Abstract
Genomic disorders contribute significantly to genetic disease and, as detection methods improve, greater numbers are being defined. Paralogous low copy repeats (LCRs) mediate many of the chromosomal rearrangements that underlie these disorders, predisposing chromosomes to recombination errors. Deletions of proximal 22q11.2 comprise the most frequently occurring microdeletion syndrome, DiGeorge/Velocardiofacial syndrome (DGS/VCFS), in which most breakpoints have been localized to a 3 Mb region containing four large LCRs. Immediately distal to this region, there are another four related but smaller LCRs that have not been characterized extensively. We used paralog-specific primers and long-range PCR to clone, sequence, and examine the distal deletion breakpoints from two patients with de novo deletions mapping to these distal LCRs. Our results present definitive evidence of the direct involvement of LCRs in 22q11 deletions and map both breakpoints to the BCRL module, common to most 22q11 LCRs, suggesting a potential region for LCR-mediated rearrangement both in the distal LCRs and in the DGS interval. These are the first reported cases of distal 22q11 deletions in which breakpoints have been characterized at the nucleotide level within LCRs, confirming that distal 22q11 LCRs can and do mediate rearrangements leading to genomic disorders.
Chromosome 22q11 shows a high frequency of de novo genomic rearrangement. This instability is attributed to the presence of several large paralogous low copy repeats (LCRs) or segmental duplications (SDs), each containing a complex modular structure and a high degree of sequence identity (>96%) over large stretches of the repeat (Shaikh et al. 2000). The LCRs apparently mediate aberrant interchromosomal exchanges during meiosis (Saitta et al. 2004), and 22q11 deletions, which occur in up to 1:4000 live births (Burn and Goodship 1996), are among the most frequent constitutional rearrangements. Other chromosomes are also known to contain similar “rearrangement-promoting” low copy repeats that are implicated in mediating genomic disorders. Examples of such well-known genetic disorders include Prader-Willi and Angelman syndromes, Williams syndrome, NF1 microdeletions, Sotos syndrome, Smith-Magenis syndrome, and the reciprocal deletions and duplications of Charcot Marie Tooth and HNPP (for reviews, see Emanuel and Shaikh 2001; Shaw and Lupski 2004).
There are a total of eight LCRs within 22q11. The four proximal LCRs have been extensively characterized, given their involvement in recurrent rearrangements of 22q11 that lead to DGS/VCFS (Edelmann et al. 1999; Shaikh et al. 2001) and Cat eye syndrome (CES) (McTaggart et al. 1998). We have previously referred to the four proximal LCRs as LCR-A through LCR-D based on their chromosomal order, with LCR-A being closest to the centromere (Shaikh et al. 2000). These proximal LCRs are larger than the distal ones and have a complex modular structure. LCR-A and LCR-D mediate the common 3 Mb deletion of DGS/VCFS and are the largest and most complex in their organization (Shaikh et al. 2000). This, combined with a high level of sequence identity between the modules (>98%), has thus far prevented the characterization of the common DGS/VCFS deletion breakpoints at the nucleotide level. The four distal LCRs, which we refer to as LCR-E to LCR-H, are smaller with fewer duplicated modules. This cluster of LCRs has rarely been associated with deletions of distal 22q11 (Rauch et al. 1999; Saitta et al. 1999; Ravnan et al. 2006), presumably because of their smaller size and a higher level of sequence variation.
We hypothesized that deletion breakpoints falling within the smaller, less complex distal LCRs would be more amenable to characterization at the nucleotide level. This data could, in turn, help identify the sequences and mechanisms involved in 22q11 LCR-mediated rearrangements. Here, we examined two de novo deletions of 22q11 each flanked by at least one smaller, less complex LCR. One deletion is flanked by LCR-D and LCR-E and confers a phenotype with features of the DGS/VCFS spectrum (Saitta et al. 1999). The second patient does not have the same phenotype and presented with developmental delays. His rearrangement is flanked instead by LCR-E and LCR-F. We used a strategy based on single nucleotide variations in the sequences of each of the eight paralogous chromosome 22q11 LCRs to isolate specific breakpoint junction fragments by long-range PCR. Subsequent sequencing of the cloned breakpoint junctions confirmed that the LCRs provide the basis for the deletion mechanism, mediating nonallelic homologous recombination (NAHR). This is the first time breakpoints of any 22q11 deletions have been characterized at the sequence level. Our finding that the deletion breakpoints in both patients localize to the BCRL module is underscored by its presence in almost all 22q11 LCRs. Since one of the deletions we studied includes a breakpoint within LCR-D that is involved in the common 3 Mb deletion of DGS/VCFS, it suggests the BCRL module as a candidate for other chromosome 22q11 deletion endpoints. Furthermore, these data show that the distal LCRs are also involved in mediating genomic disorders.
Results
The deletions are flanked by distal 22q11 LCRs
For patient CH98-18, we initially identified a de novo deletion of the area adjacent to but not overlapping with the common DGS/VCFS region (Saitta et al. 1999). We further localized the deletion by FISH to an ∼1 Mb region between LCR-D and LCR-E (Fig. 1; Saitta et al. 1999), using a series of cosmids that were taken from a chromosome 22-specific cosmid library (LL22NCO3) (Fig. 1A). The results of one of these FISH experiments using probes c11e6 and cos82 are shown in Figure 1B. Patient CH03-29 had prior clinical testing with a normal karyotype, but subtelomeric FISH testing (Telvysion; Vysis) showed a deletion of the BCR probe that was used as an interstitial control signal for the 22q telomere. He also had normal FISH results for the DGS/VCFS region using both the N25 and TUPLE probes (Vysis). Further FISH studies suggested that the ∼650 kb deletion in CH03-29 was between LCR-E and LCR-F (Fig. 1A). The FISH experiment using probes c102d1 and cos82 is shown in Figure 1C.
Figure 1.
FISH analysis of distal 22q11 deletions. (A) The region of 22q11 containing LCR-A–LCR-H is shown. The LCRs are labeled and shown as black boxes. BAC and cosmid probes used for FISH analysis are shown as thick lines labeled with clone addresses. The probes deleted in CH98-18 are shown in orange and include c45c9, c11e6, and c61e11. CH98-18 was not deleted for probes proximal to LCR-D (N25, c83c5, c87f9, and c2c9), and distal to LCR-E (c102d1, c48a11, c75c12, and BCR). The probes deleted in CH03-29 are shown in blue and include c102d1, c48a11, c75c12, and BCR. CH03-29 was not deleted for probes proximal to LCR-E including c45c9, c11e6, and c61e11 and distal to LCR-F including c27d7 and b80o7. (B,C) Metaphase spreads from deletion patients hybridized with probes labeled with digoxygenin and detected with rhodamine (red signal) or labeled with biotin and detected with FITC-avidin (green). The deleted chromosome 22 is indicated by an arrow, and the normal 22 is indicated by an arrowhead. (B) CH98-18 was probed with c11e6 (red) and control probe cos82 (green). (C) CH03-29 hybridized with c102d1 (red) and control probe cos82 (green). The control cosmid probe, cos82 (D22S39), was used to label the telomeric end of chromosome 22 (22q13.3).
