Abstract
Constitutional translocations at the same 22q11.21 low copy repeat B (LCR-B) breakpoint involved in the recurrent t(11;22) are relatively abundant. A novel 46,XY,t(8;22)(q24.13;q11.21) rearrangement was investigated to determine whether the recurrent LCR-B breakpoint is involved. Investigations demonstrated an inversion of the 3Mb region typically deleted in patients with the 22q11.2 deletion syndrome. The 22q11.21 inversion appears to be mediated by low copy repeats, and is presumed to have taken place prior to translocation with 8q24.13. Despite predictions based on inversions observed in other chromosomes harboring low copy repeats, this 22q11.2 inversion has not been observed previously. The current studies utilize novel laser microdissection and MLPA (multiplex ligation-dependent probe amplification) approaches, as adjuncts to FISH, to map the breakpoints of the complex rearrangements of 22q11.21 and 8q24.21. The t(8;22) occurs between the recurrent site on 22q11.21 and an AT-rich site at 8q24.13, making it the fifth different chromosomal locus characterized at the nucleotide level engaged in a translocation with the unstable recurrent breakpoint at 22q11.21. Like the others, this breakpoint occurs at the center of a palindromic sequence. This sequence appears capable of forming a perfect 145 bp stem–loop. Remarkably, this site appears to have been involved in a previously reported t(3;8) occurring between 8q24.13 and FRA3B on 3p14.2. Further, the fragile site-like nature of all of the breakpoint sites involved in translocations with the recurrent site on 22q11.21, suggests a mechanism based on delay of DNA replication in the initiation of these chromosomal rearrangements.
The 22q11.21 region represents a hot spot for nonrandom chromosomal aberrations, including deletions, translocations, supernumerary chromosomes, and, less frequently, interstitial duplications (Lindsay et al. 1995; Edelmann et al. 1999; Ensenauer et al. 2003; Meins et al. 2003; Hassed et al. 2004; Portnoi et al. 2005; Yobb et al. 2005). These rearrangements are associated with genetic disorders including the 22q11.21 deletion syndrome, supernumerary der(22)t(11;22) syndrome (Emanuel syndrome), cat eye syndrome (CES), and, occasionally, Opitz syndrome (OS) (for review, see Driscoll and Emanuel 1998). The breakpoints of these rearrangements are frequently localized to a class of chromosome-specific repeat sequences known as low-copy repeats (LCRs) or segmental duplications. Each LCR on 22q11 consists of cluster of sequence modules that are repeated in other chromosome 22 LCRs with 97%–98% sequence identity. LCRs differ from one another in their sequence module content and organization. A total of eight LCRs have been identified within 22q11 (LCRs A to H, proximal to distal), with most constitutional rearrangements involving LCRs A through D, or the 3 Mb typically deleted region (TDR) (Edelmann et al. 1999; Shaikh et al. 2000).
LCR-B contains a recurrent translocation breakpoint site that is not only involved in the recurrent t(11;22)(q23;q11.2) (Edelmann et al. 1999; Kurahashi et al. 2000a, b; Tapia-Paez et al. 2001) but also in two different t(17;22)(q11.2;q11.2) cases (Kehrer-Sawatzki et al. 2002; Kurahashi et al. 2003), a t(1;22)(p21.2; q11.2) (Gotter et al. 2004) and a t(4;22)(q35.1;q11.2) (Nimmakayalu et al. 2003). Remarkably, most of the 22q11 breakpoints involved in these translocations occur within 16 bp of one another, at the center of a near-perfect palindromic AT-rich repeat (PATRR) that is 595 bp long. Conversely, the breakpoints on all of the partner chromosomes involved in these translocations (11q23, 17q11.2, 1p21.2, 4q35.1) also occur at the center of inverted repeat sequences. These observations have suggested that DNA secondary structures in the form of hairpin loops or cruciforms underlie the genesis of these rearrangements. In each case there is minimal loss of sequence, the only deletions being a symmetrical loss of nucleotides from the center of the inverted repeat as if the end of hairpin loops formed by these palindromes were truncated before joining with their chromosome translocation partner. Analysis of potential secondary structures formed at all of these breakpoint sequences compared with other translocation breakpoints not involving the recurrent 22q11.21 PATRR demonstrated a unique ability of the former to form stem–loop structures. Those studies further suggested that the rate at which translocations involving the recurrent breakpoint at LCR-B occur appears to be related to the thermodynamic likelihood of forming such configurations (Gotter et al. 2004).
The distribution and composition of the LCRs responsible for the 22q11.21 deletion syndrome predict that both chromosome duplications and interstitial inversions should occur within this region (Shaikh et al. 2000, 2001; Emanuel and Shaikh 2001). During meiosis, misalignment and aberrant homologous recombination of LCRs A and D results in the loss of the TDR from one chromosome but had also been predicted to generate a reciprocal der(22) containing a duplication of this region. These duplication products have, in fact, been observed, albeit at low frequency (Ensenauer et al. 2003; Hassed et al. 2004; Yobb et al. 2005). Some of the homologous sequence modules contained within LCRs A and D are also inverted relative to one another. This suggests that these LCRs may undergo intrachromosomal homologous recombination to generate an interstitial inversion of the TDR (Shaikh et al. 2000, 2001; Emanuel and Shaikh 2001). This rearrangement, however, has so far not been observed in human genome scans designed to detect polymorphic inversions (Feuk et al. 2005; Tuzun et al. 2005; Khaja et al. 2006) or by studies that specifically evaluated 22q11.21 for this rearrangement (Gebhardt et al. 2003; Saitta et al. 2004).
In this study, we describe a complex chromosome 22 rearrangement involving both an interstitial inversion of the TDR of 22q11.2 and a subsequent translocation between the recurrent PATRR contained within LCR-B and an AT-rich sequence present at 8q24.13. Using high-resolution fluorescent in situ hybridization (FISH) and a novel application of multiplex ligation-dependent probe amplification (MLPA), the breakpoints of the 22q11.21 inversion were localized within LCRs A and D, thereby confirming predictions made previously for this rearrangement. To localize the translocation breakpoint on 8q24.13, we have used a novel laser microdissection approach to isolate derivative chromosomes for subsequent template-specific PCR mapping studies. Analysis of t(8;22) junction fragments indicate that the 8q24.13 breakpoint splits a perfect 290-bp PATRR, further substantiating the role of stem–loop structures in the genesis of translocations involving the recurrent 22q11.21 breakpoint. The specific localization of the 8q24.13 breakpoint to an AT-rich region involved in other constitutional translocations has further implications concerning the mechanism of these rearrangements.
Results
A complex rearrangement involving an inversion and translocation of sites within 22q11.2
Karyotype analysis of the proband at 2 yr of age was prompted by her developmental and language delays, camptodactyly, unilateral preauricular pit, and facial features that were different from other family members. Presently, at 10 yr of age, she has mild residual hypotonia and attention deficit disorder. Her learning is age appropriate, requiring a resource-room setting only for math. G-band analysis of metaphases derived from this patient revealed 47 chromosomes. The additional chromosome appeared to be a small, acrocentric marker chromosome. Parental chromosomal studies showed that the father carries a balanced translocation between chromosomes 8 and 22, 46,XY,t(8;22) (q24.13;q11.21), indicating that the karyotype of the proband is 47,XX,+der(22)t (8;22)(q24.13;q11.21)pat. Thus, the proband’s supernumerary der(22) was paternally derived as a result of 3:1 meiotic malsegregation, since the father carries a balanced t(8;22)(q24.1;q11.2) (Supplemental Fig. S1).
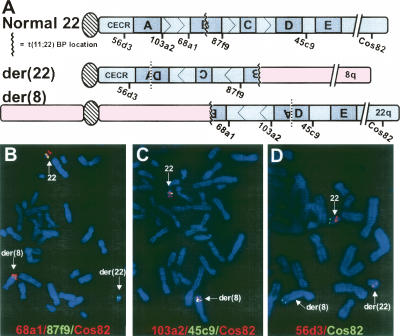
Given the 22q11.2 localization of the translocation breakpoint, we investigated whether the t(8;22) involved the recurrent LCR-B translocation breakpoint seen in the t(11;22), t(17;22), and other translocations to 22q11.2. FISH was performed on metaphase spreads from the balanced t(8;22) carrier using fluorescently labeled cosmid probes specific for regions within 22q11.2 together with a subtelomeric control probe (Cos82) (see Fig. 1A for probe locations). Initial FISH analysis using probes flanking LCR-B at the recurrent translocation breakpoint yielded unexpected results. While all three probes hybridized to the normal 22, both the proximal LCR-B probe, 68a1, as well as the subtelomeric control probe appeared to reside on the der(8), while the probe that maps distal to LCR-B, 87f9, produced a hybridization signal on the der(22) chromosome (Fig. 1B). These results indicated that the t(8;22) translocation took place within the LCR-B region of 22q11.2, since the signals from these probes are found on opposite derivative chromosomes, but also suggested that the translocation occurred in an inverted 22q11.2 region. To further map the inversion, we used additional cosmid probes flanking other known LCRs within the 22q11.2 region. In a separate experiment, probes 103a2, 45c9, and the subtelomeric Cos82 were all seen to be localized on both the normal chromosome 22 and the der(8), indicating that these 22q11.2 loci had translocated to the der(8) chromosome (Fig. 1C). A third experiment demonstrated that 56d3, a cosmid probe proximal to LCR-A in the CES chromosomal region (CECR), was found on the normal 22 and on the der(22) chromosome (Fig. 1D). Further, 106e4, a probe that is proximal and closely flanking LCR-A, is on the normal and the der(22) (data not shown). Together these results indicate that the t(8;22) carrier has a der(22) chromosome containing the proximal region of 22 adjacent to an inverted segment of chromosome 22 comprising the region from LCR-D to the distal end of LCR-B, which has rearranged at 8q24.13 (see Fig. 1A). This translocation was presumed to involve the recurrent t(11;22) translocation breakpoint in LCR-B (see Fig. 1A). The der(8), on the other hand, is predicted to contain the proximal portion of chromosome 8 (pter → q24.13) joined to the recurrent t(11;22) translocation breakpoint of an inverted segment derived from the distal end of LCR-B to the distal end of LCR-A that is then joined via an inversion breakpoint to the distal portion of 22q.
