Abstract
The prediction of gene models from genome sequence remains an unsolved problem. One hallmark of eukaryotic gene structure is the presence of introns, which are spliced out of pre-mRNAs prior to translation. The excised introns are released in the form of lariats, which must be debranched prior to their turnover. In the yeast Saccharomyces cerevisiae, the absence of the debranching enzyme causes these lariat RNAs to accumulate. This accumulation allows a comparison of tiling array signals of RNA from the debranching mutant to the wild-type parent strain, and thus the identification of lariats on a genome-wide scale. This approach identified 141 of 272 known introns, confirmed three previously predicted introns, predicted four novel introns (of which two were experimentally confirmed), and led to the reannotation of four others. In many instances, signals from the tiling array delineated the 5′ splice site and branchpoint site, confirming predicted gene structures. Nearly all introns that went undetected are present in mRNAs expressed at low levels. Overall, 97% of the significant probes could be attributed either to spliced introns or to genes up-regulated by deletion of the debranching enzyme. Because the debranching enzyme is conserved among eukaryotes, this approach could be generally applicable for the annotation of eukaryotic genes and the detection of novel and alternative splice forms.
Gene annotation remains a formidable challenge following the completion of a whole genome sequence. Annotation typically relies on available expressed sequence tags (ESTs) or other cDNA sequences, alignment to protein sequences, comparative analysis of genomes, or de novo prediction programs that use statistical models to detect codons and conserved motifs for transcription initiation, polyadenylation, and splicing (Brent 2005). Given the low gene density of many genomes and the low information content of sequences specifying gene boundaries, exon–intron junctions, and branchpoint sequences, the accuracy of gene prediction remains to be improved, especially for organisms with limited EST information. ESTs themselves suffer from 3′ bias, and often cannot distinguish between splice variants. In contrast to most eukaryotic genomes, the set of genes for the yeast Saccharomyces cerevisiae is extremely well-characterized; gene density is high, introns occur in only ∼5% of the genes and possess a near invariant UACUAAC sequence at the branchpoint, and alternative splicing and pseudogenes are not major concerns (Spingola et al. 1999). Nonetheless, even for S. cerevisiae, >10% of the ORFs were revised and several additional introns were predicted with the sequencing of related yeast species (Cliften et al. 2003; Kellis et al. 2003).
Using S. cerevisiae as a model system, we sought to develop a tiling array-based method for the genome-wide detection of pre-mRNA introns. Tiling arrays that contain overlapping oligonucleotide probes covering millions of bases have been used to measure chromosomal copy number changes (Wilson et al. 2006) and gene expression (Halasz et al. 2006; Manak et al. 2006), to identify transcript boundaries (David et al. 2006), to determine chromatin modification and accessibility (Sabo et al. 2006; Schumacher et al. 2006; Sinha et al. 2006), and to identify sequence polymorphisms (Gresham et al. 2006) for entire genomes. The S. cerevisiae tiling array from Affymetrix has ∼2.6 million 25-mer probes spaced at an average of 5 nucleotides (nt) and covering >95% of the genome.
The basis of our approach is the detection of tiling array signals corresponding to introns, due to the accumulation of a splicing intermediate in the appropriate yeast mutant. Following transcription, primary pre-mRNA transcripts are processed by the spliceosome to remove introns, which are released as lariats. Lariat RNAs must be subsequently debranched prior to their turnover by cellular exonucleases (Fig. 1A). In wild-type cells, the half-life of lariats is short; however, in yeast cells that lack the debranching RNA endonuclease Dbr1, whose activity initiates lariat degradation, lariat RNAs accumulate to high levels (Chapman and Boeke 1991) (Fig. 1B). This accumulation can allow the detection of lariat hybridization signals on an array to globally report the location of introns.
Results
In order to identify spliced introns via the detection of accumulated lariats, we isolated total RNA from diploid dbr1/dbr1 and DBR+/DBR+ yeast strains, labeled it as double-stranded cDNA, and hybridized the cDNA to S. cerevisiae tiling arrays. Signals enriched in the dbr1 strain were identified and mapped to genomic coordinates. We integrated current intron annotations, the presence of splice signals, and dbr1-specific hybridization patterns to assess the ability of the array to identify intronic regions and novel RNA splice forms. In S. cerevisiae, the great majority of introns are readily identifiable by the presence of conserved splicing signals at the 5′ and 3′ boundaries and branchpoint (Lim and Burge 2001). In order to facilitate our analysis, we compiled a reference list of 272 introns in S. cerevisiae (Supplemental Table 1) from published sources (Grate and Ares 2002; Christie et al. 2004).