Sequence analysis and characterization of LCR-E and LCR-F
LCR-A through LCR-D in 22q11 have been previously implicated in mediating its deletions. The sequence-based analysis and structural configuration of LCR-D has been described previously (Fig. 2A; Shaikh et al. 2000). We performed a similar analysis of LCR-E and LCR-F to delineate their structure and organization. LCR-E is the smallest and least complex of the eight modular LCRs (A–H) in 22q11, essentially containing only two duplicated modules, one with the BCRL marker and the other with the GGTL marker described previously (Fig. 2B; Shaikh et al. 2000). The duplicated modules in LCR-E are intrachromosomal as the paralogs are found only on chromosome 22. The total size of duplicated sequence within LCR-E is ∼35 kb, corresponding to chromosomal coordinates chr22: 21,287,634–21,322,132 (NCBI build 35, May 2004) and localizes ∼1 Mb distal to the telomeric end of LCR-D.
Figure 2.
Structure and organization of LCR-D, LCR-E, and LCR-F. The spatial arrangement of duplicated modules within LCRs D, E, and F are shown as colored boxes, and the markers within them are indicated above in the same color as the boxes. The orientation of each LCR is centromere to telomere. Arrows below the BCRL-containing modules indicate their orientation with respect to the other LCRs. Unique markers flanking the LCRs are shown in black. The locations of PCR fragments DP1 and BN1 are shown.
LCR-F is larger and more complex, containing both inter- and intrachromosomal modules interspersed with small stretches of unique sequence. It extends over 370 kb, corresponding to chromosomal coordinates chr22: 21,973,629–22,345,857. Proximally, LCR-F begins within the 3′-end of the ∼135 kb BCR gene, whose 5′-regions contain the breakpoints of the Philadelphia chromosome translocations found in chronic myeloid leukemia and acute lymphocytic leukemia (Emanuel et al. 1984; Groffen et al. 1984). An ∼14 kb fragment consisting of the 3′-end of the BCR gene (BCRL) (Fig. 2C, blue box) is found within all of the other 22q11 LCRs, except for LCR-B, which contains a gap in its sequence. LCR-F also has a duplicated module containing the marker NF1L, with paralogs in LCRs A, B, and D (Shaikh et al. 2000; Fig. 2C, green box). There are duplicated modules within LCR-F with paralogs on chromosome 22, but not within the other 22q11 LCRs (Fig. 2C, modules M1–M4). In addition, LCR-F contains modules that are duplicated on chromosomes 7, 9, and 16 (Fig. 2C).
Based on our FISH data, LCR-E appeared to be involved in the rearrangements of both CH98-18 and CH03-29. We took advantage of the simpler structure and smaller size of LCR-E to predict the likely substrates for nonallelic homologous recombination in patient CH98-18 between LCR-E and the highly complex LCR-D. Similarly, we compared the structures of LCR-E and the large, complex LCR-F to predict the likely breakpoint region for the deletion in patient CH03-29.
Cloning and molecular analysis of the deletion breakpoints
Patient CH98-18
Sequence alignments between LCR-D and LCR-E were performed using BLAST (Altschul et al. 1990). The alignments identified an ∼14 kb fragment in LCR-E, designated as BCRL-E (since it contains the BCRL marker), that shared 97% sequence identity with two regions in LCR-D designated as BCRL-D1 and BCRL-D2 (chromosomal coordinates in Supplemental Table S1). LCR-E also contains an ∼10 kb fragment, designated GGTL-E (containing the GGTL marker), which shares 96% sequence identity to a region designated GGTL-D, in LCR-D (chromosomal coordinates in Supplemental Table S1). There are three possible misalignments that could mediate NAHR: BCRL-D1 with BCRL-E, BCRL-D2 with BCRL-E, or GGTL-D with GGTL-E. The greatest sequence identity was shared between the BCRL-containing fragments of LCR-D and LCR-E, thus we predicted that these were more likely to have mediated NAHR resulting in the deletion seen in CH98-18. Furthermore, BCRL-D1 and BCRL-E are in the same orientation, whereas BCRL-D2 and BCRL-E are inverted with respect to each other.
We used a PCR-based strategy and somatic cell hybrids (S. Saitta, unpubl.) that contained either the deleted homolog (hybrid H1) or the normal homolog (hybrid H5) to determine if the proximal breakpoint of the deletion was within BCRL-D1 or in BCRL-D2 (see Supplemental Results). PCR amplimer DP1 (Fig. 2A) was amplified from both hybrids (Supplemental Fig. S1B). Amplimer BN1 (Fig. 2A) was deleted in hybrid H1 and present in hybrid H5 (Supplemental Fig. S1B), localizing the proximal deletion breakpoint to BCRL-D1.
Southern hybridization of digested genomic DNA from CH98-18 was performed with probe DP2, distal to LCR-E (Fig. 3A). DNA digested with NdeI yielded a novel ∼19 kb restriction fragment that was present in the H1 hybrid and in the total genomic DNA from CH98-18, but absent from the H5 hybrid, which contained the normal 22q homolog (Fig. 3B). The expected NdeI band hybridizing to DP2 should be 20,567 bp (Fig. 3A), and was present in the H5 hybrid, the total genomic DNA from CH98-18, and in the normal control, but was absent from the H1 hybrid (Fig. 3B), suggesting that the ∼19 kb NdeI restriction fragment represented a junction fragment resulting from NAHR, most likely between BCRL-D1 and BCRL-E.
Figure 3.
Southern hybridization analysis of CH98-18. (A) Restriction sites for NdeI that generate the expected 20.5 kb band in the LCR-E region are shown. The restriction site in LCR-D that would give rise to the ∼19 kb rearranged fragment is indicated along with the chromosomal coordinates (NCBI build 35). The location of probe DP2 used in Southern hybridization is also shown. (B) Autoradiograph showing the results of Southern hybridization of NdeI-digested genomic DNA using DP2 as a probe. (N) Normal control; (CH98-18) total genomic DNA from patient CH98-18; (H5 and H1) genomic DNA from hybrids H5 and H1, respectively. The normal 20.5 kb band is indicated by a filled arrowhead, and the ∼19 kb rearranged band is indicated by an unfilled arrowhead.