Figure 1.
The t(8;22) is part of a complex rearrangement involving an inversion of the 22q11.2 region. (A) Schematic of the location of cosmid probes used in FISH experiments relative to low copy repeats (LCRs A to E, shaded boxes) within 22q11.2. Interpreted structures of the der(8) and der(22) chromosomes are shown below the normal chromosome 22 structure. (Hatched ovals) centromeres; (CECR), cat eye syndrome minimal chromosomal region. (B–D). Dual-color FISH was performed on metaphase chromosomes from lymphoblast cells derived from the balanced t(8;22) carrier (father of the supernumerary t(8;22) der(22) proband). Differentially labeled cosmid probes were applied in the following combinations: (B) 68a1 (a.k.a. N41) (red), 87f9 (a.k.a. ZNF74) (green), and control probe Cos82 (red); (C) 103a2 (red), 45c9 (green), and Cos82 (red); (D) 56d3 (red), and a distal control probe, Cos82, (green).
Mapping inversion and translocation breakpoints by high-resolution MLPA
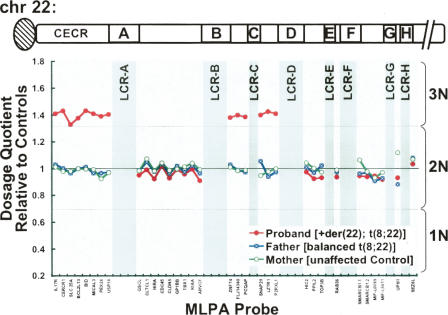
To map both the inversion and translocation sites on the der(22) at high resolution, we used MLPA to detect copy number changes at 33 loci on the long arm of chromosome 22 (Vorstman et al. 2006). In this approach, quantitative PCR is used to evaluate the quantity of ligated “hemiprobes” that have annealed to target loci present within a given DNA sample. For the present study, we evaluated the copy number at 22q11.21 loci in the proband carrying the supernumerary der(22) [t(8;22)(q24.1;q11.2)] chromosome and compared these results with those obtained from her father carrying the balanced t(8;22) and her cytogenetically normal mother. Because the proband contains two normal chromosome 22s in addition to the supernumerary der(22), this sample is expected to yield a trisomic signal, or 1.5 times that seen in controls, for loci present on the der(22). As shown in Figure 2, trisomic signal was observed in the proband sample for the eight most proximal MLPA probes distributed throughout the CECR including the USP18 probe, located just 9.6 kb away from the proximal edge of LCR-A (Fig. 2). In individuals with CES, the CECR is present in four copies. The two extra copies of these loci arise from a supernumerary bisatellited chromosome containing an inverted duplication of proximal chromosome 22 (Mears et al. 1994). Thus, this result is consistent with the phenotype of the patient, who has mild CES-like symptoms (preauricular pit, physical and cognitive developmental delays). These findings are consistent with the previously reported phenotypic variability for this disorder (Schachenmann et al. 1965; Schinzel et al. 1981), although attributing specific findings to the partial duplications of 22q and 8q in this individual will be difficult. All six of the MLPA probes located between LCRs B and D also exhibited trisomic signals, indicating that these sequences are also present on the supernumerary der(22). The nine probes corresponding to loci between LCR-A and LCR-B, however, are disomic in the proband. These results are consistent with FISH results for the balanced t(8;22) carrier, which indicated an inversion of a 22q11.2 region that includes sequences between the distal end of the CECR and LCR-D. These results also localize the 22q11.2 breakpoint for the t(8;22) translocation to a site within LCR-B. Because LCR-B of 22q11.2 harbors the common breakpoint involved in the recurrent t(11;22) and t(17;22), as well as a t(4;22) and a t(1;22) (Edelmann et al. 1999; Kurahashi et al. 2000b, 2003; Kurahashi and Emanuel 2001; Kehrer-Sawatzki et al. 2002; Nimmakayalu et al. 2003; Gotter et al. 2004), it was reasonable to predict that the breakpoint involved in this t(8;22) rearrangement occurs at the same site. This breakpoint does not disrupt any known genes.
Figure 2.
Mapping inversion and translocation breakpoints by MLPA. The relative dosage of 33 loci along 22q11 were assessed in genomic DNA samples derived from the proband (47,XX,+der(22) (q24.1;q11.2)pat) (red circles), her father (balanced t(8;22) carrier) (blue circles), and her unaffected mother (green circles) by MLPA. Dosage at each target locus is expressed as a dosage quotient relative to controls (data not shown). Dashed lines corresponding to established threshold values (Bunyan et al. 2004) are drawn such that dosage quotients between 0.3 and 0.7 are considered disomic (2N), below 0.7 haploid (1N), above 1.3 trisomic (3N). MLPA probes corresponding to 22q11 loci are listed on the X axis in relative order. The location and relative size of LCRs are indicated by gray shading. (CECR) Cat eye syndrome minimal chromosomal region.
The 8q24.1 breakpoint of the t(8;22) disrupts the RNF139 gene
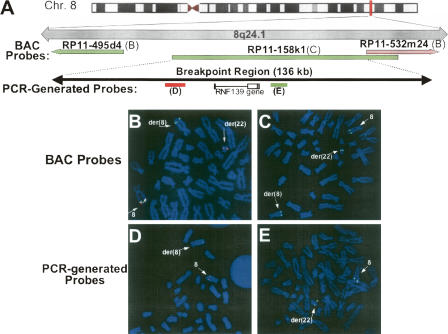
To localize the 8q24.1 breakpoint junction, FISH analysis was performed on metaphase chromosomes from the balanced t(8;22) carrier father using fully sequenced BAC clones labeled with spectrum red and spectrum green applied in a pairwise fashion (Fig. 3A). Initial mapping in this manner positioned the breakpoint to a 1-Mb region delineated by BAC clones RP11-495d4 and RP11-532m24. In the dual-labeling experiment shown in Figure 3B, hybridization signals from RP11-495d4 were seen on both the normal chromosome 8 and the der(8), while the RP11-532m24 probe recognized 8q24.1 sequence on the normal chromosome 8 as well as on the der(22). The junction was further localized to a 136-kb region corresponding to BAC clone RP11–158k1; in addition to hybridization to the normal chromosome 8, signal from this probe was seen on both the der(8) and the der(22) (Fig. 3C), indicating that the sequence contained within this BAC spans the 8q24.1 breakpoint. A portion of the sequence for this probe overlaps with that of 532m24 probe, meaning that we could further exclude the majority of this shared region from containing the breakpoint. Additional FISH experiments were also performed with a series of probes generated by long-range PCR amplification of specific RP11–158k1 sequences. Results from two of these probes (Fig. 3D,E) allowed us to further constrain the translocation junction to a 29.5-kb region of the RP11–158k1 sequence. Interestingly, this region contains the RNF139 gene that is disrupted by a t(3;8) occurring as an inherited rearrangement associated with renal cell carcinoma (Boldog et al. 1993; Ohta et al. 1996; Gemmill et al. 1998). The 8q24.1 breakpoint of that rearrangement occurs within the first intron of the RNF139 gene and is joined to a fragile site, FRA3B, found on 3p14.2. The RNF139 gene is a putative tumor suppressor gene containing a sterol-sensing domain and a RING-H2 motif essential for ubiquitin ligase activity (Brauweiler et al. 2006). However, a role for haploinsufficiency of this gene, such as would result from the disruption in the current translocation, has not been demonstrated. Nonetheless, localization of the t(8;22) breakpoint to this region raised the possibility that this translocation occurs at an unstable 8q24.1 site involved in other chromosomal rearrangements.
Figure 3.
The 8q24.1 breakpoint maps to a 29.543-kb region containing the RNF139 gene. (A) Schematic diagram depicting the location of BAC clone and PCR-generated FISH probes relative to the RNF139 gene. FISH was performed on metaphase spreads from the balanced t(8;22) carrier using fluorescently labeled BAC clones: RP11–495d4 (green) and RP11–532m24 (red) (B), RP11–158k1 (C), and PCR-generated probes labeled in red (D) and green (E).