We identified probes significantly enriched in the dbr1 sample using a window-based statistical test. Because we performed multiple, simultaneous statistical tests, we evaluated the array data using the false discovery rate (FDR), which measures the expected proportion of false positives in a set of predictions (Storey and Tibshirani 2003). We calculated the proportion of probes that fell within annotated intron regions at different FDR thresholds. At an estimated FDR of 10% (P < 5.8 × 10−4, Wilcoxon rank-sum test), the proportion of significant probes found within annotated intronic regions is 90.9% (8953 of 9851 probes). At an estimated FDR of 5% (P < 2.8 × 10−4), the proportion of probes in intronic regions increases to 92.1% (8829 of 9586 probes), and we miss probes in two annotated introns that were detected at an FDR of 10%. Finally, at an estimated FDR of 1% (P < 1 × 10−6), the proportion of probes in intronic regions increases to 95.2% (7903 of 8304 probes), and we miss probes in 10 annotated introns that were detected at an FDR of 10%. The difference between the estimated FDR and the observed proportion of probes in annotated introns could be accounted for by probes that fall within novel spliced RNAs. Because we were interested in identifying novel introns, we used an estimated FDR of 10% in subsequent analyses.
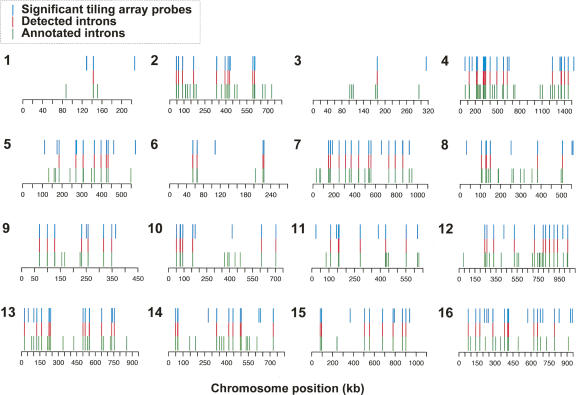
We examined the overall correspondence of dbr1-enriched signals with our annotated intronic regions (Fig. 2). At an estimated FDR of 10%, 141 of 272 annotated introns contained significant probes in the dbr1 sample. Significant tiling array signals were detected for all but three of 105 introns in ribosomal protein genes, including 89 in coding regions and 13 in 5′ untranslated regions (UTRs). Of 158 introns found in the coding regions of nonribosomal genes, 32 were detected by the tiling array, and of seven introns found in the 5′ UTRs of nonribosomal genes, five were detected on the tiling array. In addition, the two introns in snoRNAs were detected.
Figure 2.
Genome-wide view of correspondence between annotated introns in S. cerevisiae and significant regions identified using a tiling microarray. Significant probes identified at an estimated FDR of 10% on the tiling array (blue bars) and annotated introns (green) are plotted on the 16 nuclear chromosomes of S. cerevisiae. Introns in which significant probes were observed (red) are aligned in the middle. Of 272 annotated introns, 141 are identified using the tiling microarray.
In order to assess the specificity and sensitivity of the approach for identifying annotated introns, we also used a receiver–operator characteristic (ROC) plot. In this analysis, we labeled intronic regions “positive,” and all other genomic regions were labeled “negative.” With these designations, ∼13,000 probes map to 67 kb in the positive regions (introns), and 2.4 million probes map to 12 Mb in the negative regions (rest of the genome). We used the P-values assigned to each probe based on its enrichment in dbr1 versus DBR+ data sets (Supplemental Methods) as thresholds for the construction of a ROC plot (Supplemental Fig. 1). At each threshold, the proportion of significant probes found in the positive and negative regions was plotted. The area under the ROC curve is typically used to assess the quality of a classification method; a score of 1.0 indicates a perfect classifier, whereas a score of 0.5 indicates a random classifier. The tiling array data scored 0.9. We also performed a more stringent ROC analysis focused on intronic boundaries (Supplemental Fig. 1). Here, we designated intronic regions to be positives, and regions up- and downstream equal to half the size of the intervening intron to be negatives. The ROC score of the tiling array data remained 0.9, although there was a loss of sensitivity and specificity in this second classification at an estimated FDR of 10%.