We next designed long-range PCR primers to span across the putative breakpoint junction (details in Supplemental Results). Primers BDfor and BErev were used to amplify genomic DNA (Fig. 4) from CH98-18, the H1 and H5 hybrids, and from cosmid 31f3 (spans LCR-E). Fragment 1 was present only in H1 and in total genomic DNA from CH98-18, while absent in hybrid H5, consistent with a putative junction fragment. Fragments 1 and 2 from H1 and fragment 3 from H5 (Fig. 4A) were gel-purified, cloned, and sequenced. The sequence of fragment 2 was analyzed using BLAST and was shown to match a distal region on 22q11 corresponding to LCR-G, displaying 99.5% sequence identity. The sequence of fragment 3 was shown to be identical to a region within LCR-E, derived from the patient’s normal, intact 22q homolog. BLAST analysis of fragment 1 instead confirmed its sequence as the junction fragment.
Figure 4.
Long-range PCR and sequence analysis of breakpoint junction fragments in patient CH98-18. (A) Results of long-range PCR with primers BDfor and BErev on genomic DNA. (M) 1 kb DNA ladder; (N1) normal control 1; (N2) normal control 2; (NC) no template DNA, negative control; (TG) total genomic DNA from CH98-18; (H1 and H5) genomic DNA from hybrids H1 and H5, respectively. The bands that were gel-isolated from hybrid H1 are labeled “1” and “2,” and the one from H5 is labeled “3.” (B) Sequence alignment of the putative crossover region. Nucleotides 4101–4400 of the junction fragment obtained from CH98-18 were aligned with the corresponding reference sequences from BCRL-D1 and BCRL-E obtained from NCBI (build 35). The sequence variants observed are marked by gray boxes. The nucleotide positions of sequence variants are based on the coordinates of the 6083 bp junction fragment. The 109 bp region between nucleotides 4246 and 4354 is predicted to contain the crossover point (boxed).
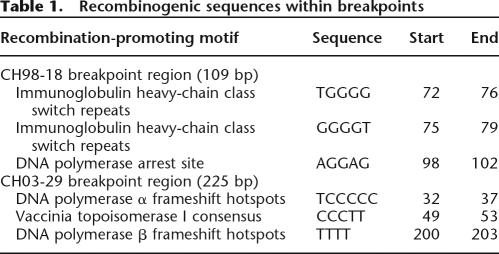
The total size of fragment 1 was 6083 bp of which the first 4354 bp shared the greatest sequence identity with BCRL-D1, while base pairs 4355–6083 shared the greatest sequence identity with nucleotides 43,880–45,609 of BAC clone b1000E4 (GenBank no. AC002308) that encompasses BCRL-E. We selected a 300 bp region (4101–4400) around the putative deletion breakpoints for further analysis, by performing multiple sequence alignments with CLUSTALW (http://www.ebi.ac.uk/clustalw; Higgins et al. 1996). A sequence variant specific to the BCRL-D1 paralog is present at position 4245 of the cloned junction fragment and BCRL-E specific sequence variants at positions 4355 and 4393 (Fig. 4B). Thus, the crossover most likely occurred in a 109 bp interval within BCRL-D1 and BCRL-E corresponding to the region between nucleotides 4246 and 4354. This putative crossover region was further assessed for recombination-promoting motifs (Table 1) using consensus sequences described previously (Badge et al. 2000; Abeysinghe et al. 2003; Visser et al. 2005). The sequence contains a putative DNA polymerase arrest site and two putative immunoglobulin heavy chain class switch repeats within the 109 bp nucleotide breakpoint region.
Table 1.
Recombinogenic sequences within breakpoints
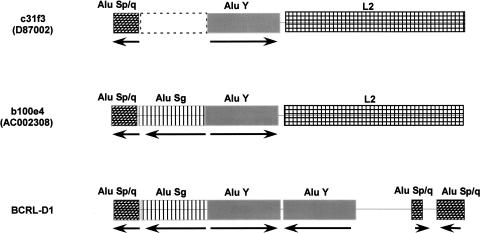
The sequence from 4355 to 6083 bp of the rearranged fragment did not match perfectly with BCRL-E of the reference human genome represented by clone c31f3 (GenBank no. D87002). An apparent genomic polymorphism exists in the sequence of BCRL-E as represented by the two clones c31f3 and b1000e4, respectively. This is an insertion/deletion polymorphism involving an Alu element that led to an extra 323 bp present in b1000e4 (43982–44304 bp of AC002308). The location of the polymorphic Alu would be in chr22: 21,317,317–21,317,318 of the reference human genome. Comparative analysis with the chimpanzee (Pan troglodytes) shows the presence of this 323 bp sequence in clone RP43-41g5 (GenBank no. AC099533), which maps to this region (T.H. Shaikh, unpubl.). Our findings suggest that the ancestral sequence may have contained the 323 bp sequence, which then apparently became deleted in humans. The deletion polymorphism is significant as described below, since its presence may destabilize the region by generating palindromes.
The breakpoint regions in BCRL-D1 and b1000e4 were further analyzed using RepeatMasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker), which demonstrated that the 323 bp polymorphism in b1000e4 was due to the presence of an Alu repeat belonging to the Sg subfamily (Fig. 5). This Alu Sg repeat was present in an inverted orientation with respect to its neighboring Alu Y repeat. Such a configuration of two highly homologous, full-length Alu repeats creates a quasi-palindrome in this region. Using Mfold (Zuker 2003; http://www.bioinfo.rpi.edu/applications/mfold/dna/form1.cgi), we performed a DNA secondary structure analysis of the region containing the Alus found in BCRL-D1 and in b1000e4, whose results suggested that these sequences could form hairpins with very high ΔG values (Supplemental Fig. S2). The palindromic Alus are in close proximity to the putative crossover point: only 168 bp away in BCRL-D1 and 182 bp away in b1000e4.
Figure 5.
Polymorphic Alu in BCRL-E. A graphical representation of the repetitive DNA elements in BCRL-E as represented by two clones: cosmid 31f3 and BAC 1000e4 represented in two different genomic libraries. Hatched and filled boxes represent the various repetitive DNA elements. The Alu element subfamilies are indicated. (L2) LINE 2 element. The orientation of the Alu elements with respect to each other is indicated by arrows. The deleted Alu Sg element in clone 31f3 is indicated by dashed lines. The corresponding region in BCRL-D1 is also shown. The repetitive DNA classification is based on the results from RepeatMasker.