Mapping the 8q24.1 breakpoint using laser microdissection of derivative chromosomes
We next sought to fine-map the 8q24.1 breakpoint using a PCR-based approach in which DNA templates derived from either the der(8) or der(22) chromosomes were used. Toward this end, we used LMPC (laser microdissection and pressure catapulting) technology to microdissect the relevant derivative chromosomes from metaphase spreads of the balanced t(8;22) carrier. Laser pulses were used to specifically catapult either the der(8) or der(22) chromosomes from over 170 metaphase spreads into separate chromosome collection pools (see Supplemental Fig. S2). DNA from these der(8) and der(22) pools was then amplified and used for template-specific PCR experiments.
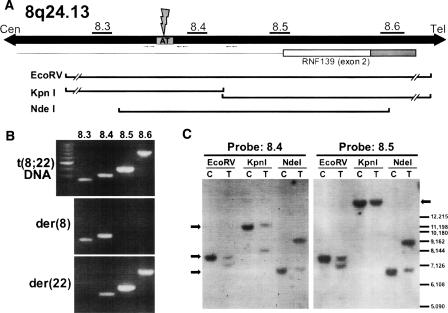
On the basis of our initial mapping results from FISH experiments that localized the breakpoint to a 29.5-kb region of 8q24.1, we designed a series of nine primer pairs to query LMPC-dissected der(8) and der(22) DNA for the presence of sequences within this breakpoint region. This method ultimately allowed us to narrow the breakpoint to a 4415-bp sequence delineated by these PCR amplicons. As shown in Figure 4B, our 8.3 amplicon was seen in reactions using DNA from the der(8) chromosome as well as total genomic DNA from the t(8;22) translocation carrier, indicating that the t(8;22) breakpoint is located distal to this PCR probe. The 8.5 and 8.6 amplicons, on the other hand, were amplified from der(22) DNA but not that from the der(8), meaning that the breakpoint lies proximal to these sequences.
Figure 4.
The 8q24.1 breakpoint localizes to a 3304-bp region within the first intron of the RNF139 gene. (A) Schematic representation of the 8q24.13 region showing the positions of the 8.3, 8.4, 8.5, and 8.6 amplicons used in mapping experiments relative to an AT-rich repeat present between the 8.3 and 8.4 primer pairs. Relative positions of restriction fragments analyzed in Southern blot experiments are also shown. The breakpoint identified by nested PCR experiments is also indicated. (B) PCR-mediated mapping of 8q24.1 sequences was performed on total genomic DNA relative to DNA template from LMPC-dissected der(8) and der(22) chromosomes. Results from the 8.3, 8.4, 8.5, and 8.6 amplicons are shown. (C) Southern blot analysis of genomic DNA from normal control sample (“C”) and the t(8;22) carrier (“T”) digested with EcoRV, KpnI, and NdeI restriction enzymes. Amplicons 8.4 and 8.5 were radiolabeled and used as probes. Arrows indicate the position of expected restriction fragments.
PCR mapping results were confirmed by Southern blotting experiments, which localized the 8q24.1 breakpoint to the same region (Fig. 4C). Amplicons 8.4 and 8.5 were separately radiolabeled and applied to Southern blots of genomic DNA from a normal control (“C”) and the t(8;22) balanced carrier (“T”). Additional bands seen in lanes from the balanced carrier on blots probed with the 8.4 probe indicate that the 8q24.1 breakpoint lies within these EcoRV, KpnI, and NdeI restriction fragments. The 8.5 probe, however, only identifies additional restriction fragments in the EcoRV and NdeI fragments, meaning that the breakpoint in the balanced carrier occurs proximal to the KpnI site. Together with the PCR results from LMPC-dissected derivative chromosome templates, these observations indicate that the 8q24.1 breakpoint is located in a 3304-bp region delineated by the 8.3 amplicon and the KpnI site. They also demonstrate that this breakpoint occurs within the first intron of the RNF139 gene, similar to that seen previously for the constitutional t(3;8) rearrangement associated with renal cell carcinoma (Ohta et al. 1996; Gemmill et al. 1998).
The 8q24.1 breakpoint occurs at the center of a PATRR capable of forming a perfect hairpin structure
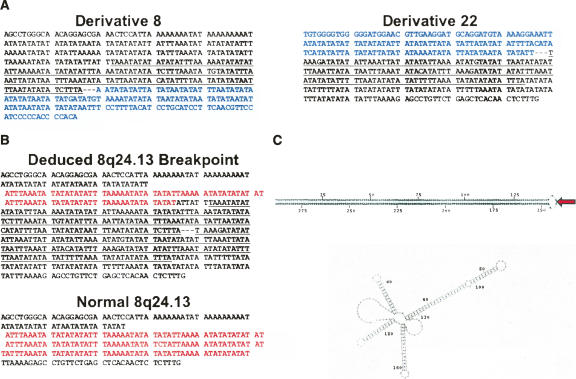
The exact genomic sequences at which these two chromosomes were joined was determined by amplifying the 8;22 translocation junction from both derivative chromosomes. Nested primer PCR was performed on DNA from the balanced t(8;22) carrier using primer sequences corresponding to the 8q24.1 region delineated by LMPC/PCR experiments along with primers derived from the recurrent breakpoint on 22q11.2. As indicated by our FISH results, however, the t(8;22) breakpoint on 22q11.2 occurs within an inverted LCR-B, such that these PCR reactions required the use of 22q11.2 primers oriented in the opposite direction from similar amplifications done previously (Kurahashi et al. 2000b; Nimmakayalu et al. 2003; Gotter et al. 2004). On the 8q24.1 side, primers were designed throughout the 3304-bp breakpoint region with particular attention paid to a 209-bp AT-rich sequence (see Fig. 4A for relative location). Indeed, forward (centromere to telomere) 8q24 primers located just proximal to this sequence successfully amplified der(8) junction fragments when used in conjunction with forward 22q11 primers (again, oppositely oriented 22q11.2 primers were used for this inverted LCR-B breakpoint region). Conversely, two different sets of reverse 8q24 primers used with reverse 22q11.2 primers amplified the der(22) junction products. Sequence analysis validated these products, where 8q24.1 sequence was seen to give way to 22q11.2 sequence just after the breakpoint in der(8) products, and vice versa for the der(22) products (Fig. 5A). The breakpoints on both chromosomes deduced from each of these junction fragments are typical of translocations at the recurrent LCR-B breakpoint; breakage occurs at the center of palindromic sequences, which are frequently AT-rich, with minimal, yet symmetrical loss of nucleotides. Relative to the longest known 22q11.2 sequence, the breakpoint within LCR-B of 22q11.2 occurs at the center of the PATRR with a symmetrical 10-bp deletion (5 bp from each inverted repeat).
Figure 5.
The 8q24.13 breakpoint occurs at the center of a perfect 290-bp AT-rich palindromic sequence. (A) Junction fragment sequence obtained by nested primer PCR using genomic DNA from the balanced t(8;22) carrier (shown centromere to telomere). 8q24 sequence is shown in plain text while that from the recurrent 22q11.2 breakpoint is shown in blue. Underlined 8q24 sequence indicates the inverted repeat sequences flanking the 8q24 breakpoint. (B) Comparison of the 8q24 breakpoint deduced from junction fragments originating from the balanced t(8;22) carrier with “normal” AT-rich sequence present in genomic databases (genomic mapping contig, NT_008046). A 51-bp repeated sequence highlighted in red comprises a potential VNTR. Palindromic sequence at the deduced 8q24 breakpoint is underlined. (C) Potential ssDNA secondary structures formed at the 8q24 breakpoint region by sequence deduced from the t(8;22) carrier (top) and normal sequence (bottom) are depicted. The 8q24 breakpoint (red arrow) is located at the tip of an undisrupted 145 base pair stem–loop structure.
Remarkably, the 8q24.1 breakpoint also appears to have occurred at the center of an AT-rich palindrome. Within both the der(8) and der(22) junction fragments, the 8q24.1 sequence immediately flanking the breakpoint includes a 145-bp sequence that is inverted and repeated on each side of the junction (Fig. 5A, underlined sequence). When these sequences are assembled to construct the 8q24.1 breakpoint sequence that was present at this locus prior to the translocation event, a perfect 290-bp AT-rich palindrome is seen, with the breakpoint precisely splitting the pair of inverted repeats (Fig. 5B, top). The amount of 8q24.1 sequence lost during the formation of this rearrangement, however, cannot be determined, since the inverted repeats seen in these junction fragment sequences are not present in “normal” sequence included in genomic databases (mapping contig, NT_008046). This suggests that the palindrome present at the deduced 8q24.1 breakpoint may represent a polymorphism that had appeared in an ancestor of the t(8;22) carrier. Both native and deduced 8q24.1 breakpoint sequences include a 51- to 52-bp repeated element potentially representing a variable number tandem repeat (VNTR) (Fig. 5B, red highlighted sequence).
Together, these results further suggest a role for stem–loop secondary structures in the genesis of translocations within the LCR-B region of 22q11.2 (Gotter et al. 2004; Kurahashi et al. 2004; Kato et al. 2006). To determine the propensity of both the normal and deduced 8q24.1 breakpoint sequences to form secondary structure within a single strand of DNA, these sequences were analyzed using the M-Fold sequence analysis package (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). As shown in Figure 5C, the most favorable structure formed by a single strand of the deduced 8q24.1 breakpoint sequence is a hairpin structure consisting of a 143-bp stem capped by a 4-nucleotide loop. This is in contrast to the normal 8q24.1 sequence, which does not form any stem–loop structures greater than 40 base pairs (Fig. 5C, bottom).