The likeliest reason for a failure to detect an intron in the dbr1-specific signals is that the primary transcript was expressed at too low a level. Alternatively, there could be a relationship between the size of an intron and its identification on the array. To address RNA expression levels, we calculated the average intensity of tiling array probes within the exons of intron-containing genes expressed in DBR+ cells and plotted these intensities versus intron length (Fig. 3A; Supplemental Table 3). Introns that were detected were likely to be either more highly expressed or larger than the mean intron length. At an estimated FDR of 10%, all of the 124 genes (131 introns) for which an intronic signal was not observed were below the mean exonic probe intensity of these genes, and all but one were below the mean intron length. Those genes not transcribed under the culture conditions used or transcribed at a level below the threshold required for significance will be missed, such as HMRa1 (a silent mating cassette that is not expressed) and SPO22 (expressed mainly during meiosis) (Primig et al. 2000). In addition, other genes whose splicing is highly regulated were missed, such as REC107, HFM1, and AMA1, which are spliced only in meiotic cells (Engebrecht et al. 1991; Spingola and Ares 2000). Finally, the intron may be due to a spurious annotation.
Figure 3.
(A) Relationship between gene expression, intron length, and intron detection. Intron-containing genes were categorized as ribosomal (105) and nonribosomal (167); (green) detected introns, (red) undetected introns; (black dashed lines) mean intron length (246 nt) and mean exon probe intensity (1269). (B) Coverage of intronic regions by tiling array probes. Normalized positions of tiling array probes are plotted. (a) 5′ splice site, (b) branchpoint, (c) 3′ splice site.
Because the lariat RNA structure could give rise to biases in the array labeling and hybridization process, we looked at the distribution of tiling array signals at annotated intron boundaries and splice signals (Fig. 3B). We calculated the position of significant probes within the 272 annotated introns by normalizing the intron loop and tail lengths to their mean values (210 and 36 nt, respectively). In order to assess signals at intron boundaries, we also considered significant probes that were found within 100-bp regions flanking the introns. At an estimated FDR of 10%, we observed a total of 8947 probes in lariat loop regions, 91 significant probes within 100 bp upstream of the 5′ splice site, and 31 significant probes downstream of 3′ splice site. One reason for this “spillover” to flanking regions is the inaccuracy of intron annotation. For example, among 91 significant probes found upstream of a 5′ splice site, 26 are upstream of the RPL26B intron, which we considered a case for reannotation. Only six significant probes were found in lariat tails. This absence of signal could be due in part to the short length of lariat tails in S. cerevisiae, which average 36 nt. Alternatively, this bias could be due to the exonucleolytic degradation of lariat tails (Chapman and Boeke 1991), which may influence their labeling efficiency and subsequent detection on the array. Because of splice site sequence degeneracy, the correct assignment of exon–intron boundaries and thus gene structures is difficult. However, with tiling array signals falling between the 5′ splice site and branch point, this approach should be useful for demarcating regions in which these signals should occur.
We examined a gene model for RPL7A, a dual intron-containing gene, in more detail (Fig. 4A). Significant tiling array probes are found within both introns, and the ratio of dbr1/DBR+ signals illustrates the correspondence of annotated introns with the accumulation of intron-specific signals. The expression of RPL7A in DBR+ cells is shown to demarcate intron–exon boundaries, and the conservation among seven related yeast species is shown to highlight the lack of conservation found in intronic regions (Cliften et al. 2003; Kellis et al. 2003).
Figure 4.
(A–G) Gene models are annotated with splicing signals (black bars), significant tiling array probes (orange bars, estimated FDR of 10%), the log2 ratio of dbr1/DBR+ signals derived from tiling array data (positive ratios in green and negative ratios in red), normalized probe intensities from DBR+ cells (blue), and conservation among seven related yeasts (gray). (Light blue) Predicted splice junctions confirmed by RT-PCR, (red arrows) proposed novel start codons.
