Abstract
It has been well documented that 22q11 contains one of the most rearrangement-prone sites in the human genome, where the breakpoints of a number of constitutional translocations cluster. This breakage-sensitive region is located within one of the remaining unclonable gaps from the human genome project, suggestive of a specific sequence recalcitrant to cloning. In this study, we cloned a part of this gap and identified a novel 595-bp palindromic AT-rich repeat (PATRR). To date we have identified three translocation-associated PATRRs. They have common characteristics: (1) they are AT-rich nearly perfect palindromes, which are several hundred base pairs in length; (2) they possess non-AT-rich regions at both ends of the PATRR; (3) they display another nearby AT-rich region on one side of the PATRR. All of these features imply a potential for DNA secondary structure. Sequence analysis of unrelated individuals indicates no major size polymorphism, but shows minor nucleotide polymorphisms among individuals and cis-morphisms between the proximal and distal arms. Breakpoint analysis of various translocations indicates that double-strand-breakage (DSB) occurs at the center of the palindrome, often accompanied by a small symmetric deletion at the center. The breakpoints share only a small number of identical nucleotides between partner chromosomes. Taken together, these features imply that the DSBs are repaired through nonhomologous end joining or single-strand annealing rather than a homologous recombination pathway. All of these results support a previously proposed paradigm that unusual DNA secondary structure plays a role in the mechanism by which palindrome-mediated translocations occur.
To elucidate the molecular etiology of genetic diseases, numerous attempts have been directed toward cloning of disease-causing genomic rearrangements. This has resulted in the identification of unusual genomic structures such as fragile sites and palindromes. The analysis of such regions has been difficult because secondary structure often causes DNA polymerase pausing in attempts at PCR/sequencing and results in instability in cloning in hosts like Escherichia coli (Yu et al. 1997; Hewett et al. 1998; Ried et al. 2000). One such example is the palindromic AT-rich repeat (PATRR). PATRRs were first identified at the 11q23 breakpoints of the constitutional t(11;22)(q23;q11), which is the only known recurrent non-Robertsonian translocation in humans (Kurahashi et al. 2000a; Edelmann et al. 2001; Tapia-Paez et al. 2001). In vitro studies indicate that the PATRR adopts an unstable cruciform structure, which is likely to contribute to its translocation breakpoint activity (Kurahashi et al. 2004). The PATRR at the 11q23 breakpoint (PATRR11) has been deleted from BAC RP11-442e11, an 11q23 sequence source in the public database. The PATRR11 was eventually cloned and sequenced by PCR of genomic DNA, but this required considerable effort (Kurahashi and Emanuel 2001a).
The exact sequence of the 22q11 breakpoint of this constitutional translocation is still unknown since it is located within one of the gaps, remaining in the human genome assembly due to challenges in cloning and sequencing of such complex regions using standard approaches (Fig. 1A) (Kurahashi et al. 2000a). The breakpoint region has been resistant even after extensive screening of YAC/BAC/PAC libraries. The breakpoints of a variety of translocations involving 22q11 cluster within this region, suggesting that this region is highly unstable in the human genome as well as in the numerous host organisms used for cloning (Budarf et al. 1996; Kehrer-Sawatzki et al. 1997; Rhodes et al. 1997; Debeer et al. 2002; Spiteri et al. 2003). To date, three translocations involving chromosome 22q11 in addition to the t(11;22) have been analyzed in detail (Kurahashi et al. 2003; Nimmakayalu et al. 2003; Gotter et al. 2004). Although the extended breakpoint regions on 22q11 have not been cloned in any of these cases, junction fragments from both translocation derivative chromosomes have been isolated. Thus, a putative structure for the breakpoint region can be inferred from the sequence information of the junction fragments of the t(11;22) and the three other translocations. These data suggest the presence of a PATRR-like sequence (PATRR22) at the breakpoints. However, since this region is known to be located within one of the low-copy repeats distributed within 22q11 (LCR22s) (Fig. 1A) (Shaikh et al. 1999, 2000), duplicated sequence may prevent the PATRR22 from being isolated owing to competing sequence, which creates excessive background during PCR amplification.
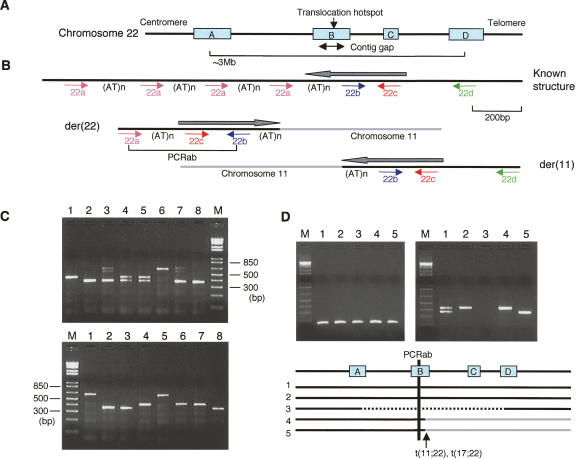
Figure 1.