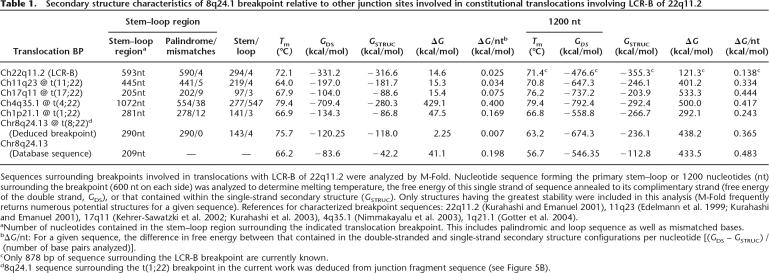
To obtain an indication of the relative propensity for these structures to form, we compared their thermodynamic stability (free energy of secondary structure, GSTRUC) to that of these sequences in the double-stranded state (free energy of double-stranded DNA, GDS) (Gotter et al. 2004). For a given sequence, the difference between GSTRUC and GDS, or ΔG, reflects the energy required for DNA to unwind and reanneal into secondary structure, such that smaller ΔG values are indicative of a greater propensity for secondary structure to form. Compared with other chromosomal loci involved in translocations with the recurrent 22q11.2 breakpoint, the ΔG for the formation of the deduced 8q24.1 stem–loop is extremely low, meaning that this sequence has an even greater penchant for forming secondary structure (Table 1). The free energy per nucleotide (ΔG/nt) for the formation of the 8q24.1 stem–loop region is a mere 0.007 kcal/mol—a value even lower than the recurrent 22q11.2 breakpoint itself. Evaluation of the 1200 nucleotides surrounding the deduced 8q24.1 breakpoint yields a ΔG/nt of 0.365 kcal/mol, which is comparable to that of the recurrent 11q23 and 17q11 breakpoints (0.334 and 0.444 kcal/mol, respectively). Native 8q24.1 sequence, however, exhibits ΔG/nt values that are much higher than the 8q24.1 sequence deduced from junction fragments of the t(8;22) carrier, suggesting that this sequence is much less likely to form secondary structure. This is primarily due to GSTRUC values that are substantially less negative than those determined for the deduced 8q24.1 breakpoint, as would be predicted from the highly unordered secondary structure formed by this sequence (see Fig. 5C, bottom). This analysis suggests that either the sequence in the database is different because of loss of the palindromic sequence during cloning in bacteria, or that an ancestor of the t(8;22) carrier harbored a palindromic polymorphism at the 8q24.1 breakpoint that predisposed that individual to form a translocation between this site and the recurrent 22q11.2 breakpoint.
Table 1.
Secondary structure characteristics of 8q24.1 breakpoint relative to other junction sites involved in constitutional translocations involving LCR-B of 22q11.2
Sequences surrounding breakpoints involved in translocations with LCR-B of 22q11.2 were analyzed by M-Fold. Nucleotide sequence forming the primary stem–loop or 1200 nucleotides (nt) surrounding the breakpoint (600 nt on each side) was analyzed to determine melting temperature, the free energy of this single strand of sequence annealed to its complimentary strand (free energy of the double strand, GDS), or that contained within the single-strand secondary structure (GSTRUC). Only structures having the greatest stability were included in this analysis (M-Fold frequently returns numerous potential structures for a given sequence). References for characterized breakpoint sequences: 22q11.2 (Kurahashi and Emanuel 2001), 11q23 (Edelmann et al. 1999; Kurahashi and Emanuel 2001), 17q11 (Kehrer-Sawatzki et al. 2002; Kurahashi et al. 2003), 4q35.1 (Nimmakayalu et al. 2003), 1q21.1 (Gotter et al. 2004).
aNumber of nucleotides contained in the stem–loop region surrounding the indicated translocation breakpoint. This includes palindromic and loop sequence as well as mismatched bases.
bΔG/nt: For a given sequence, the difference in free energy between that contained in the double-stranded and single-strand secondary structure configurations per nucleotide [(GDS – GSTRUC) / (number of base pairs analyzed)].
cOnly 878 bp of sequence surrounding the LCR-B breakpoint are currently known.
d8q24.1 sequence surrounding the t(1;22) breakpoint in the current work was deduced from junction fragment sequence (see Figure 5B).
Discussion
Novel approaches to pinpointing chromosomal breakpoints
The current study marks the first time that LMPC has been used to isolate and map the breakpoints of rearranged chromosomes. By collecting each relevant derivative chromosome in completely separate LMPC sessions, we were able to avoid contamination of template samples. Although we did observe anomalous amplification of a single PCR probe (probe 8.4, see Fig. 4B) in some PCR mapping trials, the effectiveness of this approach was confirmed by both Southern blot analysis and the subsequent amplification of derivative junction fragment sequences. LMPC-mediated isolation of derivative chromosomes offers an effective alternative to generating specific template for PCR mapping studies over more traditional approaches such as purifying DNA from somatic cell hybrids or isolating derivative chromosomes by manual microdissection.
We have also used MLPA as an adjunct in localizing the inversion and translocation breakpoints on the der(22) chromosome. MLPA is now widely used to detect and localize copy number changes associated with chromosome deletions and duplications. Its utility for rearrangements such as translocations and inversions that are not necessarily associated with a loss or gain of DNA, however, is limited. In the present study, we were able to rapidly and effectively localize all three of the rearrangement breakpoints to LCR regions on 22q11.21 in the unbalanced patient, since the inversion was disrupted by the translocation, creating a derivative chromosome with a discontinuous duplication. This study describes a new application for this technology, which is notable for its specificity, feasibility, and affordability.
Model for a complex rearrangement involving unstable regions of 22q11.2
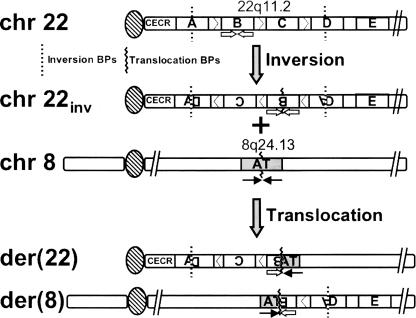
The initial event involved in this patient’s complex chromosomal rearrangement most likely involved an inversion of a region of the TDR of 22q11.2 (Fig. 6). Specifically, this includes the distal portion of LCR-A, LCR-B, LCR-C, and a proximal region of LCR-D (Fig. 6). Aberrant homologous recombination in germ cells of an ancestor of the balanced t(8;22) carrier was likely to involve misalignment of repeat sequences contained in LCR-A and LCR-D. Aberrant crossing over would then have led to an exchange of genetic material resulting in this initial inversion event. Presumably, subsequent to this rearrangement, a translocation occurred between the PATRR found at the recurrent LCR-B breakpoint and a PATRR on 8q24.1, thus giving rise to the balanced karyotype found in the father of the proband. A 3:1 meiotic malsegregation of the der(22) occurring in germ cells of the father then resulted in the unbalanced karyotype of the proband, leading to the supernumerary der(22)t(8;22). Because earlier generations (grandparents, great grandparents of the proband) are not available for analysis, we are unable to identify the individuals in whom these de novo rearrangements have occurred. Given the infrequency of such inversions and translocations, however, it is unlikely that both rearrangements occurred in the germ line of a single individual. From what is known of the mechanisms of these rearrangements, it may not be feasible for such an inversion and subsequent translocation to take place within a single germ line; inversions presumably result from aberrant homologous recombination events occurring during meiotic prophase, while the initial breakage event associated with a translocation is postulated to occur during DNA replication (see below), a process occurring at the initiation of meiosis I. It is, however, possible that the initial inversion event increased the likelihood of producing the translocation in a subsequent generation because of the potential misalignment of homologous chromosomes during meiosis.
Figure 6.
Model for the two-step genesis of the complex rearrangement observed in the supernumerary der(22)t(8;22) proband’s father. A germline inversion of a 22q11.2 region flanked by breakpoints in LCRs A and D occurred before a t(8;22) translocation, the breakpoints of which are located at the center of palindromic sequences at the recurrent LCR-B site (open arrows) and an AT-rich locus of 8q24.13 (solid arrows).