Comparative studies have identified novel introns by searching for conserved splice-donor and branchpoint signals among related yeast species (Brachat et al. 2003; Cliften et al. 2003; Kellis et al. 2003). We found correspondence between some of these predictions and several regions enriched on the tiling arrays. 5′ UTR introns predicted to be upstream of SUN4 and SIM1 (Cliften et al. 2003; Kellis et al. 2003) were identified, agreeing with previous experimental studies (Z. Zhang and F. Dietrich, unpubl.). In addition, previous studies predicted 5′ UTR introns in URA2 (Kellis et al. 2003) and BMH2 (M. Ares, pers. comm.) and a coding region intron in HRB1 (Kellis et al. 2003), and these introns were identified on the tiling arrays (Fig. 4B–D; Supplemental Table 2). We confirmed the presence of predicted introns in HRB1, URA2, and BMH2 using RT-PCR with primers that spanned the putative splice site (Supplemental Fig. 2), and sequenced these products to confirm the correct splice form.
Although the S. cerevisiae genome has been studied extensively by traditional genetics, functional genomic approaches, and comparative sequence analysis of related genomes, we identified at least four likely spliced sequences that were previously undetected. An intron in the 5′ UTR of YPR153W contains tiling array signals that lie between a canonical 5′ splice site signal and noncanonical branchpoint (5′-AACTAAC) (Fig. 4E; Supplemental Table 2). PTC7 contains an intron within its coding region (Fig. 4F). RT-PCR and sequencing of these products confirmed the presence of both of these introns (Supplemental Fig. 2). The reading frame of PTC7, encoding a mitochondrial protein phosphatase, remains intact before and after splicing, raising the possibility that the mRNA codes for two protein isoforms. We were unable to confirm putative introns in BDF2 (coding region) or YEL023C (5′ UTR) (Supplemental Fig. 3D,E).
Array data could also be used to identify misannotated introns. We found three ribosomal genes (RPL26B, RPL20A, and RPL20B) for which the array signals differed from the annotated intron regions, agreeing with other studies (M. Ares, pers. comm.). RPL26B contains a previously annotated 354-nt intron in its 5′ UTR (Fig. 4G). However, signals from the array suggest that the intron is 123 nt larger, with a different 5′ splice site that coincides with the array signals; the updated annotation for RPL26B was confirmed by RT-PCR and sequencing (Supplemental Fig. 2), and N-terminal sequencing of RPL26B is consistent with this annotation (Otaka et al. 1984). Another intron that was reannotated is YBR090C, which contains a 357-nt intron. This intron was previously suggested to be reassigned as a shorter 5′ UTR intron of NHP6B (Davis et al. 2000). However, our data support the original intron annotation (i.e., 357 nt) (Supplemental Fig. 2), but cannot conclusively assign the intron to YBR090C or NHP6B (Supplemental Fig. 3). A recent survey of full-length cDNAs in S. cerevisiae (Miura et al. 2006) assigned this intron as a 5′ UTR intron of NHP6B.
We characterized a total of 898 significant probes that fell outside of annotated intronic regions. Among them, 88 are due to reannotation (RPL26B, RPL20A, RPL20B, NHP6B/YBR090C) and 236 to newly predicted introns (URA2, BMH2, PTC7, HRB1, YPR153W). Several other genes are apparently up-regulated by DBR1 deletion, including FMP45 (172 probes), HSP12 (47 probes), and SYN8 (69 probes), accounting for 32% of the 898. Overall, 97% of the 9851 significant probes can be attributed to spliced introns or gene up-regulation in the dbr1 strain. Some of the significant probes could belong to bona fide spliced introns that remain to be experimentally verified, or could be due to spurious detection events, such as cases in which a significant dbr1/DBR+ ratio is observed for a gene expressed at a low level.
Discussion
We demonstrate an approach for identifying spliced introns on a genome-wide basis by the detection of lariat signals on a tiling array; the signals arise because of a mutation in the debranching enzyme necessary for lariat turnover. In yeast, this method is capable of identifying more than half of the known introns, providing gene models that typically allow the delineation of both the 5′ splice site and the branchpoint. Introns that were not detected lie in genes that are either not expressed or expressed at a low level under the conditions of our experiments. Despite the intense analysis of yeast introns in the decade since the genome sequence became available, our approach predicted novel introns and led to the reannotation of others.