Development of a PCR system specific for the PATRR22 region. (A) LCR22 in 22q11.2. There are at least four copies of LCR22, LCR22A, LCR22B, LCR22C, and LCR22D. One of the LCR22s, LCR22B, is the site of a genomic contig gap as well as the translocation breakpoint hotspot. (B) Location and orientation of the four PCR primers on the known structure appearing in other LCR22s and the t(11;22) translocation derivative chromosomes. There are multiple copies of primer “a” as a result of tandem repeats. Black lines indicate chromosome 22, while light-gray lines indicate chromosome 11. All of the genomic clones present in the database manifest the “known structure.” Only a der(22)-containing clone should be positive for the PCRab products. “(AT)n”s indicate the AT-rich region. Gray arrows indicate the putative PATRR22 arms. (C) Representative results of the PCRab reaction on genomic DNA. (Top) PCRab for eight normal individuals; (bottom) PCRab for patients with the 22q11.2 deletion syndrome. The additional bands originating from heteroduplexes appear as PCR artifacts in lanes 3 and 7. (D) The PCRbc (left) and PCRab (right) for somatic cell hybrids. The positive PCRbc product arises as a result of the presence of any of the LCR22s in the hybrids, while the positive PCRab product is derived only from LCR22B. (Lane 1) Normal human; (lane 2) GM10888, a human/hamster hybrid containing a normal chromosome 22 as its only intact human chromosome; (lane 3) Cl6-2/EG, a human/hamster hybrid containing a del(22q11.2) chromosome; (lane 4) Cl-4/GB, a human/hamster hybrid cell line containing the der(22) of a constitutional t(11;22); (lane 5) GM11685 (NF13/D3), a subclone of a human/mouse hybrid cell line containing the der(22) of a constitutional t(17;22). Black solid lines in the diagram below represent the chromosome 22q11 regions present in the hybrid lines analyzed.
Further, the PATRR is difficult to amplify by PCR because of strand separation due to its AT-rich composition as well as because of DNA polymerase pausing due to its palindromic nature (Inagaki et al. 2005). We recently optimized PCR conditions for such PATRRs (Inagaki et al. 2005). In the current study, we have successfully isolated an intact 595-bp PATRR22 using optimized PCR conditions. The isolated PATRR22 sequence allowed us to compare t(11;22) junction fragments with the corresponding normal sequence. Our data suggest that the 595-bp PATRR22 itself, which comprises a nearly perfect palindromic structure, contributes to its remarkable genomic instability leading to frequent translocations.
Results
Development of a PCR screening system specific for the PATRR22
In our previous studies, we identified translocation junction fragments for both the der(11) and the der(22) using translocation-specific PCR with a chromosome 11 primer and the 22c primer that is located within the putative PATRR22 (Kurahashi et al. 2000b). Primers 22b and 22c are located within the PATRR22, while primers 22a and 22d are located outside of the PATRR22 (Fig. 1B). Subsequent analysis indicated that junction fragments from all t(11;22) translocation carriers could also be amplified with primer pairs located outside of the PATRR22, junction fragments on the der(11) with a chromosome 11 proximal primer and primer 22d, and those on the der(22) with a chromosome 11 distal primer and primer 22a (data not shown).
A BLAST search with the sequence of primers 22a, 22b, 22c, or 22d identified multiple genomic clones including those that are a part of LCR22, but none has the palindromic configuration expected based on sequences of the translocation derivatives. While the sequences of primers 22a, 22b, 22c, and 22d appear in the database with the order and orientation diagrammed as “known structure” in Figure 1B, only the der(22) chromosome of the t(11;22) manifests the structure, 22a, 22c, and 22b in this order. All of the LCR22s have been sequenced except part of LCR22B. Thus, this structure is likely to be located within the unsequenced region of LCR22B, where numerous translocation breakpoints reside. Therefore, we reasoned that PCR with primers 22a and 22b (PCRab) is likely to be specific for distinguishing this PATRR22 region from the others.
To verify the specificity of the PCRab product in the LCR22B region, we performed PCR on genomic DNA from normal healthy donors, patients with the 22q11 deletion syndrome, and somatic cell hybrids derived from patients with various structural abnormalities involving 22q11 (Budarf et al. 1996). The product of PCRab in the samples manifested size polymorphisms presumably due to variation in the length of the AT-rich region between 22a and 22b (Fig. 1B). Although three different length PCR products can be identified, no normal healthy donor yielded three PCR products. Further, all patients with the 22q11 deletion or somatic cell hybrids that carry only one human chromosome 22 yield a single PCR product (Fig. 1C,D). These data are consistent with the idea that PCRab is specifically located within the sequence gap within LCR22B. Hybrids containing the der(22) of either the t(11;22) or the t(17;22) as their only relevant chromosome are positive for the PCR product, which also locates PCRab just proximal to the 22q11 translocation breakpoints. Since PATRR22 is extremely rearrangement prone, it is suspected that there may be more than two copies of this PATRR22 within the critical region of the LCR22B (Kurahashi and Emanuel 2001a). However, these data demonstrate that PATRR22 is likely to be present as a single copy in a normal chromosome 22 (Fig. 1D). Obviously, it is still possible that more than one copy of the PATRR22 exists on a given chromosome if they contain AT-rich regions of the same size.
Molecular cloning of the hotspot for translocation breakpoints
To amplify the breakpoint region, we performed PCR using primers 22a and 22d (PCRad) in conditions optimized to amplify a known PATRR on chromosome 17 (Inagaki et al. 2005). The expected product, including the PATRR22, should be greater than 1 kb, as estimated by the junction fragments. All of the other LCR22s contain a similar structure, but the size of the PCRad product should be ∼500 bp (“known structure” in Fig. 1B). However, we obtained an extensive ladder of multiple PCR products (Fig. 2A). We cloned the PCR products that produced prominent bands, but none of them corresponded to the LCR-B-associated PATRR22.
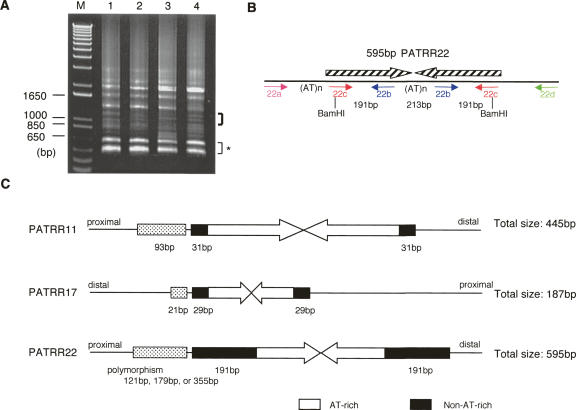
Figure 2.
