsky H., Pyntikova T., Minx P.J., Graves T., Rozen S., Wilson R.K., Page D.C., Pyntikova T., Minx P.J., Graves T., Rozen S., Wilson R.K., Page D.C., Minx P.J., Graves T., Rozen S., Wilson R.K., Page D.C., Graves T., Rozen S., Wilson R.K., Page D.C., Rozen S., Wilson R.K., Page D.C., Wilson R.K., Page D.C., Page D.C. Conservation of Y-linked genes during human evolution revealed by comparative sequencing in chimpanzee. Nature. 2005;437:101–104. doi: 10.1038/nature04101. [DOI] [PubMed] [Google Scholar]
- Iwase M., Satta Y., Hirai Y., Hirai H., Imai H., Takahata N., Satta Y., Hirai Y., Hirai H., Imai H., Takahata N., Hirai Y., Hirai H., Imai H., Takahata N., Hirai H., Imai H., Takahata N., Imai H., Takahata N., Takahata N. Differentiation of sex chromosomal amelogenin genes in primates and other mammals. Proc. Natl. Acad. Sci. 2003;100:5258–5263. [Google Scholar]
- Jakubiczka S., Schnieders F., Schmidtke J., Schnieders F., Schmidtke J., Schmidtke J. A bovine homologue of the human TSPY gene. Genomics. 1993;17:732–735. doi: 10.1006/geno.1993.1394. [DOI] [PubMed] [Google Scholar]
- Jukes T.H., Cantor C.R., Cantor C.R. Evolution of protein molecules. In: Munro H.N., editor. Mammalian protein metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Kumar S., Hedges S.B., Hedges S.B. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T., Skaletsky H., Brown L.G., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Skaletsky H., Brown L.G., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Brown L.G., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Minx P.J., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Cordum H.S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Waterston R.H., Wilson R.K., Silber S., Oates R., Rozen S., Wilson R.K., Silber S., Oates R., Rozen S., Silber S., Oates R., Rozen S., Oates R., Rozen S., Rozen S., et al. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat. Genet. 2001;29:279–286. doi: 10.1038/ng757. [DOI] [PubMed] [Google Scholar]
- Kuroki Y., Toyoda A., Noguchi H., Taylor T.D., Itoh T., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Toyoda A., Noguchi H., Taylor T.D., Itoh T., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Noguchi H., Taylor T.D., Itoh T., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Taylor T.D., Itoh T., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Itoh T., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Kim D.S., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Kim D.W., Choi S.H., Kim I.C., Choi H.H., Choi S.H., Kim I.C., Choi H.H., Kim I.C., Choi H.H., Choi H.H., et al. Comparative analysis of chimpanzee and human Y chromosomes unveils complex evolutionary pathway. Nat. Genet. 2006;38:158–167. doi: 10.1038/ng1729. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. Functional coherence of the human Y chromosome. Science. 1997;278:675–680. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. Retroposition of autosomal mRNA yielded testis-specific gene family on human Y chromosome. Nat. Genet. 1999a;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. Four evolutionary strata on the human X chromosome. Science. 1999b;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lahn B.T., Page D.C., Page D.C. A human sex-chromosomal gene family expressed in male germ cells and encoding variably charged proteins. Hum. Mol. Genet. 2000;9:311–319. doi: 10.1093/hmg/9.2.311. [DOI] [PubMed] [Google Scholar]
- Lingenfelter P.A., Delbridge M.L., Thomas S., Hoeskstra H.E., Mitchell M.J., Graves J.A.M., Disteche C.M., Delbridge M.L., Thomas S., Hoeskstra H.E., Mitchell M.J., Graves J.A.M., Disteche C.M., Thomas S., Hoeskstra H.E., Mitchell M.J., Graves J.A.M., Disteche C.M., Hoeskstra H.E., Mitchell M.J., Graves J.A.M., Disteche C.M., Mitchell M.J., Graves J.A.M., Disteche C.M., Graves J.A.M., Disteche C.M., Disteche C.M. Expression and conservation of processed copies of the RBMX gene. Mamm. Genome. 2001;12:538–545. doi: 10.1007/s00335001-0003-z. [DOI] [PubMed] [Google Scholar]