Previous studies have used microarrays to study mRNA splicing and its regulation (Clark et al. 2002; Johnson et al. 2003). These studies employed oligonucleotide probes that are designed to hybridize at the exon–exon boundaries of spliced mRNAs. Although useful for studying many aspects of pre-mRNA splicing, this strategy requires detailed knowledge of each splicing event in order to design exon–exon probes, and thus would miss some of the noncanonical sequences in yeast and could miss many more splicing events in higher eukaryotes. By contrast, the analysis of lariat RNA provides verification of numerous gene structure models from a single experiment, enabling the confirmation of 5′ splice sites and branchpoints with near base-pair resolution, and identifies novel mRNA splice forms. A recent study (Miura et al. 2006) used whole-genome full-length cDNA sequencing for the annotation of S. cerevisiae, confirming several of our findings by identifying novel introns in BMH2, YPR153W, and NHP6B, and correcting the intron annotations of RPL26B, RPL20A, and RPL20B.
Spliced intronic lariats serve as markers for transcriptional as well as spliceosomal activity. This utility is in marked contrast with total RNA hybridization to tiling arrays (Kapranov et al. 2002; Kampa et al. 2004; David et al. 2006), which revealed abundant antisense transcription but could not distinguish between functional and spurious transcripts. Yeast tiling arrays were previously used to provide a global view of transcription, and were effective in discerning many exon–intron boundaries (David et al. 2006). However, our method allows the identification of a number of introns that were missed when assessing transcription alone.
Because splicing in metazoans is significantly more complicated than in yeast, the tiling array method could have limitations when applied to other organisms. For example, alternative splicing would give rise to complex tiling array signals; however, alternative splice forms might be addressed by the identification of subsets of introns within a gene that have distinct dbr1/DBR+ signals. Another concern would be the assignment of 5′ and 3′ splice sites and branchpoints, which can be highly degenerate (Lim and Burge 2001). This method should provide high-resolution definition of the 5′ splice site and branchpoint. However, because stabilization of intron lariats results in the degradation of their 3′ tails, the identification of 3′ splice sites may be difficult, precluding the assignment of alternative 3′ splice sites (e.g., NAGNAG motifs; Hiller et al. 2004). Finally, intron size and expression level may confound the use of this approach. In humans, introns average 87 nt in length (Lander et al. 2001), and in Caenorhabditis elegans they average 47 nt (C. elegans Sequencing Consortium 1998). Although we identified several short introns in yeast (as small as 93 nt), most of those we identified are >200 nt. Genes expressed at low levels will be difficult to detect, but the method could be used on tissue-specific samples from a variety of tissue types to encompass cases in which certain genes are more highly expressed.
Our approach may therefore be applicable to at least a subset of genes in most eukaryotes, and it should be complementary to other approaches to identify introns and annotate genes. The regulation of intron turnover in metazoans via debranching is largely unknown. The Pfam database (Finn et al. 2006) indicates that invertebrates have a single, highly conserved DBR1 ortholog, but that vertebrates may have two or more DBR1 orthologs, possibly as a result of alternative splicing. However, the activity of DBR1 genes has been successfully reduced in more complex eukaryotes by RNAi, resulting in the manipulation of lariat RNA levels. Targeting of the debranching enzyme via siRNA in Drosophila stabilizes intron lariats (Conklin et al. 2005), and targeting of this enzyme in human cell culture affects the efficiency of HIV retroviral replication (Ye et al. 2005). Thus, this approach could prove useful for genome annotation in many organisms.
Methods
Strains and culturing
S. cerevisiae BY4743 (MATa/MATα his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 lys2Δ0/+ met15Δ0/+ ura3Δ0/ura3Δ0) and its corresponding dbr1 double-deletion strain were obtained from Open Biosystems. Yeast was cultured in rich medium (YPD) at 30°C. Escherichia coli strain DH5α was used in cDNA cloning.
RNA preparation and hybridization
Total RNA from exponential phase (OD660 = 1.0) S. cerevisiae cultures was purified using an RNeasy Mini kit (Qiagen) after treatment with Turbo DNase (Ambion). Probe labeling was done using the GeneChip WT Double-Stranded cDNA Synthesis Kit (Affymetrix) following the manufacturer’s protocol. Labeled probes were hybridized to the GeneChip S. cerevisiae Tiling 1.0R Array (Affymetrix) using the manufacturer’s protocol. These arrays contain ∼2.6 million 25-mer perfect match (PM) probes and ∼2.6 million corresponding mismatch (MM) probes, which have a one-base mismatch at the thirteenth nucleotide in the sequence, overlapped with each other at an average of 5 nt offset, covering >95% of the genome. Scanning and data collection were done by GeneChip Scanner 3000 7G (Affymetrix) and GeneChip Operating Software. Three independent cultures of the BY4743 and dbr1 strains were applied to the tiling arrays. Raw data for array experiments have been deposited in the NCBI Gene Expression Omnibus under accession GSE5470 (http://www.ncbi.nlm.nih.gov/geo/).