Inversion of the unstable 22q11.21 region is mediated by LCRs
Although this is the first report of an inversion of the 22q11.21 region delineated by LCRs A and D, this particular rearrangement has been predicted by analysis of the genomic architecture that surrounds the previously observed deletions and duplications of this region (Shaikh et al. 2000, 2001; Emanuel and Shaikh 2001). These observations suggested that a homologous recombination-mediated inversion of the 22q11.21 TDR might also be possible. The mechanism for this rearrangement would involve an intrachromosomal recombination event occurring between homologous sequence modules contained within LCRs A and D that are inverted relative to one another. Inverse alignment of LCRs would lead to the formation of a looped intermediate in which a single crossover event at homologous LCR sequences would generate a paracentric inversion of sequences between the paired sites (Emanuel and Shaikh 2001). Similar LCR-mediated mechanisms appear to underlie polymorphic interstitial inversions of both the 1.5-Mb region of 7q11.23 involved in Williams-Beuren syndrome (Osborne et al. 2001) and a 4.7-Mb region containing an olfactory receptor gene cluster at 8p23.1 (Giglio et al. 2001). The higher inversion frequency at the 7q and 8p loci relative to that observed at 22q11.21 is probably the result of the sequence module organization contained within these LCRs. For example, the LCRs mediating the 7q11.23 inversion contain ∼300 kb of sequence that is repeated and inverted in relation to each other (Osborne et al. 2001), whereas LCRs A and D of 22q11.21 share only about 75 kb of inverted repeated sequence (Shaikh et al. 2000). On the other hand, LCRs A and D share 130 kb of direct repeat sequence, which explains the higher frequency of 22q11.21 deletions relative to that seen at 7q11.23. Thus, our discovery of an inversion of the 22q11.21 TDR flanked by LCRs A and D fulfills the prediction suggested by Emanuel and Shaikh (2001) and further substantiates the proposed LCR-mediated looped intermediate mechanism for the generation of interstitial chromosomal inversions.
Palindromic sequences drive translocations involving the recurrent 22q11.21 breakpoint
The 8q24.1 junction described here marks the fifth different chromosomal locus involved in a translocation with the recurrent LCR-B breakpoint on 22q11.21 to be characterized at the nucleotide level. These include over 40 cases of the t(11;22) (Kurahashi et al. 2000b), two instances of the t(17;22) (Kehrer-Sawatzki et al. 2002; Kurahashi et al. 2003), and individual t(1;22) and t(4;22) cases (Nimmakayalu et al. 2003; Gotter et al. 2004). Remarkably, all of these 22q11.21 breakpoints occur within 16 base pairs of each other at the center of the PATRR within LCR-B. The only differences in breakpoint location are small, <10-bp deletions occurring at symmetrical distances from the center of the inverted repeat. Conversely, all of the other chromosomal sites that form translocations with the PATRR of 22q11.21 also contain palindromic sequences, suggesting that stem–loop structures are involved in the genesis of these translocations. The stability of these secondary structures is reflected in the calculated free energy for their formation, where the difference between the inherent stability of the double-stranded sequence and that of the potential secondary structure formed at these sites (ΔG) is significantly less than those values for translocation breakpoints on chromosome 22 not involving the recurrent 22q11.21 site (Gotter et al. 2004). Because the palindrome at the 8q24.13 breakpoint described in the current work is capable of forming a perfect stem–loop structure, the ΔG for this sequence is extremely low (see Table 1) and is comparable or even lower than the other breakpoints involved in translocations with the 22q11.21 PATRR. For these structures to form in vivo, DNA at these sites must first go through a single-stranded DNA (ssDNA) intermediate. For this reason, AT-rich sequences having lower melting temperatures have a smaller activation energy to overcome and are predicted to form these structures more readily. AT-rich palindromes should, therefore, be better substrates for translocations with the PATRR at 22q11.21. This too is reflected by smaller ΔG values, since the free energy inherent in the double-stranded state of these sequences is low compared with the relative stability of secondary structure formed by these sites. The 8q24.13 breakpoint site described here is almost entirely composed of AT-rich sequence and, together with its palindromic nature, is predicted to be an excellent candidate for 22q11.21-mediated translocations.
If the 8q24.13 site is such an excellent substrate for translocation with 22q11.21, then why has it not been observed before? The ΔG values seen in Table 1 suggest that it may be an even better substrate than sequences contained at the recurrent 11q23 and the 17q11 breakpoints. One possibility is that these chromosomal loci may not be favorably oriented relative to one another during meiosis. Ashley et al. (2006) have demonstrated that 22q11.21 and 11q23 are positioned in proximity to one another during meiosis and that this is likely to at least partially underlie the recurrent nature of the t(11;22). The lack of a recurrent t(8;22) may be explained by unfavorable chromosomal spatial localization or organization during meiosis. A second possibility is that the palindrome present at the 8q24.13 site uncovered in this study is a rare polymorphism that is not normally present. In this case, the presence of this PATRR in an ancestor of the proband may have predisposed that individual to generation of the t(8;22). The “normal” sequence for this site found in genomic databases does not contain this extended inverted repeat but does include three copies of a 51- to 52-bp repeat. This sequence, which potentially represents a VNTR, supports the idea that this site may undergo frequent sequence changes. VNTR expansion and contraction due to DNA replication stalling can occur at such sequence motifs, especially those that are AT rich. This type of mechanism is also thought to underlie fragile site instability and chromosomal rearrangements (Yunis and Soreng 1984; Cimprich 2003).
An alternative possibility is that the palindrome at the deduced 8q24.1 breakpoint more accurately reflects normal genomic sequence than that which is found in databases. Genomic databases are constructed from shotgun sequencing of overlapping bacterial and yeast artificial chromosomes (BACs and YACs, respectively). Human sequences containing large palindromes, however, are either unclonable in these systems or are resolved by deletion of variable amounts of the inverted repeat sequence (Leach 1994). In fact, it has been demonstrated that the 11q23 breakpoint, consisting of 441 bp of palindromic sequence, is also partially deleted from genomic database contigs (Kurahashi and Emanuel 2001). In the present studies, PCR amplification and sequence analysis of the 8q24.13 site in three different CEPH controls and a chromosome 8 somatic cell hybrid generally confirmed the shorter sequence seen in genomic databases (data not shown). We did, however, observe larger PCR products that were unclonable, even in bacteria mutant for DNA repair pathways responsible for resolving some secondary structure (uvrC− and umuC−), and that did not yield reliable results from direct sequencing. Thus, it remains possible that the extended PATRR at the 8q24.13 breakpoint does normally exist within the genome. On the other hand, the 290-bp palindrome at this site with no mismatches suggests that if it is normally present, it is likely to be a relatively new addition to the human genome.
Insights into translocation mechanisms
The initial breakage event involved in many constitutional and somatic translocations is thought to be the result of stalled DNA replication. Genomic instability at stalled replication forks is likely to be related to the presence of unreplicated ssDNA existing between the helicase unwinding activity at the leading edge of the replisome and DNA polymerases responsible for synthesizing the second strand of DNA. The 8q24.13 breakpoint further suggests that hairpin–loop secondary structure may be involved in the mechanism underlying translocations with the recurrent 22q11.21 site. Not only does this site contain a perfect tandem inverted repeat, but it is also potentially involved in a previously described constitutional t(3;8)(p14.2;q24.13) (Ohta et al. 1996; Gemmill et al. 1998). This latter rearrangement involves an 8q24.13 breakpoint contained within the 10.5-kb intron of the RNF139 gene and the FRA3B fragile site at 3p14.2. Although the precise genomic location of these breakpoint sequences has not been determined, this latter translocation is likely to involve similar secondary structure formed at each of these loci. Like the 22q11.2 site involved with 8q24.13 in the current work, FRA3B is highly AT rich and has been predicted to form numerous secondary structures capable of impeding replication (Arlt et al. 2006). Given their propensity for secondary structure, all three of these sites, 8q24.13, 3p14.2, and 22q11.21, as well as those involved in other translocations with the recurrent 22q11.21 site (11q23, 17q11, 1p21.1, 4p35.1), are likely to slow replication to a similar degree, thus making them susceptible to similar structure-specific nucleases and subsequent translocation.
Common secondary structure characteristics shared by translocation breakpoints may not be unique to breakpoints involved in translocations with the recurrent 22q11.21 site. Our previous analysis of other translocation breakpoints indicated that, in general, these rearrangements tend to occur between sequences of similar melting temperature and propensity for secondary structure, even though none of these other breakpoints were capable of specifically forming stem–loop structures (Gotter et al. 2004). This may indicate that replication stalling at different times during S phase occurs at sites having a similar propensity for secondary structure, thus making them better substrates for translocations with one another. Many fragile sites including FRA3B, for example, are replicated late in S phase, much as one would expect for sites involved in translocations with the recurrent 22q11.21 site, since these sites are capable of stable secondary structures impeding replication. Other sites forming less stable secondary structures may impede replication to a lesser degree and coexist with one another at earlier times during S phase. Further inhibition of replication by DNA damage or metabolic stress may then lock the replication site at these sites allowing nucleases to create a double-stranded intermediate capable of forming a translocation between these sites. Further studies on the role of replication stalling in this process will enhance our understanding of the sequence specificity of these chromosomal rearrangements.
Methods
FISH
Transformed lymphoblastoid cell lines derived from the supernumerary der(22) proband, her father carrying the balanced t(8;22), and her unaffected mother were prepared using standard methodology. Cosmid probes specific for chromosome 22 had been isolated from the LL22NCO3 library (Shaikh et al. 2000), while BAC clones corresponding to chromosome 8 sequences were selected on the basis of their location from the UCSC database (http://genome.ucsc.edu/) and purchased from BACPAC resources (http://bacpac.chori.org/). All probes used for FISH were labeled by nick translation and applied to metaphase spreads using pre- and posthybridization conditions described previously with minor modifications (Trask 1991; Holmes et al. 1997).