Molecular cloning and characterization of the PATRR22. (A) PCRad for four individuals. PCR products appearing as prominent bands did not contain the PATRR22. The DNA fraction indicated by the thick bracket contains the authentic PATRR22. Bands indicated by a thin bracket with an asterisk are derived from LCR22s with the standard structure. (B) Structure of the PATRR22. Hatched arrows indicate each unit of inverted repeats. Small arrows depict PCR primers. (C) Characteristics of the three translocation-related PATRRs. Arrows indicate each unit of inverted repeats of the PATRR. Black boxes indicate relatively non-AT-rich regions, while stippled boxes indicate the AT-rich regions flanking the PATRRs.
One reason for multiple PCR products is that, despite optimizing the PCR conditions for amplification of the PATRR, the efficiency is still low relative to nonspecific products originating from the other LCR22s. Another reason is the presence of an AT-rich region-HSAT1-Alu cassette repeating three or four times in every 45-kb module of the LCR (Shaikh et al. 1999; Babcock et al. 2003) (Fig. 1B). Due to the repeated nature of primer 22a as well as size variation of the AT-rich region among the LCR22s, PCRad yields a highly complex ladder.
Performing PCRab on the PCRad product allowed us to identify the desired PCR products from the multiple PCRad products. We size fractionated the PCRad products by standard agarose gel and then applied PCRab to each fraction, resulting in the identification of a single positive fraction (Fig. 2A). We generated a plasmid library using the PCR products derived from the PCRab-positive fraction. The size of the PCR products (around 1 kb) in the positive fraction is reasonable for the size estimated for the translocation-derivative chromosomes, indicating that the translocation occurs without the loss of any large genomic segment. We screened a significant number of clones with the PCRab primers, and finally identified clones positive for the correct PCR products.
Plasmids positive for the PCRab product were expanded and sequenced. In our previous studies, a considerable portion of the insert was inevitably deleted from PATRR11-containing plasmids even in SURE cells. Based on the underrepresentation of clones containing the PATRR22 in genomic libraries and the fact that PATRR22 is more rearrangement prone in humans than is PATRR11, maintenance of the PATRR22 insert was predicted to be difficult. However, in contrast to the plasmids containing the PATRR11, PATRR22 clones could be stably expanded in SURE cells.
Structure of the PATRR22
We sequenced the putative PATRR22 clones using RNA polymerase-mediated sequencing and confirmed that they contained the PATRR22. The size of the PATRR22 is 595 bp. When compared with the PATRR22 sequence that we inferred from our compilation of der(11) and der(22) sequence, the assembled sequence has no large gaps and only 3 bp of additional sequence was identified at the center of the palindrome (Fig. 2B). The overall AT-content is 74.1% (441/595 nt). PATRR22 has a non-AT-rich region at both ends, which was previously referred to as the NF1L region (Shaikh et al. 1999). The AT-content of the central AT-rich region is strikingly high, 99.5% (371/373 nt). The homology between the proximal and distal arms is 98.7%, conferring a nearly perfect palindromic structure.
To date, we have identified a total of three translocation-associated PATRRs (Fig. 2C). We found three common features among the PATRR22, PATRR11, and PATRR17. (1) They comprise nearly perfect palindromes, whose lengths are several hundred base pairs. (2) An AT-rich region is located at the center, while there are non-AT-rich segments on both sides. (3) Another AT-rich region resides on one side of the PATRR. Further, in spite of their AT-rich base content, no substantial homology has been observed between the PATRR11 and the PATRR22 (Identities: 58% between PATRR11 and PATRR22).
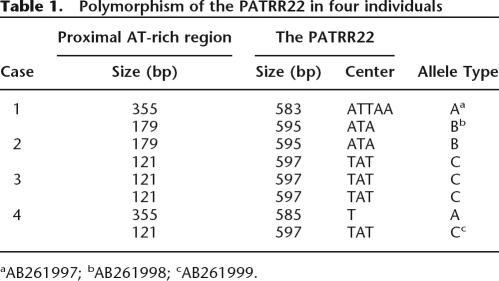
We cloned the PATRR22 from eight chromosomes derived from four individuals. In contrast to the PATRR11 or the PATRR17, no large deletion polymorphisms are observed within the PATRR22. Nonetheless, some sequence variations have been observed. All of the PATRR22s manifest nearly perfect palindromes with subtle sequence differences between proximal and distal arms (cis-morphisms). Further, sequence variation resulting in polymorphism is more prominent than are cis-morphisms. We have classified these polymorphisms into three types according to the size and characteristics of the center of the PATRR (Table 1). Percent identity of pairwise comparison between each type is 97% for A and B, 97% for B and C, and 95% for A and C. These polymorphisms of the PATRR22 appear to be linked to size polymorphisms of the flanking AT-rich region. Representatives of these three types have been deposited in GenBank as accession numbers AB261997, AB261998, and AB261999.
Table 1.
Polymorphism of the PATRR22 in four individuals
Breakpoint analysis of translocations involving the PATRR22
We compared the sequence of junction fragments obtained by translocation-specific PCR of t(11;22) derivative chromosomes with sequence derived from authentic PATRR22s (A, B, or C). We used the PATRR22 sequence from one chromosome (C, AB261999) as a reference, since we do not know which original sequence gave rise to the translocation. We analyzed sequences of junction fragments for the der(11) and the der(22) from 43 unrelated t(11;22) cases (Kurahashi et al. 2000b) as well as from those reported in the literature (Edelmann et al. 2001; Tapia-Paez et al. 2001). The majority of t(11;22) breakpoints map near the center of the PATRR22, whereas a significant number of breakpoints are distributed in the AT-rich region of the PATRR arms, suggesting small central deletions (Fig. 3A; Table 2). It is noteworthy that the sequences of der(11)s and der(22)s appear to be quite similar to one another, particularly for sequence near or at the breakpoint. In other words, the breakpoint locations are similar for the der(11) and the der(22) for individual reciprocal translocations. This had been previously predicted from the same junction fragment data in the absence of the complete sequence for the normal PATRR22, and were suggestive of symmetrical central deletions (Kurahashi and Emanuel 2001a).