Patient CH03-29
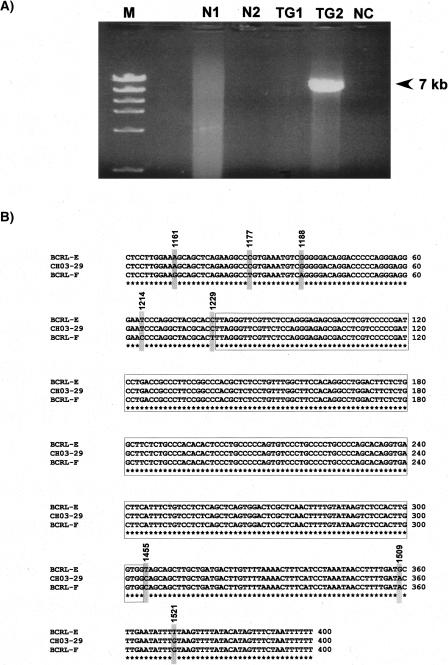
Sequence alignment between LCR-E and LCR-F was performed using BLAST (Altschul et al. 1990), which demonstrated that BCRL-E shared 97% sequence identity with BCRL-F. These two modules are in the same direct orientation to each other, and are the only duplicated module shared between LCR-E and LCR-F. This became the best candidate region to mediate NAHR, resulting in the deletion found in CH03-29. There were no somatic cell hybrids available for this patient, thus we used long-range PCR to directly amplify the junction fragment in CH03-29 using total genomic DNA. We designed PCR primers to amplify a rearranged fragment potentially generated by NAHR between BCRL-E and BCRL-F. Primers BEfor1 and BFrev were designed to generate an ∼15 kb PCR product unique to patient CH03-29 representing the deletion junction fragment (primer details in Supplemental Results). In addition, we designed a nested primer, BEfor2, that would generate an ∼7 kb PCR product when used with primer BFrev representing the junction fragment.
Long-range PCR with BEfor1 and BFrev did not yield a visually detectable PCR fragment, presumably because of the large (15 kb) product size. Instead, BEfor2 and BFrev, when used as PCR primers, yielded the expected ∼7 kb fragment from CH03-29 (Fig. 6A). This 6834 bp PCR product was cloned, sequenced, and determined to be a junction fragment (details in Supplemental Results). Sequence alignments showed that the first 1228 bp of the junction fragment shared greatest sequence identity with BCRL-E and 1229–6834 shared the greatest sequence identity with BCRL-F. In order to refine the analysis and pinpoint the exact crossover point between BCRL-E and BCRL-F, we selected a 400 bp region (1151–1550) around the putative deletion breakpoints for further analysis that suggested the crossover most likely occurred in a 225 bp region within BCRL-E and BCRL-F, corresponding to a region between nucleotides 1230 and 1454 of the junction fragment. The recombination-promoting motifs identified in this region are shown in Table 1. There is one putative DNA polymerase α frameshift hotspot, one putative topoisomerase I consensus sequence, and one potential DNA polymerase β frameshift hotspot within the 225 nucleotide breakpoint region.
Figure 6.
Long-range PCR and sequence analysis of the breakpoint junction fragment in patient CH03-29. (A) Results of long-range PCR with primers BEfor2 and BFrev on 250 ng of genomic DNA. (M) 1 kb DNA ladder; (N1) normal control 1; (N2) normal control 2; (TG1) total genomic DNA from CH98-18; (TG2) total genomic DNA from CH03-29; (NC) no template DNA, negative control. (B) Sequence alignment of the putative crossover region. Nucleotides 1151–1550 of the junction fragment obtained from CH03-29 were aligned with the corresponding reference sequences in BCRL-E and BCRL-F obtained from NCBI (build 35). The sequence variants observed are marked by gray boxes. The nucleotide positions of sequence variants are based on the coordinates of the 6834 bp junction fragment. The 225 bp region predicted to contain the crossover point is boxed.
Discussion
LCRs of chromosome 22q11 have been implicated in the rearrangements associated with DGS/VCFS and CES (Edelmann et al. 1999; Shaikh et al. 2000; McDermid and Morrow 2002). The complexity and large size of the LCRs responsible for these disorders have prevented the precise localization of the deletion breakpoints within the proximal 22q11 LCRs. Our strategy to analyze deletions of 22q11 mediated by the distal, lower complexity LCRs enabled us to successfully identify breakpoints within 22q11 LCRs. The deletion breakpoints we analyzed are each located within a duplicated module containing the BCRL marker, suggesting that the sequences within this module may predispose the region to rearrangement. Interestingly, the region corresponding to the BCRL module within the 22q11 LCRs was previously predicted to be a potential rearrangement hotspot within LCR-A (LCR22-2) and LCR-D (LCR22-4) (Pavlicek et al. 2005). This interval, designated ΨBCR, is enriched in shared polymorphic sites (SPSs) and poor in paralogous sequence variants (PSVs), suggesting it as a region for gene conversion and consequently for meiotic crossover and rearrangement (Hurles et al. 2004; Pavlicek et al. 2005). This congruence between the predicted region and our reported breakpoint regions is highly suggestive.
The BCRL module is the only duplicated module common to almost all (7/8) 22q11 LCRs in the reference human genome sequence (Fig. 7). The relative orientation of the BCRL modules may further predict which LCRs will mediate NAHR, since it has been suggested that LCRs in a direct orientation mediate deletions and duplications (Shaffer and Lupski 2000). Predicted substrates for 22q11 LCR-mediated NAHR would include BCRL-A, BCRL-C, BCRL-D2, and BCRL-G in one group (Fig. 7) and BCRL-D1, BCRL-E, BCRL-F, and BCRL-H in the other group (Fig. 7). The deletions in CH98-18 and CH03-29, which are mediated by BCRL-D1 and BCRL-E and BCRL-E and BCRL-F, respectively, appear to correlate well with the predicted NAHR substrates. Our data suggest that the BCRL module should be considered a strong candidate for the crossover region in the more common rearrangements associated with DGS/VCFS and CES that are mediated by the larger, more complex proximal LCRs, including LCR-D. Previous breakpoint analysis in other genomic disorders has suggested the presence of preferred crossover regions or rearrangement “hotspots” within larger LCRs. These include regions within the CMT1A-REPs in chromosome 17p11.2–p12 (Reiter et al. 1998); the NF1-REPs in chromosome 17q11 (Lopez-Correa et al. 2001); the SMS-REPs in chromosome 17p11.2 (Bi et al. 2003); the 7q11.23 LCRs (Bayes et al. 2003); and the SoS-REPs in chromosome 5q35 (Visser et al. 2005).
Figure 7.
BCRL modules within the 22q11 LCRs. The region of 22q11 containing LCR-A–LCR-H is shown. The LCRs are labeled and shown as white boxes. The BCRL modules within each LCR are shown as gray boxes. The gap within the sequence of LCR-B is indicated. The “?” denotes the unknown status of the presence or absence of a BCRL module within LCR-B. The two copies of BCRL in LCR-D are denoted as “1” and “2.” The relative orientation of the BCRL modules with respect to each other is indicated with directional arrows.