MLPA
MLPA was performed on genomic DNA purified from lymphoblast cell lines derived from the proband carrying the supernumerary der(22), the balanced t(8;22) carrier (father of the proband), and the unaffected mother essentially as described previously (Schouten et al. 2002; Vorstman et al. 2006). Briefly, genomic DNA isolated from lymphoblast cell lines (20–500 ng) was denatured at 98°C for 5 min and hybridized to MLPA hemiprobes included with a modified version of the P023 probe set (DiGeorge Syndrome; MRC Holland) according to the manufacturer’s protocol. PCR amplification was performed on an ABI 9700 thermal cycler (Applied Biosystems) and separation and detection of variable length amplicons contained in single reactions was performed by capillary electrophoresis on an ABI 3700 DNA analyzer (Applied Biosystems). Analysis was carried out with GeneMarker analysis Software (SoftGenetics, LLC). GeneMarker, in the MLPA mode, converts data taken from the sequencer, provides two normalization and analysis methods, and corrects for amplicon size using a binomial regression model. In addition to the test samples presented, each experiment includes six normal control samples for baseline determination and normalization. Genetic dosage quotients near 1.0 are described as disomic while those above a threshold value of 1.3 are considered trisomic and those below 0.7 haploid in accordance with empirically derived and commonly accepted values (Bunyan et al. 2004).
Laser microdissection of derivative chromosomes and template-specific PCR
Fixed chromosome preparations were made according to standard protocols and were G-banded on cleaned irradiated glass coverslips. Well-spread metaphases were selected and derivative chromosomes 8 and 22 were captured separately using a Zeiss Axiovert 200M inverted microscope equipped with a P.A.L.M microlaser system (Zeiss) in accordance with published protocols with modifications (Rook et al. 2004). Dissection of selected chromosomes was accomplished with brief pulses from the microlaser, which effectively removed the genetic material from the metaphase-containing coverslip and simultaneously collected them in the cap of a sterile reaction tube containing 40 μl of sterile water. In practice, glass slides were found to yield superior optics and were therefore more efficient in collecting chromosomes than specialized ultra-thin membranes provided by the manufacturer as previously observed (Fominaya et al. 2005). For each derivative, more than 170 chromosomes were captured separately from multiple coverslips and were pooled. Pooled chromosomal DNA was dried by vacuum centrifugation and resuspended in water. A 1- to 2-μl aliquot of resuspended DNA was then nonspecifically amplified using Amersham’s ø29 polymerase-based GenomiPhi kit (Amersham Biosciences) according to manufacturer’s instructions.
Amplified derivative chromosome DNA and whole genomic DNA purified from lymphoblast cells derived from the balanced t(8;22) carrier were used for template-specific PCR analysis. A total of 9 primer pairs amplifying 200- to 400-bp sequences were designed within the 29.5-kb region delineated by prior FISH mapping studies, the spacing of these amplicons being ∼3.5 kb apart (primer pairs 8.1–8.9; only primer pairs 8.3–8.6 shown). PCR reactions were carried out with the Accuprime Taq DNA polymerase system (Invitrogen) using the following step annealing protocol for enhanced specificity: 2 min at 94°C; 14 cycles of: 94°C (30 sec), 68–55°C (−1°C/cycle) (45 sec), and 68°C (1 min); 30 cycles of: 94°C (30 sec), 55°C (45 sec), and 68°C (1 min); 68°C (7 min). Products were visualized using standard agarose gel electrophoresis.
Southern blot analysis
Genomic DNA purified from lymphoblast cell lines derived from the balanced t(8;22) carrier and a CEPH control were restriction digested with EcoRV, KpnI, and NdeI, run on agarose gel electrophoresis, transferred to Hybond-N+ nylon membranes (Amersham Pharmacia Biotech), and probed according to standard procedures. To confirm results from PCR mapping studies using template from laser microdissected chromosomes, two probes, corresponding to the 8.4 and 8.5 amplicons seen in Fig. 4B, were [α-32P]dCTP labeled using the DECA prime II random primed labeling kit (Ambion Inc.) and hybridized to membranes according to manufacturer’s recommendations (Amersham).
Amplification and sequence analysis of t(8;22) junction sequences
Nested primer PCR was used to amplify the t(8;22) junction from both the der(8) and der(22) chromosomes using genomic DNA purified from the lymphoblast cell line derived from the balanced t(8;22) carrier similar to procedures and chromosome 22 primers described previously (Gotter et al. 2004). Primers corresponding to 8q24.13 sequences were designed based on the results derived from laser-microdissected derivative chromosome mapping studies. Der(8) products were amplified using the following nested primers: Primary amplification: 8q24–7F, CAGTG AGCCAAGATCACGCCATTGCACT with Ch22-B3, GGGGGTGG GGGATGGAACGTTGAAGGATC. Secondary amplification: 8q24–8F, AGCCTGGGCAACAGGAGCGAAACTCCAT with Ch22-B4R, TGTGGGGTGGGGGATGGAACGTTGAAGGATG. Two different der(22) products were amplified with identical chromosome 22 primers and two different sets of 8q24.13 nested primers. Sequences of smaller amplicons not only exactly matched one another but also matched the overlapping region of larger products (because of space considerations, only small products are shown; Fig. 5B). Primary amplification of smaller der(22) products: Ch22-B2R, GGAAGGGAAAAACATGTTAAAAA CAAAGAGAGGTAC with 8q24–7R, CCTGATTCCCCAAGAAAA TGGGCCACTA. Secondary amplifications: Ch22-B4R (see above) with 8q24–8R, CAAAGAGTTGTGAGCTCAGAACAGGCTCT. Primary amplification of larger products: Ch22-B2R with 8q24-R4, CTGCAAATCTCCAAAGTTCTGAAACTCGA. Secondary amplification of larger products: Ch22-B4R with 8q24-R5, GCCTTAAAC TAGAATCAGCTTTTCCTTGTG. Primary and secondary PCR amplifications were carried out with the Advantage 2 Polymerase Mix system (Clontech) using a derivation of the step annealing protocol described above (annealing temperatures were empirically determined for each primer pair and 2 min extensions at 72°C were performed). Products appearing in reactions using t(8;22) carrier DNA as template, but not from CEPH controls, were excised from agarose gels, QiaQuick (Qiagen) purified, cloned into the pCRII-TopoTA vector (Invitrogen), and sequenced using vector-specific primers. For each amplicon, 3 PCR products were used to corroborate junction fragment sequence. Sequencer 4.6 software (Gene Codes Corp.) was used to compile sequences, and potential secondary structures along with their thermodynamic free energy determinations were obtained using the Mfold server (http://bioweb.pasteur.fr/seqanal/interfaces/mfold-simple.html). Free energy for the formation of secondary structures was determined as previously described (Gotter et al. 2004) (see also legend to Table 1).
Acknowledgments
We thank Alexander R. Judkins, M.D., Dan Martinez, Julia Brown, and David Tomkins for technical assistance. MRC-Holland is acknowledged for providing extended MLPA probes. This work was supported by the Florence R.C. Murray Program (Children’s Hospital of Philadelphia) (A.L.G.), the Charles E.H. Upham chair in Pediatrics (B.S.E.), and grants HL74731 and CA39926 (B.S.E.) from the National Institutes of Health.