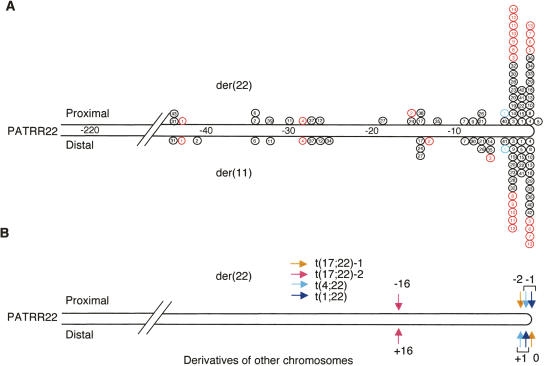
Figure 3.
Localization of the translocation breakpoints within the PATRR22. (A) Mapping of the t(11;22) breakpoints on the PATRR22. Each circle represents the breakpoints of the der(22) (top) and the der(11) (bottom). The scale indicates nucleotide number from the center of the PATRR22. The numbers in the circles indicate each individual translocation carrier. Black circles are from Kurahashi et al. (2000b), red from Edelmann et al. (2001), and blue from Tapia-Paez et al. (2001). Samples 38, 39, 43 (black), and 14 (red) have only the der(22) breakpoint, since they are from patients with the supernumerary-der(22) syndrome. It is noteworthy that the majority of balanced carriers show similar breakpoint locations on the PATR22 for the der(11) and the der(22). (B) Mapping the breakpoints of other translocations involving the 22q11 PATRR.
Table 2.
Location of the t(11;22) breakpoints on the PATRR22
aNumber of samples that carry the breakpoints at a given interval from the center of the PATRR22.
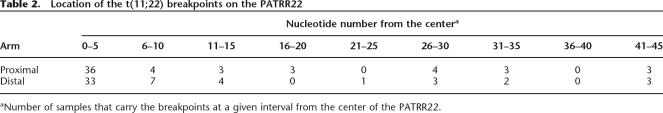
To elucidate the DSB-repair mechanism underlying PATRR-mediated translocations, junction fragments were examined for identical nucleotides at the point where the original two sequences were joined at a breakpoint. The majority of t(11;22)s manifest only a small number of identical nucleotides (<10 nt) at the breakpoints (Fig. 4A). When compared with a random simulation model for nonhomologous end joining (NHEJ), the distribution of identical nucleotides is shifted slightly to a larger number than that of in silico translocations, suggesting that a small stretch of microhomology is utilized for this DSB-repair (Fig. 4B). Some of the t(11;22)s possess nine identical nucleotides at the breakpoints, indicating the involvement of these short repeats close to the center of both PATRRs (Fig. 4C). Based on these results, combined with the fact that PATRR11 and PATRR22 do not share significant homology, the expected mechanism for the translocation is NHEJ or single-strand annealing rather than homologous recombination. These results are also supported by results of our recent meiotic analysis that indicate that these regions of 11q23 and 22q11 are not recombination hotspots (Ashley et al. 2006).
Figure 4.
Analysis of microhomologies at the breakpoints. (A) The number of identical nucleotides at the breakpoints of the t(11;22). The dotted line indicates the putative sample numbers from simulation of NHEJ. (B) A random simulation of the translocation. The details of the procedure are given in the Methods. The scale of the vertical axis is adjusted to reflect sample number relative to that shown in the graph (A). (C) Detail of one junction showing nine identical nucleotides (Case 3). The top indicates the junction fragment, while the middle and bottom indicate original breakpoint sequences. The arrows indicate each unit of the inverted repeats, while the arrowheads indicate the center of the PATRR.
We also examined the location of breakpoints for translocations whose partner chromosomes are other than chromosome 11. Similar observations were obtained for these breakpoint locations (Fig. 3B). A symmetrical deletion was also observed for one case with the t(17;22) (Kurahashi et al. 2003).
Structure of the region surrounding the PATRR22
Since the PATRR22 is the only known hotspot for constitutional translocation breakpoints, it represents one of the most rearrangement-prone sites in the human germ-cell lineage. The total size of a typical PATRR22 is 595 bp, which is just 150 bp longer than that of PATRR11. Given the remarkable instability of the region, we had anticipated finding a much longer PATRR on chromosome 22 than on chromosome 11. To examine whether some other genomic structures have a detrimental influence on the instability of this region, the sequence of the region surrounding the PATRR22 was examined.
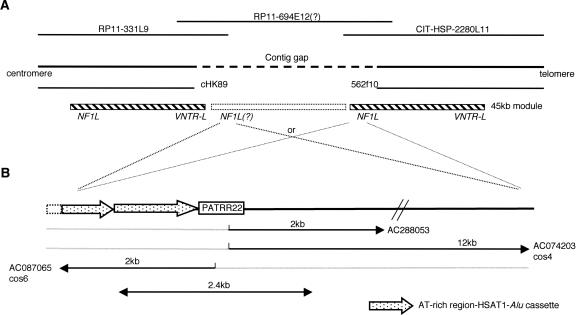
In the Ensembl public database from the Sanger Center (http://www.ensembl.org/Homo_sapiens/index.html), three BACs, RP11-331L9, RP11-694E12, and CIT-HSP-2280L11, are depicted as tiling clones for this unsequenced gap (Fig. 5A). BAC RP11-694E12 has not been entirely sequenced, while the sequences of RP11-331L9 and CIT-HSP-2280L11 that are present in the database do not include the PATRR22 sequence. We obtained BAC RP11-694E12 to test for the presence of PATRR22 by using PATRR22-specific PCR, PCRab. We selected multiple clones and performed PCRab on each clone as well as pooled clones, but all were negative for the authentic PCR product. We also performed PCRad, which yielded a 500-bp product derived from a previously known structure, but not from the PATRR22. These results suggest that either the contig is incorrect or that the RP11-694E12 clones examined had sustained a deletion of the relevant region during bacterial culture.