- Manz E., Schnieders F., Brechlin A.M., Schmidtke J., Schnieders F., Brechlin A.M., Schmidtke J., Brechlin A.M., Schmidtke J., Schmidtke J. TSPY-related sequences represent a microheterogeneous gene family organized as constitutive elements in DYZ5 tandem repeat units on the human Y chromosome. Genomics. 1993;17:726–731. doi: 10.1006/geno.1993.1393. [DOI] [PubMed] [Google Scholar]
- Martin R.D. Primate origins: Plugging the gaps. Nature. 1993;363:223–234. doi: 10.1038/363223a0. [DOI] [PubMed] [Google Scholar]
- Mazeyrat S., Mitchell M.J., Mitchell M.J. Rodent Y chromosome TSPY gene is functional in rat and non-functional in mouse. Hum. Mol. Genet. 1998;7:557–562. doi: 10.1093/hmg/7.3.557. [DOI] [PubMed] [Google Scholar]
- Ohno S. Sex chromosomes and sex-linked genes. Springer; Berlin-Heidelberg-New York: 1967. [Google Scholar]
- Ross M.T., Grafham D.V., Coffey A.J., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Grafham D.V., Coffey A.J., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Coffey A.J., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Coffey A.J., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Scherer S., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., McLay K., Muzny D., Platzer M., Howell G.R., Burrows C., Muzny D., Platzer M., Howell G.R., Burrows C., Platzer M., Howell G.R., Burrows C., Howell G.R., Burrows C., Burrows C., et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–336. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., Marszalek J.D., Minx P.L., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Skaletsky H., Marszalek J.D., Minx P.L., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Marszalek J.D., Minx P.L., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Minx P.L., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Cordum H.S., Waterston R.H., Wilson R.K., Page D.C., Waterston R.H., Wilson R.K., Page D.C., Wilson R.K., Page D.C., Page D.C. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature. 2003;423:873–876. doi: 10.1038/nature01723. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M., Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saxena R., Brown L.G., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Brown L.G., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Hawkins T., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Alagappan R.K., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Skaletsky H., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Reeve M.P., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Reijo R., Rozen S., Dinulos M.B., Disteche C.M., Rozen S., Dinulos M.B., Disteche C.M., Dinulos M.B., Disteche C.M., Disteche C.M., et al. The DAZ gene cluster on the human Y chromosome arose from an autosomal gene that was transposed, repeatedly amplified and pruned. Nat. Genet. 1996;14:292–299. doi: 10.1038/ng1196-292. [DOI] [PubMed] [Google Scholar]
- Saxena R., de Vries J.W.A., Repping S., Alagappan R.K., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., de Vries J.W.A., Repping S., Alagappan R.K., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., Repping S., Alagappan R.K., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., Alagappan R.K., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., Skaletsky H., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., Brown L.G., Ma P., Chen E., Hoovers J.M., Page D.C., Ma P., Chen E., Hoovers J.M., Page D.C., Chen E., Hoovers J.M., Page D.C., Hoovers J.M., Page D.C., Page D.C. Four DAZ genes in two clusters found in the AZFc region of the human Y chromosome. Genomics. 2000;67:256–267. doi: 10.1006/geno.2000.6260. [DOI] [PubMed] [Google Scholar]