Footnotes
[Supplemental material is available online at www.genome.org]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6130907
References
- Arlt M.F., Durkin S.G., Ragland R.L., Glover T.W., Durkin S.G., Ragland R.L., Glover T.W., Ragland R.L., Glover T.W., Glover T.W. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst.) 2006;5:1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- Ashley T., Gaeth A.P., Inagaki H., Seftel A., Cohen M.M., Anderson L.K., Kurahashi H., Emanuel B.S., Gaeth A.P., Inagaki H., Seftel A., Cohen M.M., Anderson L.K., Kurahashi H., Emanuel B.S., Inagaki H., Seftel A., Cohen M.M., Anderson L.K., Kurahashi H., Emanuel B.S., Seftel A., Cohen M.M., Anderson L.K., Kurahashi H., Emanuel B.S., Cohen M.M., Anderson L.K., Kurahashi H., Emanuel B.S., Anderson L.K., Kurahashi H., Emanuel B.S., Kurahashi H., Emanuel B.S., Emanuel B.S. Meiotic recombination and spatial proximity in the etiology of the recurrent t(11;22) Am. J. Hum. Genet. 2006;79:524–538. doi: 10.1086/507652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldog F.L., Gemmill R.M., Wilke C.M., Glover T.W., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Gemmill R.M., Wilke C.M., Glover T.W., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Wilke C.M., Glover T.W., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Glover T.W., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Nilsson A.S., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Chandrasekharappa S.C., Brown R.S., Li F.P., Drabkin H.A., Brown R.S., Li F.P., Drabkin H.A., Li F.P., Drabkin H.A., Drabkin H.A. Positional cloning of the hereditary renal carcinoma 3;8 chromosome translocation breakpoint. Proc. Natl. Acad. Sci. 1993;90:8509–8513. doi: 10.1073/pnas.90.18.8509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauweiler A., Lorick K.L., Lee J.P., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M., Lorick K.L., Lee J.P., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M., Lee J.P., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M., Tsai Y.C., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M., Chan D., Weissman A.M., Drabkin H.A., Gemmill R.M., Weissman A.M., Drabkin H.A., Gemmill R.M., Drabkin H.A., Gemmill R.M., Gemmill R.M. RING-dependent tumor suppression and G2/M arrest induced by the TRC8 hereditary kidney cancer gene. Oncogene. 2006 doi: 10.1038/sj.onc.1210017. doi: 10.1038/sj.onc.1210017. [DOI] [PubMed] [Google Scholar]
- Bunyan D.J., Eccles D.M., Sillibourne J., Wilkins E., Thomas N.S., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Eccles D.M., Sillibourne J., Wilkins E., Thomas N.S., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Sillibourne J., Wilkins E., Thomas N.S., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Wilkins E., Thomas N.S., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Thomas N.S., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Shea-Simonds J., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Duncan P.J., Curtis C.E., Robinson D.O., Harvey J.F., Curtis C.E., Robinson D.O., Harvey J.F., Robinson D.O., Harvey J.F., Harvey J.F., et al. Dosage analysis of cancer predisposition genes by multiplex ligation-dependent probe amplification. Br. J. Cancer. 2004;91:1155–1159. doi: 10.1038/sj.bjc.6602121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimprich K.A. Fragile sites: Breaking up over a slowdown. Curr. Biol. 2003;13:R231–R233. doi: 10.1016/s0960-9822(03)00158-1. [DOI] [PubMed] [Google Scholar]
- Driscoll D.A., Emanuel B.S., Emanuel B.S. The 22q11 deletion. DiGeorge and velocardiofacial syndrome. In: Jameson J.L., editor. Principles of Molecular Medicine. Humana Press; Totowa, NJ: 1998. pp. 1079–1085. [Google Scholar]
- Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Magenis E., Shprintzen R.J., Morrow B.E., Shprintzen R.J., Morrow B.E., Morrow B.E. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Emanuel B.S., Shaikh T.H., Shaikh T.H. Segmental duplications: an ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- Ensenauer R.E., Adeyinka A., Flynn H.C., Michels V.V., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Adeyinka A., Flynn H.C., Michels V.V., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Flynn H.C., Michels V.V., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Michels V.V., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Lindor N.M., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Dawson D.B., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Thorland E.C., Lorentz C.P., Goldstein J.L., McDonald M.T., Lorentz C.P., Goldstein J.L., McDonald M.T., Goldstein J.L., McDonald M.T., McDonald M.T., et al. Microduplication 22q11.2, an emerging syndrome: clinical, cytogenetic, and molecular analysis of thirteen patients. Am. J. Hum. Genet. 2003;73:1027–1040. doi: 10.1086/378818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feuk L., MacDonald J.R., Tang T., Carson A.R., Li M., Rao F., Khaja R., Scherer S.W., MacDonald J.R., Tang T., Carson A.R., Li M., Rao F., Khaja R., Scherer S.W., Tang T., Carson A.R., Li M., Rao F., Khaja R., Scherer S.W., Carson A.R., Li M., Rao F., Khaja R., Scherer S.W., Li M., Rao F., Khaja R., Scherer S.W., Rao F., Khaja R., Scherer S.W., Khaja R., Scherer S.W., Scherer S.W. Discovery of human inversion polymophisms by comparative analysis of human and chimpanzee DNA sequence assemblies. PLoS Genet. 2005;1:489–498. doi: 10.1371/journal.pgen.0010056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fominaya A., Linares C., Loarce Y., Ferrer E., Linares C., Loarce Y., Ferrer E., Loarce Y., Ferrer E., Ferrer E. Microdissection and microcloning of plant chromosomes. Cytogenet. Genome Res. 2005;109:8–14. doi: 10.1159/000082376. [DOI] [PubMed] [Google Scholar]
- Gebhardt G.S., Devriendt K., Thoelen R., Swillen A., Pijkels E., Fryns J.P., Vermeesch J.R., Gewillig M., Devriendt K., Thoelen R., Swillen A., Pijkels E., Fryns J.P., Vermeesch J.R., Gewillig M., Thoelen R., Swillen A., Pijkels E., Fryns J.P., Vermeesch J.R., Gewillig M., Swillen A., Pijkels E., Fryns J.P., Vermeesch J.R., Gewillig M., Pijkels E., Fryns J.P., Vermeesch J.R., Gewillig M., Fryns J.P., Vermeesch J.R., Gewillig M., Vermeesch J.R., Gewillig M., Gewillig M. No evidence for a parental inversion polymorphism predisposing to rearrangements at 22q11.2 in the DiGeorge/Velocardiofacial syndrome. Eur. J. Hum. Genet. 2003;11:109–111. doi: 10.1038/sj.ejhg.5200930. [DOI] [PubMed] [Google Scholar]
- Gemmill R.M., West J.D., Boldog F., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A., West J.D., Boldog F., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A., Boldog F., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A., Tanaka N., Robinson L.J., Smith D.I., Li F., Drabkin H.A., Robinson L.J., Smith D.I., Li F., Drabkin H.A., Smith D.I., Li F., Drabkin H.A., Li F., Drabkin H.A., Drabkin H.A. The hereditary renal cell carcinoma 3;8 translocation fuses FHIT to a patched-related gene, TRC8. Proc. Natl. Acad. Sci. 1998;95:9572–9577. doi: 10.1073/pnas.95.16.9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglio S., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Broman K.W., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Matsumoto N., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Calvari V., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Gimelli G., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Neumann T., Ohashi H., Voullaire L., Larizza D., Giorda R., Ohashi H., Voullaire L., Larizza D., Giorda R., Voullaire L., Larizza D., Giorda R., Larizza D., Giorda R., Giorda R., et al. Olfactory receptor-gene clusters, genomic-inversion polymorphisms and common chromosome rearrangements. Am. J. Hum. Genet. 2001;68:874–883. doi: 10.1086/319506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S., Budarf M.L., Rhodes C.H., Emanuel B.S., Rhodes C.H., Emanuel B.S., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassed S.J., Hopcus-Niccum D., Zhang L., Li S., Mulvihill J.J., Hopcus-Niccum D., Zhang L., Li S., Mulvihill J.J., Zhang L., Li S., Mulvihill J.J., Li S., Mulvihill J.J., Mulvihill J.J. A new genomic duplication syndrome complementary to the velocardiofacial (22q11 deletion) syndrome. Clin. Genet. 2004;65:400–404. doi: 10.1111/j.0009-9163.2004.0212.x. [DOI] [PubMed] [Google Scholar]
- Holmes S.E., Riazi M.A., Gong W., McDermid H.E., Sellinger B.T., Hua A., Chen F., Wang Z., Zhang G., Roe B., Riazi M.A., Gong W., McDermid H.E., Sellinger B.T., Hua A., Chen F., Wang Z., Zhang G., Roe B., Gong W., McDermid H.E., Sellinger B.T., Hua A., Chen F., Wang Z., Zhang G., Roe B., McDermid H.E., Sellinger B.T., Hua A., Chen F., Wang Z., Zhang G., Roe B., Sellinger B.T., Hua A., Chen F., Wang Z., Zhang G., Roe B., Hua A., Chen F., Wang Z., Zhang G., Roe B., Chen F., Wang Z., Zhang G., Roe B., Wang Z., Zhang G., Roe B., Zhang G., Roe B., Roe B., et al. Disruption of the clathrin heavy chain-like gene (CLTCL) associated with features of DGS/VCFS: a balanced (21;22)(p12;q11) translocation. Hum. Mol. Genet. 1997;6:357–367. doi: 10.1093/hmg/6.3.357. [DOI] [PubMed] [Google Scholar]
- Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H., Taniguchi M., Emanuel B.S., Kurahashi H., Emanuel B.S., Kurahashi H., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H., Assum G., Hameister H., Assum G., Hameister H., Hameister H. Molecular characterisation of t(17;22)(q11.2;q11.2) is not consistent with NF1 gene duplication. Hum. Genet. 2002;111:465–467. 468–469. doi: 10.1007/s00439-002-0794-3. ; author reply . [DOI] [PubMed] [Google Scholar]