Figure 5.
Structure of the region surrounding the PATRR22. (A) Predicted structure of LCR22B. There is still a contig gap in LCR22B; the end of the proximal contig is the telomeric end of cHK89, whereas the end of the more distal contig is the centromeric end of BAC RP11-562f10. Ensembl indicates BAC RP11-694E12 as a bridging clone possibly using partial sequence data. Two 45-kb modules, major components of LCR22, are indicated by hatched boxes. Both 45-kb modules contain segments previously designated “NF1L” and “VNTR-L” in the same orientation (Shaikh et al. 2000). Another 45-kb module could potentially reside in the middle. (B) The region surrounding PATRR22. AC288053 and AC074203 are derived from the der(11), whereas AC087065 is derived from the der(22). The bidirectional arrow indicates the longest PCR product identified in this study. Thick dotted arrows indicate the AT-rich region-HSAT1-Alu cassette.
We continued our attempt to clone larger PCR products encompassing the PATRR22 region utilizing the same PCR primers. We eventually obtained 2.4-kb PCR products spanning PATRR22 from samples from four individuals. To further analyze the surrounding region, sequence of junction fragments can be used as a source of information. We had previously isolated a 4.2-kb der(11) junction fragment from a genomic library generated from DNA obtained from a balanced carrier (GenBank accession no. AF288053) (Kurahashi et al. 2000a). Later, larger genomic clones including the t(11;22) junction fragments for both derivative chromosomes were reported by others (cos4: AC074203, cos6: AC087065) (Tapia-Paez et al. 2001). Combined with these data from the databases, we reconstructed a ∼10-kb region surrounding the PATRR22 (Fig. 5B).
The sequence of the ∼10-kb region is similar to part of a 45-kb module of LCR22 (Shaikh et al. 2000). The distal contig (RP11-562f10, AC007731) ends within the 45-kb module, but it does not contain the NF1L end (Fig. 5A). On the other hand, the end of the proximal contig (cHK89, AF024070) is located within the 45-kb module containing the standard NF1L module. Altogether, LCR22-B has at least two copies of the 45-kb module in the same orientation. The proximal copy of the 45-kb module might harbor the PATRR22, or it is possible that another copy of the 45-kb module contains the PATRR22 and is located between these two modules.
Based on compiled sequence, the 45-kb module harboring the PATRR22 has the same structure as the ones appearing in other LCR22s, with no other distinct characteristic structure except for the presence of the PATRR22. This unique structure appears only in LCR22-B, which might explain why translocation breakpoints cluster in LCR22-B. Therefore, the presence of the 595-bp PATRR22 alone appears to be responsible for this remarkable instability, acting as a hotspot for translocation breakpoints.
Discussion
Although >99% of the euchromatic region of the human genome has been represented as high-quality genomic sequence, a significant number of regions remain unsequenced (Eichler et al. 2004). These regions constitute unclonable or unsequenceable gaps using standard approaches. Most of these gaps in the sequence are composed of some kind of repetitive units. Because of their susceptibility to rearrangement, such regions are often associated with human diseases. Thus, it is important to clone and analyze these regions. However, their susceptibility to rearrangement is also observed in the host organisms used for cloning, creating obstacles that introduce resistance to cloning. In this study, the PATRR22—a hotspot for translocation breakpoints and one of the most rearrangement-prone sites in the human genome—has been successfully cloned.
There is now ample evidence that palindrome-mediated translocation is a common pathway for human genomic rearrangement (Kurahashi et al. 2003; Nimmakayalu et al. 2003; Gotter et al. 2004). Since it has been demonstrated that the t(14;18) observed in follicular lymphoma results from instability of triplex DNA at the breakpoint, translocations mediated by noncanonical DNA structures represent an emerging topic in human genetics (Raghavan et al. 2004, 2005). The PATRR22 has been shown to be a structure similar to other reported translocation-associated PATRRs. Such PATRRs share three features in common with one another; several hundred base pairs of a nearly perfect palindrome, an AT-rich center with non-AT-rich ends, and the presence of another flanking AT-rich region. It has been suggested that for palindromic sequences in double-stranded DNA, the interstrand base pairs might convert to intrastrand pairs, producing a set of hairpin structures described as a cruciform (Kurahashi et al. 2000a, 2004). We proposed that PATRRs adopt a cruciform configuration creating a chromosomal context that induces genomic instability leading to translocations. The central AT-richness might contribute to the initiation of strand separation, while the nearly perfect palindrome might contribute to the susceptibility for cruciform extrusion. The size of both the palindrome and the non-AT-rich region within the palindrome might contribute to the degree of stability of the resulting cruciform. Further, the flanking AT-rich region might facilitate unwinding of the chromosomal DNA (Sullivan and Lilley 1986). The size differences of the flanking AT-rich regions among the observed polymorphic PATRR22s might affect their susceptibility for sustaining a translocation. Hence, these three features are likely to play a pivotal role in creating DNA secondary structure, which is permissive for induction of the translocation. All of the known PATRR-mediated translocations involve the PATRR22 as a partner, indicative of the extreme degree of instability of the PATRR22, not only among other PATRRs, but also among other DSB-susceptible regions of the human genome. The fact that the PATRR22 is the longest of the three known PATRRs, has the highest AT-content in its central region, and has the longest flanking AT-rich region, is likely to contribute to its propensity for adopting secondary structure leading to its extreme instability in humans.