- Schnieders F., Dork T., Arnemann J., Vogel T., Werner M., Schmidtke J., Dork T., Arnemann J., Vogel T., Werner M., Schmidtke J., Arnemann J., Vogel T., Werner M., Schmidtke J., Vogel T., Werner M., Schmidtke J., Werner M., Schmidtke J., Schmidtke J. Testis-specific protein, Y-encoded (TSPY) expression in testicular tissues. Hum. Mol. Genet. 1996;5:1801–1807. doi: 10.1093/hmg/5.11.1801. [DOI] [PubMed] [Google Scholar]
- Skaletsky H., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Kuroda-Kawaguchi T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Minx P.J., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Cordum H.S., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Hillier L., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Brown L.G., Repping S., Pyntikova T., Ali J., Bieri T., Repping S., Pyntikova T., Ali J., Bieri T., Pyntikova T., Ali J., Bieri T., Ali J., Bieri T., Bieri T., et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Stouffs K., Lissens W., Landuyt L.V., Tournaye H., Steirteghem A.V., Liebaers I., Lissens W., Landuyt L.V., Tournaye H., Steirteghem A.V., Liebaers I., Landuyt L.V., Tournaye H., Steirteghem A.V., Liebaers I., Tournaye H., Steirteghem A.V., Liebaers I., Steirteghem A.V., Liebaers I., Liebaers I. Characterization of the genomic organization, localization and expression of four PRY genes (PRY1, PRY2, PRY3 and PRY4) Mol. Hum. Reprod. 2001;17:603–610. doi: 10.1093/molehr/7.7.603. [DOI] [PubMed] [Google Scholar]
- Takahata N. Tobias P.V., et al. Humanity from African Naissance to coming millennia. Witwatersrand University Press; Johannesburg, South Africa: 2001. Molecular phylogeny and demographic history of humans; pp. 299–305. [Google Scholar]
- Takezaki N., Rzhetsky A., Nei M., Rzhetsky A., Nei M., Nei M. Phylogenetic test of the molecular clock and linearized tree. Mol. Biol. Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- Tessari A., Salata E., Ferlin A., Bartoloni L., Slongo M.L., Foresta C., Salata E., Ferlin A., Bartoloni L., Slongo M.L., Foresta C., Ferlin A., Bartoloni L., Slongo M.L., Foresta C., Bartoloni L., Slongo M.L., Foresta C., Slongo M.L., Foresta C., Foresta C. Characterization of HSPY, a novel AZFb gene on the Y chromosome with a possible role in human spermatogenesis. Mol. Hum. Reprod. 2004;10:253–258. doi: 10.1093/molehr/gah036. [DOI] [PubMed] [Google Scholar]
- Waters P.D., Delbridge M.L., Deakin J.E., EI-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Delbridge M.L., Deakin J.E., EI-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Deakin J.E., EI-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., EI-Mogharbel N., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Kirby P.J., Carvalho-Silva D.R., Graves J.A.M., Carvalho-Silva D.R., Graves J.A.M., Graves J.A.M. Autosomal location of genes from the conserved mammalian X in the platypus (Ornithorhynchus abatinus): Implication for mammalian sex chromosome evolution. Chromosome Res. 2005;13:401–410. doi: 10.1007/s10577-005-0978-5. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Gerstein M., Gerstein M. Large-scale analysis of pseudogenes in the human genome. Curr. Opin. Genet. Dev. 2004;14:328–335. doi: 10.1016/j.gde.2004.06.003. [DOI] [PubMed] [Google Scholar]
- Zou S.W., Zhang J.C., Zhang X.D., Miao S.Y., Zong S.H., Sheng Q., Wang L.F., Zhang J.C., Zhang X.D., Miao S.Y., Zong S.H., Sheng Q., Wang L.F., Zhang X.D., Miao S.Y., Zong S.H., Sheng Q., Wang L.F., Miao S.Y., Zong S.H., Sheng Q., Wang L.F., Zong S.H., Sheng Q., Wang L.F., Sheng Q., Wang L.F., Wang L.F. Expression and localization of VCX/Y proteins and their possible involvement in regulation of ribosome assembly during spermatogenesis. Cell Res. 2003;13:171–177. doi: 10.1038/sj.cr.7290161. [DOI] [PubMed] [Google Scholar]