- Khaja R., Zhang J., MacDonald J.R., He Y., Joseph-George A.M., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., Zhang J., MacDonald J.R., He Y., Joseph-George A.M., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., MacDonald J.R., He Y., Joseph-George A.M., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., He Y., Joseph-George A.M., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., Joseph-George A.M., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., Wei J., Rafiq M.A., Qian C., Shago M., Pantano L., Rafiq M.A., Qian C., Shago M., Pantano L., Qian C., Shago M., Pantano L., Shago M., Pantano L., Pantano L., et al. Genome assembly comparison identifies structural variants in the human genome. Nat. Genet. 2006;38:1413–1418. doi: 10.1038/ng1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Emanuel B.S., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10:2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Roe B.A., Emanuel B.S., Budarf M.L., Emanuel B.S., Budarf M.L., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000a;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S., Driscoll D.A., Budarf M.L., Emanuel B.S., Budarf M.L., Emanuel B.S., Emanuel B.S. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000b;67:763–768. doi: 10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T., Takata M., Toda T., Emanuel B.S., Shaikh T., Takata M., Toda T., Emanuel B.S., Takata M., Toda T., Emanuel B.S., Toda T., Emanuel B.S., Emanuel B.S. The constitutional t(17;22): Another translocation mediated by palindromic AT-rich repeats. Am. J. Hum. Genet. 2003;72:733–738. doi: 10.1086/368062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Ohye T., Taniguchi M., Emanuel B.S., Toda T., Taniguchi M., Emanuel B.S., Toda T., Emanuel B.S., Toda T., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- Lindsay E.A., Shaffer L.G., Carrozzo R., Greenberg F., Baldini A., Shaffer L.G., Carrozzo R., Greenberg F., Baldini A., Carrozzo R., Greenberg F., Baldini A., Greenberg F., Baldini A., Baldini A. De novo tandem duplication of chromosome segment 22q11-q12: Clinical, cytogenetic, and molecular characterization. Am. J. Med. Genet. 1995;56:296–299. doi: 10.1002/ajmg.1320560316. [DOI] [PubMed] [Google Scholar]
- Mears A.J., Duncan A.M., Budarf M.L., Emanuel B.S., Sellinger B., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Duncan A.M., Budarf M.L., Emanuel B.S., Sellinger B., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Budarf M.L., Emanuel B.S., Sellinger B., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Emanuel B.S., Sellinger B., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Sellinger B., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Siegel-Bartelt J., Greenberg C.R., McDermid H.E., Greenberg C.R., McDermid H.E., McDermid H.E. Molecular characterization of the marker chromosome associated with cat eye syndrome. Am. J. Hum. Genet. 1994;55:134–142. [PMC free article] [PubMed] [Google Scholar]
- Meins M., Burfeind P., Motsch S., Trappe R., Bartmus D., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Burfeind P., Motsch S., Trappe R., Bartmus D., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Motsch S., Trappe R., Bartmus D., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Trappe R., Bartmus D., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Bartmus D., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Langer S., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Speicher M.R., Muhlendyck H., Bartels I., Zoll B., Muhlendyck H., Bartels I., Zoll B., Bartels I., Zoll B., Zoll B. Partial trisomy of chromosome 22 resulting from an interstitial duplication of 22q11.2 in a child with typical cat eye syndrome. J. Med. Genet. 2003;40:e62. doi: 10.1136/jmg.40.5.e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmakayalu M.A., Gotter A.L., Shaikh T.H., Emanuel B.S., Gotter A.L., Shaikh T.H., Emanuel B.S., Shaikh T.H., Emanuel B.S., Emanuel B.S. A novel sequence-based approach to localize translocation breakpoints identifies the molecular basis of a t(4;22) Hum. Mol. Genet. 2003;12:2817–2825. doi: 10.1093/hmg/ddg301. [DOI] [PubMed] [Google Scholar]
- Ohta M., Inoue H., Cotticelli M.G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Inoue H., Cotticelli M.G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Cotticelli M.G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Siprashvili Z., Mori M., McCue P., Druck T., Mori M., McCue P., Druck T., McCue P., Druck T., Druck T., et al. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84:587–597. doi: 10.1016/s0092-8674(00)81034-x. [DOI] [PubMed] [Google Scholar]
- Osborne L.R., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Li M., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Pober B., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Chitayat D., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Bodurtha J., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Mandel A., Costa T., Grebe T., Cox S., Tsui L.C., Costa T., Grebe T., Cox S., Tsui L.C., Grebe T., Cox S., Tsui L.C., Cox S., Tsui L.C., Tsui L.C., et al. A 1.5 million-base pair inversion polymorphism in families with Williams-Beuren syndrome. Nat. Genet. 2001;29:321–325. doi: 10.1038/ng753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portnoi M.F., Lebas F., Gruchy N., Ardalan A., Biran-Mucignat V., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Lebas F., Gruchy N., Ardalan A., Biran-Mucignat V., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Gruchy N., Ardalan A., Biran-Mucignat V., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Ardalan A., Biran-Mucignat V., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Biran-Mucignat V., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Malan V., Finkel L., Roger G., Ducrocq S., Gold F., Finkel L., Roger G., Ducrocq S., Gold F., Roger G., Ducrocq S., Gold F., Ducrocq S., Gold F., Gold F., et al. 22q11.2 duplication syndrome: Two new familial cases with some overlapping features with DiGeorge/velocardiofacial syndromes. Am. J. Med. Genet. A. 2005;137:47–51. doi: 10.1002/ajmg.a.30847. [DOI] [PubMed] [Google Scholar]
- Rook M.S., Delach S.M., Deyneko G., Worlock A., Wolfe J.L., Delach S.M., Deyneko G., Worlock A., Wolfe J.L., Deyneko G., Worlock A., Wolfe J.L., Worlock A., Wolfe J.L., Wolfe J.L. Whole genome amplification of DNA from laser capture-microdissected tissue for high-throughput single nucleotide polymorphism and short tandem repeat genotyping. Am. J. Pathol. 2004;164:23–33. doi: 10.1016/S0002-9440(10)63092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta S.C., Harris S.E., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Harris S.E., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Chakraborty P.K., Hacker A.M., Zackai E.H., Hacker A.M., Zackai E.H., Zackai E.H., et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet. 2004;13:417–428. doi: 10.1093/hmg/ddh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachenmann G., Schmid W., Fraccaro M., Mannini A., Tiepolo L., Perona G.P., Sartori E., Schmid W., Fraccaro M., Mannini A., Tiepolo L., Perona G.P., Sartori E., Fraccaro M., Mannini A., Tiepolo L., Perona G.P., Sartori E., Mannini A., Tiepolo L., Perona G.P., Sartori E., Tiepolo L., Perona G.P., Sartori E., Perona G.P., Sartori E., Sartori E. Chromosomes in coloboma and anal atresia. Lancet. 1965;II:290. doi: 10.1016/s0140-6736(65)92415-3. [DOI] [PubMed] [Google Scholar]
- Schinzel A., Schmid W., Fraccaro M., Tiepolo L., Zuffardi O., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Schmid W., Fraccaro M., Tiepolo L., Zuffardi O., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Fraccaro M., Tiepolo L., Zuffardi O., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Tiepolo L., Zuffardi O., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Zuffardi O., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Opitz J.M., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Lindsten J., Zetterqvist P., Enell H., Baccichetti C., Zetterqvist P., Enell H., Baccichetti C., Enell H., Baccichetti C., Baccichetti C., et al. The ′cat eye syndrome': Decentric small marker chromosome probably derived from a 22 (tetrasomy 22pter;q11) associated with a characteristic phenotype. Report of 11 patients and delineation of the clinical picture. Hum. Genet. 1981;57:148–158. doi: 10.1007/BF00282012. [DOI] [PubMed] [Google Scholar]
- Schouten J.P., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G., McElgunn C.J., Waaijer R., Zwijnenburg D., Diepvens F., Pals G., Waaijer R., Zwijnenburg D., Diepvens F., Pals G., Zwijnenburg D., Diepvens F., Pals G., Diepvens F., Pals G., Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Saitta S.C., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Kurahashi H., Saitta S.C., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Saitta S.C., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Zackai E.H., Budarf M.L., Budarf M.L., et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Emanuel B.S., Kurahashi H., Emanuel B.S., Emanuel B.S. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet. Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Tapia-Paez I., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Blennow E., Fedorova L., Imreh S., Dumanski J.P., Fedorova L., Imreh S., Dumanski J.P., Imreh S., Dumanski J.P., Dumanski J.P. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- Trask B.J. DNA sequence localization in metaphase and interphase cells by fluorescence in situ hybridization. Methods Cell Biol. 1991;35:3–35. doi: 10.1016/s0091-679x(08)60567-1. [DOI] [PubMed] [Google Scholar]
- Vorstman J.A., Jalali G.R., Rappaport E.F., Hacker A.M., Scott C., Emanuel B.S., Jalali G.R., Rappaport E.F., Hacker A.M., Scott C., Emanuel B.S., Rappaport E.F., Hacker A.M., Scott C., Emanuel B.S., Hacker A.M., Scott C., Emanuel B.S., Scott C., Emanuel B.S., Emanuel B.S. MLPA: A rapid, reliable, and sensitive method for detection and analysis of abnormalities of 22q. Hum. Mutat. 2006;27:814–821. doi: 10.1002/humu.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobb T.M., Somerville M.J., Willatt L., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Somerville M.J., Willatt L., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Willatt L., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Firth H.V., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Harrison K., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., MacKenzie J., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Gallo N., Morrow B.E., Shaffer L.G., Babcock M., Morrow B.E., Shaffer L.G., Babcock M., Shaffer L.G., Babcock M., Babcock M., et al. Microduplication and triplication of 22q11.2: A highly variable syndrome. Am. J. Hum. Genet. 2005;76:865–876. doi: 10.1086/429841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunis J.J., Soreng A.L., Soreng A.L. Constitutive fragile sites and cancer. Science. 1984;226:1199–1204. doi: 10.1126/science.6239375. [DOI] [PubMed] [Google Scholar]