We narrowed down the putative crossover regions in CH98-18 and CH03-29 to 109 bp and 225 bp, respectively. The breakpoint sequences were tested further to determine if the genomic architecture or repetitive DNA content within each BCRL module predisposed the region to rearrangement. Alu-mediated recombination has been shown to cause deletions that lead to many human diseases (Deininger and Batzer 1999). We found no repetitive elements including Alus within the putative crossover regions in CH98-18 and CH03-29. However, while Alu repeats are not directly involved at the deletion breakpoints, analysis of the junction fragment in CH98-18 suggests that an apparent Alu-based insertion/deletion polymorphism may play a role in creating genomic instability. We identified the Alu polymorphism within the sequence of BCRL-E, based on sequences of clones obtained from different genomic libraries. The presence of such a polymorphism in close proximity to a deletion breakpoint is intriguing, but more importantly, the polymorphism leads to the juxtaposition of two highly homologous Alus in an inverted orientation with respect to each other. Inverted Alu elements are rare within the human genome (Stenger et al. 2001) but are known to be hotspots for genomic instability (Gebow et al. 2000; Lobachev et al. 2000; Stenger et al. 2001). This Alu polymorphism may represent a genomic variant that predisposes BCRL-E to genomic instability resulting in deletions.
Computational analysis with Mfold predicted the formation of cruciform structures resulting from the inverted Alus. Palindrome-mediated cruciforms have been shown to be involved in several constitutional translocation breakpoints (Kurahashi et al. 2000; Gotter et al. 2004). Based on similar observations at deletion breakpoints, it has been suggested that sequences that can form non-B DNA structures, including cruciforms, may be predisposed to instability and breakage (Bacolla et al. 2004). In our patient, the palindromic Alus are in close proximity to the putative crossover point: only 168 bp away in BCRL-D1 and 182 bp away in BCRL-E. Thus, genomic architecture that leads to the formation of unstable DNA configuration may play a role in creating instability near the endpoints of deletions and other chromosomal rearrangements.
The deletion breakpoint sequences were also tested for the presence of recombination promoting motifs that have been previously identified at or near deletion and translocation breakpoints (Badge et al. 2000; Abeysinghe et al. 2003). We identified a putative DNA polymerase arrest site within the breakpoint sequence of CH98-18. Polymerase arrest sites have been shown to trigger both homologous and nonhomologous recombination leading to DNA rearrangement (Stary and Sarasin 1992; Hyrien 2000; Michel 2000). In CH03-29, we identified a putative topoisomerase I consensus sequence in the 225 nucleotide breakpoint sequence. Topoisomerase I consensus cleavage sites have been previously reported in the vicinity of deletion breakpoints (Cao et al. 2001). The presence of these sequences within the breakpoint region does not definitively predict a role in the rearrangement mechanism since none of these consensus motifs have been verified experimentally to directly cause rearrangements; however, many deletion hotspots are enriched for these motifs (Shaw et al. 2004; Visser et al. 2005). It is clear that understanding the mechanism underlying the deletions mediated by the 22q11 LCRs will require further work to determine not only the role of the nucleotide sequences involved, but also of the biochemical factors that recognize and bind these sequences to mediate recombination.
Our results provide confirmation that distal 22q11 LCRs (LCR-E–LCR-H) can and do mediate rearrangements leading to genomic disorders. More patients may exist who harbor microdeletions or microduplications in 22q11 mediated by the distal LCRs that are undetected by current techniques used in clinical testing. Recently, microarray-based CGH has been used to detect copy number alterations in 22q11 and across the entire genome (Mantripragada et al. 2004; Ming et al. 2006; Urban et al. 2006), which might have identified the rearrangements in both CH98-18 as well as in CH03-29. These array-based methods would allow more sensitive and rapid breakpoint localization without the need for multiple FISH experiments as was done previously (Saitta et al. 1999; Shaikh et al. 2000). This has recently been highlighted by the use of CGH arrays for the identification of a new microdeletion syndrome mediated by the LCRs of chromosome 17q21.3 (Koolen et al. 2006; Sharp et al. 2006; Shaw-Smith et al. 2006). The application of these techniques will allow identification of other distal deletions and also facilitate breakpoint identification in the proximal 22q11.2 deletions as well. Our ability to localize, clone, and sequence breakpoints within the LCRs of chromosome 22q has also allowed the identification of structural variation within the genome that may predispose a given chromosome to NAHR. This not only has implications for the deletion mechanisms of chromosome 22q in particular, but also provides important information on the role of genomic architecture in rearrangements, chromosome evolution, and in human disease.
Methods
Patient samples
Clinical histories and family data were obtained from medical records. Each patient was evaluated by a clinical geneticist, and each had a normal karyotype and normal chromosome 22q11.2 FISH using commercial DiGeorge syndrome probes. Patient CH98-18 has been reported previously (Saitta et al. 1999) and had microcephaly, hypertelorism, cardiac defect, and bifid uvula. Patient CH03-29 displayed developmental delay, speech apraxia, short stature, hypertelorism, down-turned mouth, and pectus excavatum.
PCR
Unless otherwise indicated, all PCR reagents were part of the GeneAmp PCR system, purchased from Applied Biosystems. Standard PCR reactions were carried out in 25 μL volumes using final concentrations of dNTPs at 200 μM and primers at 200 μM. Amplitaq (Applied Biosystems) Taq polymerase was used at 2.5 U per reaction. All primers designed for PCR and/or DNA sequencing were purchased from Integrated DNA Technologies.
Southern blots
Using standard protocols (Sambrook and Russell 2001), high-molecular-weight DNA was extracted from each patient’s peripheral blood lymphocytes, normal controls, and from the H1 and H5 hybrids. The DNA was digested with several restriction enzymes based on an in silico digest of the LCR-D and LCR-E regions using WebCutter software (http://www.firstmarket.com/cutter/cut2.html). For analysis of CH98-18, 10 μg of each sample was digested overnight with NdeI according to the manufacturer’s protocol and supplied buffers (New England Biolabs). The digested DNA was electrophoresed in an 0.8% agarose gel using a pulsed field apparatus (FIGE Mapper; Bio-Rad). The gel was run for 22 h with switch times of 0.1–0.4 sec for a separation range between 1 and 25 kb. The gel was transferred, and the blot hybridized with a 32P-labeled probe prepared using a random priming method (Amersham). The bands were visualized by autoradiography. The probe used in the blots was DP2, a 1047 bp PCR product generated using DNA from cosmid 31f3 as a template. PCR included 40 cycles of denaturation at 94°C for 30 sec, annealing at 58°C for 30 sec, and extension at 72°C for 1 min. Approximately 100 ng of the amplicon was used for radiolabeling.