Based on observations regarding the instability of PATRR22 in humans, it was not unreasonable to predict that the PATRR22 might also be quite unstable and lost during its culture in E. coli. However, in contrast to the other PATRRs, the PATRR22 is, in fact, rather stable in E. coli. It is formally possible that the palindromic sequence is deleted in bacteria in a replication-dependent manner (Leach 1994). Combined with our data that de novo t(11;22)s are detected only in human sperm, not in blood cells or fibroblasts, PATRR-mediated translocation is clearly dictated by a mechanism different from replication-dependent deletion of palindromic sequences in cloning hosts (Kurahashi and Emanuel 2001b). It is still an enigma why the PATRR22 is underrepresented in the publicly available genomic libraries. The utilization of SURE cells might have been responsible for the stable cloning and propagation of the PATRR22. It is conceivable that construction of genomic libraries using SURE cells might enable the cloning of such unstable regions.
Although the inference is based on sequence data from a small number of chromosomes, the PATRR22 does not appear to display a large deletion polymorphism. This finding is in contrast to what has been observed for the PATRR11 or PATRR17 (Inagaki et al. 2005; Kato et al. 2006). One possibility is that the PATRR22 emerged more recently in the history of primate evolution. Alu–Alu inverted repeats are underrepresented relative to the number of direct repeats in the human genome database, suggesting that similarly sized palindromic sequences are likely to disappear rapidly from the human genome (Stenger et al. 2001). Alternatively, it is possible that the PATRR22 is resistant to deletion in human as well as in E. coli. This hypothesis also implies that PATRR-mediated translocation is governed by a mechanism different from that of deletion within the PATRR.
Since full sequence information of the entire breakpoint region of 22q11 had not been available, we were previously not able to analyze the precise location of translocation breakpoints. In this study, acquisition of the complete sequence of one example of the PATRR22 has provided a new opportunity for determining the precise breakpoint location based on the knowledge of the normal sequences for both of the breakpoint regions. Breakpoints were consistently located close to the center of the palindrome, although some were located in the arms. The differences of breakpoint location in different examples of the t(11;22) are interpreted as: (1) the translocation was derived from a center breakage of a polymorphic short PATRR22, or (2) the breakpoint took place precisely at the center, followed by nucleotide resection creating a small deletion. However, the high similarity of the PATRR22 in the eight chromosomes we analyzed suggests that a DSB gets processed prior to the translocation, generating a small deletion. This is also the case for the small central deletion of the t(17;22), in which a deleted PATRR17 is unlikely to be responsible for the translocation (Fig. 3B) (Inagaki et al. 2005). In either event, our results suggest that the center of the palindrome is susceptible to DSBs that induce this illegitimate chromosomal rearrangement (Edelmann et al. 2001; Kurahashi and Emanuel 2001a).
In addition, precise breakpoint analysis has provided us with valuable information about the mechanism of the repair process of the translocation. In the junction fragments from either the der(11) or the der (22), only a few identical nucleotides are observed between the end points of the original sequences. No strong homology is observed between the PATRR11 and PATRR22 sequences except for their shared AT-richness. These results, combined with the observation that there is often a microdeletion at the center of the PATRR, implicate a homology-independent pathway for the repair instead of homologous recombination. The extent of observed microhomology was always <10 bp, suggesting that the ends of the DSBs are repaired either by NHEJ or a single-strand annealing pathway (Richardson and Jasin 2000).
Finally, we examined the surrounding region for other specific structures that might contribute to the etiology of the translocation at this specific chromosome 22 LCR. However, it is similar to other LCR22s except for the presence of the PATRR22. Hence, the ∼590-bp PATRR22 alone is likely to be both necessary and sufficient for induction of the recurrent translocation. The biology that drives palindrome-mediated translocation encompasses several interesting topics in the field of human genetics. Although noncanonical DNA conformation has been proposed as an etiology for palindrome-mediated translocation, it is still a “smoking gun.” It has been suggested, but not determined for certain, whether the translocations occur during meiosis at the time of recombination or specifically only in male germ cells (Kurahashi and Emanuel 2001b). To directly examine all of the processes involved in generation of the translocation, we need to establish an experimental model for palindrome-mediated translocation using a yeast or mouse system. The cloned PATRR22 as well as the previously isolated PATRR11 will facilitate generating model systems to further investigate the mechanism of this recurrent genomic rearrangement.
Methods
Human samples and cell lines
Human samples were provided from individual volunteers after obtaining appropriate informed consent. Genomic DNA was extracted from peripheral blood samples using PureGene (Gentra). The study was approved by the Ethical Review Boards for Human Genome Studies at Fujita Health University.
We used the following somatic cell hybrids: GM10888, a human/hamster hybrid containing a normal chromosome 22 as its only intact human chromosome; Cl6-2/EG is a human/hamster hybrid made from cells of a patient with the 22q11.2 deletion syndrome; Cl-4/GB is a human/hamster hybrid cell line with a constitutional t(11;22), retaining the der(22) as the only relevant human chromosome; GM11685 (NF13/D3) is a subclone of a human/mouse hybrid cell line made using cells from a patient with a constitutional t(17;22)(q11.2;q11.2) and neurofibromatosis type 1. GM10888 and GM11685 were obtained from Coriell Cell Repository, and the others were established in our laboratory (Budarf et al. 1996).
PCR
PCR primers were designed as described before and slightly modified for this study (Kehrer-Sawatzki et al. 1997; Kurahashi et al. 2000b). (22a) 5′-CCCAGTGTGAATTGGGATTCAG-3′, (22b) 5′-CTGCATCCTTCAACGTTCCATC-3′, (22c) 5′-CCTCCAACG GATCCATACTACTG-3′, (22d) 5′-GTTGGGTGATTGACTGTGATT GAC-3′. PCR was performed using conditions optimized for amplification of the PATRR (Inagaki et al. 2005).