Tiling array data analysis
Raw tiling array data were analyzed using the Tiling Analysis Software (TAS) package (Affymetrix). Data from triplicate arrays were combined and quantile normalized prior to analysis. For expression measurements (i.e., signals derived from BY4743 total RNA), signals obtained from PM and MM probes from three biological array replicates were compared in ∼11 probe windows using a Wilcoxon rank-sum test. The center probe of the window was assigned the resulting P-value, and the calculation was repeated for each probe on the array. In order to assess dbr1-specific enrichment, we looked at the overall gene expression correspondence between the BY4743 and dbr1 strains. The Pearson correlation of probes within exonic regions from two strains is 0.93, indicating an overall correspondence between probe intensities from each strain (Supplemental Fig. 4). The Wilcoxon rank-sum test was used to obtain a P-value for 41-bp windows (i.e., 24 probes from BY7473 replicates and 24 probes from dbr1 replicates). Data were visualized using the UCSC Genome Browser, and supplemental files used for all analyses are available on the Web (http://depts.washington.edu/sfields/supplemental_data/intron_tiling_supplement/).
False discovery rate estimation
The QVALUE software (Storey and Tibshirani 2003) was used to estimate the false discovery rate (FDR) in the analysis of dbr1/DBR+ enrichment. Default values were used for the analysis, and the overall proportion of true null hypotheses (π0) was estimated to be 0.637.
RT-PCR
A total of 5 μg of total RNA from S. cerevisiae BY4743 and dbr1 strains were reverse transcribed using an anchored oligo-dT primer (5′-T21VN-3′) using SuperScript III Reverse Transcriptase (Invitrogen). Gene-specific primer pairs were used to amplify cDNA potentially spanning the novel introns (Supplemental Methods). Amplified fragments were gel purified and cloned into the pCR2.1 TA vector (Invitrogen) and sequenced to confirm predicted splice junctions. Sequences of novel splice forms have been deposited in the NCBI GenBank under accessions DQ881448 (BMH2), DQ881449 (HRB1), DQ881450 (PTC7), DQ881451 (RPL26B), DQ881452 (URA2), DQ881453 (YPR153W), EF138821 (RPL20A), and EF138822 (RPL20B). Details of novel or modified introns are listed in Supplemental Table 2.
Acknowledgments
We thank W. Noble and P. Green for helpful discussions, and M. Ares for access to unpublished data. J.H. was supported by an NIH NRSA (F32HG003439-02), P41 RR11823, and a Rosetta Fellowship provided to the University of Washington by Merck Research Laboratories. S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
[Supplemental material is available online at www.genome.org and at http://depts.washington.edu/sfields/supplemental_data/intron_tiling_supplement/.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6049107
References
- Brachat S., Dietrich F.S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P., Dietrich F.S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P., Lerch A., Gates K., Gaffney T., Philippsen P., Gates K., Gaffney T., Philippsen P., Gaffney T., Philippsen P., Philippsen P. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brent M.R. Genome annotation past, present, and future: How to define an ORF at each locus. Genome Res. 2005;15:1777–1786. doi: 10.1101/gr.3866105. [DOI] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chapman K.B., Boeke J.D., Boeke J.D. Isolation and characterization of the gene encoding yeast debranching enzyme. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- Christie K.R., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Weng S., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Balakrishnan R., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Costanzo M.C., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Dolinski K., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Dwight S.S., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Engel S.R., Feierbach B., Fisk D.G., Hirschman J.E., Feierbach B., Fisk D.G., Hirschman J.E., Fisk D.G., Hirschman J.E., Hirschman J.E., et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark T.A., Sugnet C.W., Ares M., Jr., Sugnet C.W., Ares M., Jr., Ares M., Jr. Genomewide analysis of mRNA processing in yeast using splicing-specific microarrays. Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- Cliften P., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Sudarsanam P., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Desikan A., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton L., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Fulton B., Majors J., Waterston R., Cohen B.A., Johnston M., Majors J., Waterston R., Cohen B.A., Johnston M., Waterston R., Cohen B.A., Johnston M., Cohen B.A., Johnston M., Johnston M. Finding functional features in Saccharomyces genomes by phylogenetic footprinting. Science. 2003;301:71–76. doi: 10.1126/science.1084337. [DOI] [PubMed] [Google Scholar]