DP2for, 5′-ATGGTTTGCTGACCCCAGCT-3′
DP2rev, 5′-AGACATCGTCTCTCTGCTCA-3′
Long-range PCR
Long-range PCR amplifications were performed using reagents and guidelines included in the Expand PCR System (Roche). Primers for the fragments were obtained from Integrated DNA Technologies.
Patient CH 98-18
BDfor, 5′-CCCTGTTGCATCTCTCTTACTAGGAGCAAG-3′
BErev, 5′-AACTCTGAATGTCTGTCTGCCTGGTCCTCA-3′
The amplification profile included 2 min of initial denaturation at 94°C, followed by 10 cycles of 94°C for 10 sec, 64°C for 30 sec, and 68°C for 7 min. A subsequent 18 cycles of the same segments was performed followed by a 7 min extension at 68°C. The PCR products were electrophoresed on a 1% agarose gel in 0.5× TBE using a pulsed-field apparatus (CHEF Mapper; Bio-Rad).
Patient CH 03-29
BEfor1, 5′-GTACCTCTCTGCCTCCTATGCCTTTAAGCA-3′
BEfor2, 5′-CAGGTGTTTGAGATCAGTCTGGGCAATGCA-3′
BFrev, 5′-GTGGGTCGTGCTTGTAATCCTAGCACATTG-3′
The amplification profile for the BEfor2 and BFrev primers included 2 min of initial denaturation at 94°C, followed by 10 cycles of 94°C for 10 sec, 66°C for 30 sec, and 68°C for 7 min. A subsequent 18 cycles of the same segments was performed followed by a 7 min extension at 68°C. The PCR products were electrophoresed on a 1% agarose gel in 1× TBE using a standard gel electrophoresis apparatus.
Acknowledgments
This work was supported by the NIH, K08 HL04487 (S.C.S.), the Physician Scientist award program, P30 HD28815 (S.C.S.), and by R01 GM64725 (T.H.S.).
Footnotes
[Supplemental material is available online at www.genome.org. The sequence data have been submitted to GenBank under accession nos. EF025176–EF025177.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.5986507
References
- Abeysinghe S.S., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N., Chuzhanova N., Krawczak M., Ball E.V., Cooper D.N., Krawczak M., Ball E.V., Cooper D.N., Ball E.V., Cooper D.N., Cooper D.N. Translocation and gross deletion breakpoints in human inherited disease and cancer I: Nucleotide composition and recombination-associated motifs. Hum. Mutat. 2003;22:229–244. doi: 10.1002/humu.10254. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J., Gish W., Miller W., Myers E.W., Lipman D.J., Miller W., Myers E.W., Lipman D.J., Myers E.W., Lipman D.J., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D., O’Connell C.D., Cooper D.N., Wells R.D., Cooper D.N., Wells R.D., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badge R.M., Yardley J., Jeffreys A.J., Armour J.A., Yardley J., Jeffreys A.J., Armour J.A., Jeffreys A.J., Armour J.A., Armour J.A. Crossover breakpoint mapping identifies a subtelomeric hotspot for male meiotic recombination. Hum. Mol. Genet. 2000;9:1239–1244. doi: 10.1093/hmg/9.8.1239. [DOI] [PubMed] [Google Scholar]
- Bayes M., Magano L.F., Rivera N., Flores R., Perez Jurado L.A., Magano L.F., Rivera N., Flores R., Perez Jurado L.A., Rivera N., Flores R., Perez Jurado L.A., Flores R., Perez Jurado L.A., Perez Jurado L.A. Mutational mechanisms of Williams-Beuren syndrome deletions. Am. J. Hum. Genet. 2003;73:131–151. doi: 10.1086/376565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W., Park S.S., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R., Park S.S., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R., Shaw C.J., Withers M.A., Patel P.I., Lupski J.R., Withers M.A., Patel P.I., Lupski J.R., Patel P.I., Lupski J.R., Lupski J.R. Reciprocal crossovers and a positional preference for strand exchange in recombination events resulting in deletion or duplication of chromosome 17p11.2. Am. J. Hum. Genet. 2003;73:1302–1315. doi: 10.1086/379979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burn J., Goodship J., Goodship J. Developmental genetics of the heart. Curr. Opin. Genet. Dev. 1996;6:322–325. doi: 10.1016/s0959-437x(96)80009-8. [DOI] [PubMed] [Google Scholar]
- Cao X., Eu K.W., Seow-Choen F., Zhao Y., Cheah P.Y., Eu K.W., Seow-Choen F., Zhao Y., Cheah P.Y., Seow-Choen F., Zhao Y., Cheah P.Y., Zhao Y., Cheah P.Y., Cheah P.Y. Topoisomerase-I- and Alu-mediated genomic deletions of the APC gene in familial adenomatous polyposis. Hum. Genet. 2001;108:436–442. doi: 10.1007/s004390100492. [DOI] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A., Batzer M.A. Alu repeats and human disease. Mol. Genet. Metab. 1999;67:183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Edelmann L., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Pandita R.K., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Spiteri E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Funke B., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Goldberg R., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Palanisamy N., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Chaganti R.S., Magenis E., Shprintzen R.J., Morrow B.E., Magenis E., Shprintzen R.J., Morrow B.E., Shprintzen R.J., Morrow B.E., Morrow B.E. A common molecular basis for rearrangement disorders on chromosome 22q11. Hum. Mol. Genet. 1999;8:1157–1167. doi: 10.1093/hmg/8.7.1157. [DOI] [PubMed] [Google Scholar]
- Emanuel B.S., Shaikh T.H., Shaikh T.H. Segmental duplications: An ‘expanding’ role in genomic instability and disease. Nat. Rev. Genet. 2001;2:791–800. doi: 10.1038/35093500. [DOI] [PubMed] [Google Scholar]
- Emanuel B.S., Selden J.R., Wang E., Nowell P.C., Croce C.M., Selden J.R., Wang E., Nowell P.C., Croce C.M., Wang E., Nowell P.C., Croce C.M., Nowell P.C., Croce C.M., Croce C.M. In situ hybridization and translocation breakpoint mapping. I. Nonidentical 22q11 breakpoints for the t(9;22) of CML and the t(8;22) of Burkitt lymphoma. Cytogenet. Cell Genet. 1984;38:127–131. doi: 10.1159/000132044. [DOI] [PubMed] [Google Scholar]
- Gebow D., Miselis N., Liber H.L., Miselis N., Liber H.L., Liber H.L. Homologous and nonhomologous recombination resulting in deletion: Effects of p53 status, microhomology, and repetitive DNA length and orientation. Mol. Cell. Biol. 2000;20:4028–4035. doi: 10.1128/mcb.20.11.4028-4035.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S., Budarf M.L., Rhodes C.H., Emanuel B.S., Rhodes C.H., Emanuel B.S., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groffen J., Stephenson J.R., Heisterkamp N., de Klein A., Bartram C.R., Grosveld G., Stephenson J.R., Heisterkamp N., de Klein A., Bartram C.R., Grosveld G., Heisterkamp N., de Klein A., Bartram C.R., Grosveld G., de Klein A., Bartram C.R., Grosveld G., Bartram C.R., Grosveld G., Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. 1984;36:93–99. doi: 10.1016/0092-8674(84)90077-1. [DOI] [PubMed] [Google Scholar]