The PCR products were cloned into pBluescript vector (Stratagene). The PATRRs are highly unstable, and the plasmid often loses the palindromic insert during bacterial culture. We used the SURE strain (Stratagene) to maintain the unstable insert. The plasmid inserts were sequenced by the RNA-sequencing method using T3/T7 RNA polymerases (Inagaki et al. 2005).
In silico translocation
We cut the PATRR11 and the PATRR22 sequences at random in silico within a 50-bp region located at the center, and then connected the proximal part of PATRR11 and the distal part of PATRR22 into artificial translocation products. We attempted to determine, assuming that NHEJ occurs between complete blunt ends, how many nucleotides appear as microhomology. We compared the end sequence of the proximal PATRR11 fragment with that of the proximal PATRR22 fragment. The number of identical nucleotides in the same orientation at the ends was recorded. We counted the total number of the putative translocation events for each number of microhomologies. All of the analyses were performed using Microsoft Excel (Microsoft).
Acknowledgments
We thank Dr. T.H. Shaikh for helpful discussion and Miss H. Kowa, K. Nagaoka, and T. Mori for technical assistance. These studies were supported by a grant-in-aid for Scientific Research and 21st Century COE program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (H.K.). These studies were also supported by NIH grant CA39926 (B.S.E) as well as funds from the Charles E.H. Upham Chair in Pediatrics (B.S.E.).
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5769507
References
- Ashley T., Gaeth A.P., Inagaki H., Seftel A., Cohen M.M., Anderson L., Kurahashi H., Emanuel B.S., Gaeth A.P., Inagaki H., Seftel A., Cohen M.M., Anderson L., Kurahashi H., Emanuel B.S., Inagaki H., Seftel A., Cohen M.M., Anderson L., Kurahashi H., Emanuel B.S., Seftel A., Cohen M.M., Anderson L., Kurahashi H., Emanuel B.S., Cohen M.M., Anderson L., Kurahashi H., Emanuel B.S., Anderson L., Kurahashi H., Emanuel B.S., Kurahashi H., Emanuel B.S., Emanuel B.S. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am. J. Hum. Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babcock M., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Pavlicek A., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Spiteri E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Kashork C.D., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Ioshikhes I., Shaffer L.G., Jurka J., Morrow B.E., Shaffer L.G., Jurka J., Morrow B.E., Jurka J., Morrow B.E., Morrow B.E. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 2003;13:2519–2532. doi: 10.1101/gr.1549503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budarf M.L., Eckman B., Michaud D., McDonald T., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Eckman B., Michaud D., McDonald T., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Michaud D., McDonald T., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., McDonald T., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Gavigan S., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Buetow K.H., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Tatsumura Y., Liu Z., Hilliard C., Driscoll D., Liu Z., Hilliard C., Driscoll D., Hilliard C., Driscoll D., Driscoll D., et al. Regional localization of over 300 loci on human chromosome 22 using a somatic cell hybrid mapping panel. Genomics. 1996;35:275–288. doi: 10.1006/geno.1996.0358. [DOI] [PubMed] [Google Scholar]
- Debeer P., Mols R., Huysmans C., Devriendt K., Van de Ven W.J., Fryns J.P., Mols R., Huysmans C., Devriendt K., Van de Ven W.J., Fryns J.P., Huysmans C., Devriendt K., Van de Ven W.J., Fryns J.P., Devriendt K., Van de Ven W.J., Fryns J.P., Van de Ven W.J., Fryns J.P., Fryns J.P. Involvement of a palindromic chromosome 22-specific low-copy repeat in a constitutional t(X;22)(q27;q11) Clin. Genet. 2002;62:410–414. doi: 10.1034/j.1399-0004.2002.620510.x. [DOI] [PubMed] [Google Scholar]
- Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E., Bialer M.G., Shanske A., Goldberg R., Morrow B.E., Shanske A., Goldberg R., Morrow B.E., Goldberg R., Morrow B.E., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler E.E., Clark R.A., She X., Clark R.A., She X., She X. An assessment of the sequence gaps: Unfinished business in a finished human genome. Nat. Rev. Genet. 2004;5:345–354. doi: 10.1038/nrg1322. [DOI] [PubMed] [Google Scholar]
- Gotter A.L., Shaikh T.H., Chieffo C., Budarf M.L., Rhodes C.H., Emanuel B.S., Shaikh T.H., Chieffo C., Budarf M.L., Rhodes C.H., Emanuel B.S., Chieffo C., Budarf M.L., Rhodes C.H., Emanuel B.S., Budarf M.L., Rhodes C.H., Emanuel B.S., Rhodes C.H., Emanuel B.S., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewett D.R., Handt O., Hobson L., Mangelsdorf M., Eyre H.J., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Handt O., Hobson L., Mangelsdorf M., Eyre H.J., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Hobson L., Mangelsdorf M., Eyre H.J., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Mangelsdorf M., Eyre H.J., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Eyre H.J., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Baker E., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Sutherland G.R., Schuffenhauer S., Mao J.I., Richards R.I., Schuffenhauer S., Mao J.I., Richards R.I., Mao J.I., Richards R.I., Richards R.I. FRA10B structure reveals common elements in repeat expansion and chromosomal fragile site genesis. Mol. Cell. 1998;1:773–781. doi: 10.1016/s1097-2765(00)80077-5. [DOI] [PubMed] [Google Scholar]