- Conklin J.F., Goldman A., Lopez A.J., Goldman A., Lopez A.J., Lopez A.J. Stabilization and analysis of intron lariats in vivo. Methods. 2005;37:368–375. doi: 10.1016/j.ymeth.2005.08.002. [DOI] [PubMed] [Google Scholar]
- David L., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Huber W., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Granovskaia M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Toedling J., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Palm C.J., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Bofkin L., Jones T., Davis R.W., Steinmetz L.M., Jones T., Davis R.W., Steinmetz L.M., Davis R.W., Steinmetz L.M., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.A., Grate L., Spingola M., Ares M., Jr., Grate L., Spingola M., Ares M., Jr., Spingola M., Ares M., Jr., Ares M., Jr. Test of intron predictions reveals novel splice sites, alternatively spliced mRNAs and new introns in meiotically regulated genes of yeast. Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engebrecht J.A., Voelkel-Meiman K., Roeder G.S., Voelkel-Meiman K., Roeder G.S., Roeder G.S. Meiosis-specific RNA splicing in yeast. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- Finn R.D., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Mistry J., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Schuster-Bockler B., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Griffiths-Jones S., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Hollich V., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Lassmann T., Moxon S., Marshall M., Khanna A., Durbin R., Moxon S., Marshall M., Khanna A., Durbin R., Marshall M., Khanna A., Durbin R., Khanna A., Durbin R., Durbin R., et al. Pfam: Clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grate L., Ares M., Jr., Ares M., Jr. Searching yeast intron data at Ares lab Web site. Methods Enzymol. 2002;350:380–392. doi: 10.1016/s0076-6879(02)50975-7. [DOI] [PubMed] [Google Scholar]
- Gresham D., Ruderfer D.M., Pratt S.C., Schacherer J., Dunham M.J., Botstein D., Kruglyak L., Ruderfer D.M., Pratt S.C., Schacherer J., Dunham M.J., Botstein D., Kruglyak L., Pratt S.C., Schacherer J., Dunham M.J., Botstein D., Kruglyak L., Schacherer J., Dunham M.J., Botstein D., Kruglyak L., Dunham M.J., Botstein D., Kruglyak L., Botstein D., Kruglyak L., Kruglyak L. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- Halasz G., van Batenburg M.F., Perusse J., Hua S., Lu X.J., White K.P., Bussemaker H.J., van Batenburg M.F., Perusse J., Hua S., Lu X.J., White K.P., Bussemaker H.J., Perusse J., Hua S., Lu X.J., White K.P., Bussemaker H.J., Hua S., Lu X.J., White K.P., Bussemaker H.J., Lu X.J., White K.P., Bussemaker H.J., White K.P., Bussemaker H.J., Bussemaker H.J. Detecting transcriptionally active regions using genomic tiling arrays. Genome Biol. 2006;7:R59. doi: 10.1186/gb-2006-7-7-r59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiller M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M., Huse K., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M., Szafranski K., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M., Jahn N., Hampe J., Schreiber S., Backofen R., Platzer M., Hampe J., Schreiber S., Backofen R., Platzer M., Schreiber S., Backofen R., Platzer M., Backofen R., Platzer M., Platzer M. Widespread occurrence of alternative splicing at NAGNAG acceptors contributes to proteome plasticity. Nat. Genet. 2004;36:1255–1257. doi: 10.1038/ng1469. [DOI] [PubMed] [Google Scholar]
- Johnson J.M., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Castle J., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Garrett-Engele P., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Kan Z., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Loerch P.M., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Armour C.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Santos R., Schadt E.E., Stoughton R., Shoemaker D.D., Schadt E.E., Stoughton R., Shoemaker D.D., Stoughton R., Shoemaker D.D., Shoemaker D.D. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- Kampa D., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cheng J., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Kapranov P., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Yamanaka M., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Brubaker S., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Cawley S., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Drenkow J., Piccolboni A., Bekiranov S., Helt G., Piccolboni A., Bekiranov S., Helt G., Bekiranov S., Helt G., Helt G., et al. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Cawley S.E., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Drenkow J., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Bekiranov S., Strausberg R.L., Fodor S.P., Gingeras T.R., Strausberg R.L., Fodor S.P., Gingeras T.R., Fodor S.P., Gingeras T.R., Gingeras T.R. Large-scale transcriptional activity in chromosomes 21 and 22. Science. 2002;296:916–919. doi: 10.1126/science.1068597. [DOI] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S., Patterson N., Endrizzi M., Birren B., Lander E.S., Endrizzi M., Birren B., Lander E.S., Birren B., Lander E.S., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Devon K., Dewar K., Doyle M., FitzHugh W., Dewar K., Doyle M., FitzHugh W., Doyle M., FitzHugh W., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Burge C.B., Burge C.B. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., Cheng J., Bell I., Ghosh S., Piccolboni A., Bell I., Ghosh S., Piccolboni A., Ghosh S., Piccolboni A., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T., Sese J., Toyoda A., Hattori M., Morishita S., Ito T., Toyoda A., Hattori M., Morishita S., Ito T., Hattori M., Morishita S., Ito T., Morishita S., Ito T., Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otaka E., Higo K., Itoh T., Higo K., Itoh T., Itoh T. Yeast ribosomal proteins: VIII. Isolation of two proteins and sequence characterization of twenty-four proteins from cytoplasmic ribosomes. Mol. Gen. Genet. 1984;195:544–546. doi: 10.1007/BF00341461. [DOI] [PubMed] [Google Scholar]
- Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E., Hwang S.Y., Davis R.W., Esposito R.E., Davis R.W., Esposito R.E., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- Sabo P.J., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Kuehn M.S., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Thurman R., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Johnson B.E., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Johnson E.M., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Cao H., Yu M., Rosenzweig E., Goldy J., Haydock A., Yu M., Rosenzweig E., Goldy J., Haydock A., Rosenzweig E., Goldy J., Haydock A., Goldy J., Haydock A., Haydock A., et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- Schumacher A., Kapranov P., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Kapranov P., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Kaminsky Z., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Flanagan J., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Assadzadeh A., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Yau P., Virtanen C., Winegarden N., Cheng J., Gingeras T., Virtanen C., Winegarden N., Cheng J., Gingeras T., Winegarden N., Cheng J., Gingeras T., Cheng J., Gingeras T., Gingeras T., et al. Microarray-based DNA methylation profiling: Technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha I., Wiren M., Ekwall K., Wiren M., Ekwall K., Ekwall K. Genome-wide patterns of histone modifications in fission yeast. Chromosome Res. 2006;14:95–105. doi: 10.1007/s10577-005-1023-4. [DOI] [PubMed] [Google Scholar]
- Spingola M., Ares M., Jr., Ares M., Jr. A yeast intronic splicing enhancer and Nam8p are required for Mer1p-activated splicing. Mol. Cell. 2000;6:329–338. doi: 10.1016/s1097-2765(00)00033-2. [DOI] [PubMed] [Google Scholar]
- Spingola M., Grate L., Haussler D., Ares M., Jr., Grate L., Haussler D., Ares M., Jr., Haussler D., Ares M., Jr., Ares M., Jr. Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae. RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey J.D., Tibshirani R., Tibshirani R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson G.M., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Flibotte S., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Missirlis P.I., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Marra M.A., Jones S., Thornton K., Clark A.G., Holt R.A., Jones S., Thornton K., Clark A.G., Holt R.A., Thornton K., Clark A.G., Holt R.A., Clark A.G., Holt R.A., Holt R.A. Identification by full-coverage array CGH of human DNA copy number increases relative to chimpanzee and gorilla. Genome Res. 2006;16:173–181. doi: 10.1101/gr.4456006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., De Leon J., Yokoyama N., Naidu Y., Camerini D., De Leon J., Yokoyama N., Naidu Y., Camerini D., Yokoyama N., Naidu Y., Camerini D., Naidu Y., Camerini D., Camerini D. DBR1 siRNA inhibition of HIV-1 replication. Retrovirology. 2005;2:63. doi: 10.1186/1742-4690-2-63. [DOI] [PMC free article] [PubMed] [Google Scholar]