- Higgins D.G., Thompson J.D., Gibson T.J., Thompson J.D., Gibson T.J., Gibson T.J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- Hurles M.E., Willey D., Matthews L., Hussain S.S., Willey D., Matthews L., Hussain S.S., Matthews L., Hussain S.S., Hussain S.S. Origins of chromosomal rearrangement hotspots in the human genome: Evidence from the AZFa deletion hotspots. Genome Biol. 2004;5:R55. doi: 10.1186/gb-2004-5-8-r55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- Koolen D.A., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Vissers L.E., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Pfundt R., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., de Leeuw N., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Knight S.J., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Regan R., Kooy R.F., Reyniers E., Romano C., Fichera M., Kooy R.F., Reyniers E., Romano C., Fichera M., Reyniers E., Romano C., Fichera M., Romano C., Fichera M., Fichera M., et al. A new chromosome 17q21.31 microdeletion syndrome associated with a common inversion polymorphism. Nat. Genet. 2006;38:999–1001. doi: 10.1038/ng1853. [DOI] [PubMed] [Google Scholar]
- Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Hu P., Roe B.A., Emanuel B.S., Budarf M.L., Roe B.A., Emanuel B.S., Budarf M.L., Emanuel B.S., Budarf M.L., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9:1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- Lobachev K.S., Stenger J.E., Kozyreva O.G., Jurka J., Gordenin D.A., Resnick M.A., Stenger J.E., Kozyreva O.G., Jurka J., Gordenin D.A., Resnick M.A., Kozyreva O.G., Jurka J., Gordenin D.A., Resnick M.A., Jurka J., Gordenin D.A., Resnick M.A., Gordenin D.A., Resnick M.A., Resnick M.A. Inverted Alu repeats unstable in yeast are excluded from the human genome. EMBO J. 2000;19:3822–3830. doi: 10.1093/emboj/19.14.3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Correa C., Dorschner M., Brems H., Lazaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Dorschner M., Brems H., Lazaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Brems H., Lazaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Lazaro C., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Clementi M., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Upadhyaya M., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Dooijes D., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Moog U., Kehrer-Sawatzki H., Rutkowski J.L., Kehrer-Sawatzki H., Rutkowski J.L., Rutkowski J.L., et al. Recombination hotspot in NF1 microdeletion patients. Hum. Mol. Genet. 2001;10:1387–1392. doi: 10.1093/hmg/10.13.1387. [DOI] [PubMed] [Google Scholar]
- Mantripragada K.K., Tapia-Paez I., Blennow E., Nilsson P., Wedell A., Dumanski J.P., Tapia-Paez I., Blennow E., Nilsson P., Wedell A., Dumanski J.P., Blennow E., Nilsson P., Wedell A., Dumanski J.P., Nilsson P., Wedell A., Dumanski J.P., Wedell A., Dumanski J.P., Dumanski J.P. DNA copy-number analysis of the 22q11 deletion-syndrome region using array-CGH with genomic and PCR-based targets. Int. J. Mol. Med. 2004;13:273–279. [PubMed] [Google Scholar]
- McDermid H.E., Morrow B.E., Morrow B.E. Genomic disorders on 22q11. Am. J. Hum. Genet. 2002;70:1077–1088. doi: 10.1086/340363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTaggart K.E., Budarf M.L., Driscoll D.A., Emanuel B.S., Ferreira P., McDermid H.E., Budarf M.L., Driscoll D.A., Emanuel B.S., Ferreira P., McDermid H.E., Driscoll D.A., Emanuel B.S., Ferreira P., McDermid H.E., Emanuel B.S., Ferreira P., McDermid H.E., Ferreira P., McDermid H.E., McDermid H.E. Cat eye syndrome chromosome breakpoint clustering: Identification of two intervals also associated with 22q11 deletion syndrome breakpoints. Cytogenet. Cell Genet. 1998;81:222–228. doi: 10.1159/000015035. [DOI] [PubMed] [Google Scholar]
- Michel B. Replication fork arrest and DNA recombination. Trends Biochem. Sci. 2000;25:173–178. doi: 10.1016/s0968-0004(00)01560-7. [DOI] [PubMed] [Google Scholar]
- Ming J.E., Geiger E., James A.C., Ciprero K.L., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., Geiger E., James A.C., Ciprero K.L., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., James A.C., Ciprero K.L., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., Ciprero K.L., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., Nimmakayalu M., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., Zhang Y., Huang A., Vaddi M., Rappaport E., Zackai E.H., Huang A., Vaddi M., Rappaport E., Zackai E.H., Vaddi M., Rappaport E., Zackai E.H., Rappaport E., Zackai E.H., Zackai E.H., et al. Rapid detection of sub-microscopic chromosomal rearrangements in children with multiple congenital anomalies using high density oligonucleotide arrays. Hum. Mutat. 2006;27:467–473. doi: 10.1002/humu.20322. [DOI] [PubMed] [Google Scholar]
- Pavlicek A., House R., Gentles A.J., Jurka J., Morrow B.E., House R., Gentles A.J., Jurka J., Morrow B.E., Gentles A.J., Jurka J., Morrow B.E., Jurka J., Morrow B.E., Morrow B.E. Traffic of genetic information between segmental duplications flanking the typical 22q11.2 deletion in velo-cardio-facial syndrome/DiGeorge syndrome. Genome Res. 2005;15:1487–1495. doi: 10.1101/gr.4281205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Pfeiffer R.A., Leipold G., Singer H., Tigges M., Hofbeck M., Pfeiffer R.A., Leipold G., Singer H., Tigges M., Hofbeck M., Leipold G., Singer H., Tigges M., Hofbeck M., Singer H., Tigges M., Hofbeck M., Tigges M., Hofbeck M., Hofbeck M. A novel 22q11.2 microdeletion in DiGeorge syndrome. Am. J. Hum. Genet. 1999;64:659–666. doi: 10.1086/302235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravnan J.B., Tepperberg J.H., Papenhausen P., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L., Tepperberg J.H., Papenhausen P., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L., Papenhausen P., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L., Lamb A.N., Hedrick J., Eash D., Ledbetter D.H., Martin C.L., Hedrick J., Eash D., Ledbetter D.H., Martin C.L., Eash D., Ledbetter D.H., Martin C.L., Ledbetter D.H., Martin C.L., Martin C.L. Subtelomere FISH analysis of 11,688 cases: An evaluation of the frequency and pattern of subtelomere rearrangements in individuals with developmental disabilities. J. Med. Genet. 2006;43:478–489. doi: 10.1136/jmg.2005.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter L.T., Hastings P.J., Nelis E., De Jonghe P., Van Broeckhoven C., Lupski J.R., Hastings P.J., Nelis E., De Jonghe P., Van Broeckhoven C., Lupski J.R., Nelis E., De Jonghe P., Van Broeckhoven C., Lupski J.R., De Jonghe P., Van Broeckhoven C., Lupski J.R., Van Broeckhoven C., Lupski J.R., Lupski J.R. Human meiotic recombination products revealed by sequencing a hotspot for homologous strand exchange in multiple HNPP deletion patients. Am. J. Hum. Genet. 