- Inagaki H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H., Shaikh T.H., Emanuel B.S., Kurahashi H., Emanuel B.S., Kurahashi H., Kurahashi H. Palindromic AT-rich repeat in NF1 gene is hypervariable in human and evolutionally conserved among primates. Hum. Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Taniguchi M., Emanuel B.S., Kurahashi H., Emanuel B.S., Kurahashi H., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Haussler J., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G., Haussler J., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G., Krone W., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G., Bode H., Jenne D.E., Mehnert K.U., Tummers U., Assum G., Jenne D.E., Mehnert K.U., Tummers U., Assum G., Mehnert K.U., Tummers U., Assum G., Tummers U., Assum G., Assum G. The second case of a t(17;22) in a family with neurofibromatosis type 1: Sequence analysis of the breakpoint regions. Hum. Genet. 1997;99:237–247. doi: 10.1007/s004390050346. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001a;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S., Emanuel B.S. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001b;29:139–140. doi: 10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Roe B.A., Emanuel B.S., Budarf M.L., Emanuel B.S., Budarf M.L., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000a;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Driscoll D.A., Budarf M.L., Emanuel B.S., Budarf M.L., Emanuel B.S., Emanuel B.S. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000b;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Takata M., Toda T., Emanuel B.S., Shaikh T.H., Takata M., Toda T., Emanuel B.S., Takata M., Toda T., Emanuel B.S., Toda T., Emanuel B.S., Emanuel B.S. The constitutional t(17;22): Another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Taniguchi M., Emanuel B.S., Toda T., Emanuel B.S., Toda T., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Nimmakayalu M.A., Gotter A.L., Shaikh T.H., Emanuel B.S., Gotter A.L., Shaikh T.H., Emanuel B.S., Shaikh T.H., Emanuel B.S., Emanuel B.S. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R., Wu X., Hsieh C.L., Lieber M.R., Hsieh C.L., Lieber M.R., Lieber M.R. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428:88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- Raghavan S.C., Chastain P., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Chastain P., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R., Hsieh C.L., Haworth I.S., Lieber M.R., Haworth I.S., Lieber M.R., Lieber M.R. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J. Biol. Chem. 2005;280:22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- Rhodes C.H., Call K.M., Budarf M.L., Barnoski B.L., Bell C.J., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Call K.M., Budarf M.L., Barnoski B.L., Bell C.J., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Budarf M.L., Barnoski B.L., Bell C.J., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Barnoski B.L., Bell C.J., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Bell C.J., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Emanuel B.S., Bigner S.H., Park J.P., Mohandas T.K., Bigner S.H., Park J.P., Mohandas T.K., Park J.P., Mohandas T.K., Mohandas T.K. Molecular studies of an ependymoma-associated constitutional t(1;22)(p22;q11.2) Cytogenet. Cell Genet. 1997;78:247–252. doi: 10.1159/000134667. [DOI] [PubMed] [Google Scholar]
- Richardson C., Jasin M., Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- Ried K., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Finnis M., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Hobson L., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Mangelsdorf M., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Dayan S., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Nancarrow J.K., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Woollatt E., Kremmidiotis G., Gardner A., Venter D., Kremmidiotis G., Gardner A., Venter D., Gardner A., Venter D., Venter D., et al. Common chromosomal fragile site FRA16D sequence: Identification of the FOR gene spanning FRA16D and homozygous deletions and translocation breakpoints in cancer cells. Hum. Mol. Genet. 2000;9:1651–1663. doi: 10.1093/hmg/9.11.1651. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Budarf M.L., Celle L., Zackai E.H., Emanuel B.S., Budarf M.L., Celle L., Zackai E.H., Emanuel B.S., Celle L., Zackai E.H., Emanuel B.S., Zackai E.H., Emanuel B.S., Emanuel B.S. Clustered 11q23 and 22q11 breakpoints and 3:1 meiotic malsegregation in multiple unrelated t(11;22) families. Am. J. Hum. Genet. 1999;65:1595–1607. doi: 10.1086/302666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Zackai E.H., Budarf M.L., Budarf M.L., et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Spiteri E., Babcock M., Kashork C.D., Wakui K., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Babcock M., Kashork C.D., Wakui K., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Kashork C.D., Wakui K., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Wakui K., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Gogineni S., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Lewis D.A., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Williams K.M., Minoshima S., Sasaki T., Shimizu N., Minoshima S., Sasaki T., Shimizu N., Sasaki T., Shimizu N., Shimizu N., et al. Frequent translocations occur between low copy repeats on chromosome 22q11.2 (LCR22s) and telomeric bands of partner chromosomes. Hum. Mol. Genet. 2003;12:1823–1837. doi: 10.1093/hmg/ddg203. [DOI] [PubMed] [Google Scholar]
- Stenger J.E., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Darden T.A., Jurka J., Resnick M.A., Jurka J., Resnick M.A., Resnick M.A. Biased distribution of inverted and direct Alus in the human genome: Implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- Sullivan K.M., Lilley D.M., Lilley D.M. A dominant influence of flanking sequences on a local structural transition in DNA. Cell. 1986;47:817–827. doi: 10.1016/0092-8674(86)90524-6. [DOI] [PubMed] [Google Scholar]
- Tapia-Paez I., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Fedorova L., Imreh S., Dumanski J.P., Imreh S., Dumanski J.P., Dumanski J.P. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- Yu S., Mangelsdorf M., Hewett D., Hobson L., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Mangelsdorf M., Hewett D., Hobson L., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Hewett D., Hobson L., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Hobson L., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Baker E., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Eyre H.J., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Lapsys N., Le Paslier D., Doggett N.A., Sutherland G.R., Le Paslier D., Doggett N.A., Sutherland G.R., Doggett N.A., Sutherland G.R., Sutherland G.R., et al. Human chromosomal fragile site FRA16B is an amplified AT-rich minisatellite repeat. Cell. 1997;88:367–374. doi: 10.1016/s0092-8674(00)81875-9. [DOI] [PubMed] [Google Scholar]