1998;62:1023–1033. doi: 10.1086/301827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta S.C., McGrath J.M., Mensch H., Shaikh T.H., Zackai E.H., Emanuel B.S., McGrath J.M., Mensch H., Shaikh T.H., Zackai E.H., Emanuel B.S., Mensch H., Shaikh T.H., Zackai E.H., Emanuel B.S., Shaikh T.H., Zackai E.H., Emanuel B.S., Zackai E.H., Emanuel B.S., Emanuel B.S. A 22q11.2 deletion that excludes UFD1L and CDC45L in a patient with conotruncal and craniofacial defects. Am. J. Hum. Genet. 1999;65:562–566. doi: 10.1086/302514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitta S.C., Harris S.E., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Harris S.E., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Gaeth A.P., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Driscoll D.A., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., McDonald-McGinn D.M., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Maisenbacher M.K., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Yersak J.M., Chakraborty P.K., Hacker A.M., Zackai E.H., Chakraborty P.K., Hacker A.M., Zackai E.H., Hacker A.M., Zackai E.H., Zackai E.H., et al. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum. Mol. Genet. 2004;13:417–428. doi: 10.1093/hmg/ddh041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D.W., Russell D.W. Molecular cloning: A laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2001. [Google Scholar]
- Shaffer L.G., Lupski J.R., Lupski J.R. Molecular mechanisms for constitutional chromosomal rearrangements in humans. Annu. Rev. Genet. 2000;34:297–329. doi: 10.1146/annurev.genet.34.1.297. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Kurahashi H., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Saitta S.C., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., O’Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., Zackai E.H., Budarf M.L., Budarf M.L., et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: Genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- Shaikh T.H., Kurahashi H., Emanuel B.S., Kurahashi H., Emanuel B.S., Emanuel B.S. Evolutionarily conserved low copy repeats (LCRs) in 22q11 mediate deletions, duplications, translocations, and genomic instability: An update and literature review. Genet. Med. 2001;3:6–13. doi: 10.1097/00125817-200101000-00003. [DOI] [PubMed] [Google Scholar]
- Sharp A.J., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hansen S., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Selzer R.R., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Cheng Z., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Regan R., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Hurst J.A., Stewart H., Price S.M., Blair E., Hennekam R.C., Stewart H., Price S.M., Blair E., Hennekam R.C., Price S.M., Blair E., Hennekam R.C., Blair E., Hennekam R.C., Hennekam R.C., et al. Discovery of previously unidentified genomic disorders from the duplication architecture of the human genome. Nat. Genet. 2006;38:1038–1042. doi: 10.1038/ng1862. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Lupski J.R., Lupski J.R. Implications of human genome architecture for rearrangement-based disorders: The genomic basis of disease. Hum. Mol. Genet. 2004;13:R57–R64. doi: 10.1093/hmg/ddh073. [DOI] [PubMed] [Google Scholar]
- Shaw C.J., Withers M.A., Lupski J.R., Withers M.A., Lupski J.R., Lupski J.R. Uncommon deletions of the Smith-Magenis syndrome region can be recurrent when alternate low-copy repeats act as homologous recombination substrates. Am. J. Hum. Genet. 2004;75:75–81. doi: 10.1086/422016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Pittman A.M., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Willatt L., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Martin H., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Rickman L., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Gribble S., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Curley R., Cumming S., Dunn C., Kalaitzopoulos D., Cumming S., Dunn C., Kalaitzopoulos D., Dunn C., Kalaitzopoulos D., Kalaitzopoulos D., et al. Microdeletion encompassing MAPT at chromosome 17q21.3 is associated with developmental delay and learning disability. Nat. Genet. 2006;38:1032–1037. doi: 10.1038/ng1858. [DOI] [PubMed] [Google Scholar]
- Stary A., Sarasin A., Sarasin A. Molecular analysis of DNA junctions produced by illegitimate recombination in human cells. Nucleic Acids Res. 1992;20:4269–4274. doi: 10.1093/nar/20.16.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger J.E., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Lobachev K.S., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Gordenin D., Darden T.A., Jurka J., Resnick M.A., Darden T.A., Jurka J., Resnick M.A., Jurka J., Resnick M.A., Resnick M.A. Biased distribution of inverted and direct Alus in the human genome: Implications for insertion, exclusion, and genome stability. Genome Res. 2001;11:12–27. doi: 10.1101/gr.158801. [DOI] [PubMed] [Google Scholar]
- Urban A.E., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Korbel J.O., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Selzer R., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Richmond T., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Hacker A., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Popescu G.V., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Cubells J.F., Green R., Emanuel B.S., Gerstein M.B., Green R., Emanuel B.S., Gerstein M.B., Emanuel B.S., Gerstein M.B., Gerstein M.B., et al. High-resolution mapping of DNA copy alterations in human chromosome 22 using high-density tiling oligonucleotide arrays. Proc. Natl. Acad. Sci. 2006;103:4534–4539. doi: 10.1073/pnas.0511340103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser R., Shimokawa O., Harada N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N., Shimokawa O., Harada N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N., Harada N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N., Kinoshita A., Ohta T., Niikawa N., Matsumoto N., Ohta T., Niikawa N., Matsumoto N., Niikawa N., Matsumoto N., Matsumoto N. Identification of a 3.0-kb major recombination hotspot in patients with Sotos syndrome who carry a common 1.9-Mb microdeletion. Am. J. Hum. Genet. 2005;76:52–67. doi: 10.1086/426950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]