Abstract
Molecular genetic analysis of Borrelia burgdorferi, the cause of Lyme disease, has been hampered by the absence of any means of efficient generation, identification, and complementation of chromosomal and plasmid null gene mutants. The similarity of borrelial G + C content to that of Gram-positive organisms suggested that a wide-host-range plasmid active in Gram-positive bacteria might also be recognized by borrelial DNA replication machinery. One such plasmid, pGK12, is able to propagate in both Gram-positive and Gram-negative bacteria and carries erythromycin and chloramphenicol resistance markers. pGK12 propagated extrachromosomally in B. burgdorferi B31 after electroporation but conferred only erythromycin resistance. pGK12 was used to express enhanced green fluorescent protein in B31 under the control of the flaB promoter. Escherichia coli transformed with pGK12 DNA extracted from B31 expressing only erythromycin resistance developed both erythromycin and chloramphenicol resistance, and plasmid DNA isolated from these transformed E. coli had a restriction pattern similar to the original pGK12. Our data indicate that the replicons of pGK12 can provide the basis to continue developing efficient genetic systems for B. burgdorferi together with the erythromycin resistance and reporter egfp genes.
The ability to propagate and express native and foreign recombinant DNA in Borrelia has, in general, been hindered by the lack of knowledge of the structure of borrelial replication origins and replicon functions in the genome (1), the paucity of readily available antibiotic markers for use in selection of Borrelia containing potential cloning vectors (2), the difference in codon usage in Borrelia as compared with other well characterized organisms (3), and the dearth of information on regulatory DNA sequences and other factors regulating gene expression in this pathogen (4).
Nonreplicative plasmid constructs containing Borrelia promoters fused to the chloramphenicol acetyltransferase gene have been used to study transient gene expression in Borrelia (5). After electroporation of mutant-containing linear borrelial DNA conferring coumermycin A1 resistance into B. burgdorferi, coumermycin A1-resistant chromosomal borrelial recombinants could be isolated (6–8). Heterologous DNA containing a coumermycin A1 resistance marker has also been shown to be integrated into the Borrelia genome after electroporation (9). However, the low frequency of recombination at the desirable chromosome site of this linear DNA, the large background of spontaneous mutants on coumermycin A1-selective plates, and the high frequency of recombination of incoming DNA into the coumermycin A1 resistance locus has hindered widespread application of this method in Borrelia genetics.
The similarity of borrelial G + C content (28.6%) to that of Gram-positive organisms suggested that a wide-host-range plasmid active in Gram-positive bacteria might be recognized by borrelial DNA replication machinery. The use of plasmids from bacteria whose DNA had a G + C content close to that of Borrelia might also facilitate expression of antibiotic resistance genes contained in these plasmids. On the other hand, a similar G + C content between Gram-positive organisms and Borrelia might facilitate homologous recombination of the cloning vector into the borrelial chromosome. Because the efficiency of electroporation of linear DNA into Borrelia is low (6), large amounts of plasmid DNA are needed for electroporation (10). It was therefore decided to employ covalently closed DNA from small cloning vectors able to replicate in Escherichia coli to facilitate production of such large amounts of plasmid DNA (11, 12). We report here that we have been able to introduce a wide-host-range plasmid, pGK12, into the high-passage B. burgdorferi strain B31 (B31). This plasmid contains replication functions of Lactococcus lactis pWV01 and antibiotic resistance genes to erythromycin (ermC) and chloramphenicol (cm) from Staphylococcus aureus plasmids pE194 and pC194 (11, 13, 14), and is able to replicate in Staphylococcus, Streptococcus, Bacillus, and E. coli (15). This plasmid replicated extrachromosomally in B31 for several generations while conferring only erythromycin resistance, replicated in E. coli after extraction from B31 (conferring both erythromycin and chloramphenicol resistance), and permitted cloning and expression of enhanced green fluorescent protein (EGFP) in B31. These results extend to Borrelia the paradigm that natural wide-host-range plasmids or multifunctional replicons generated in vitro can be used to develop molecular genetic systems in this genus (16).
Materials and Methods
Bacterial Strains, Plasmids, and Growth Conditions.
The B31 used in these experiments is a high-passage, noninfectious B. burgdorferi strain. It was grown at 32°C in Barbour–Stoenner–Kelly (BSK-H) medium (Sigma) supplemented with 7% rabbit sera (Sigma). E. coli strains DH5α and MM294 were used to propagate plasmids before and after B31 electroporation. Plasmids used in this work were the wide-host-range plasmids pGK12 and pHP13 encoding erythromycin and chloramphenicol resistance (11, 12). Plasmid DNA was purified from E. coli strain MM294 grown in LB broth by using CsCl gradient centrifugation (17). Recombinant plasmid pMS1 was propagated in E. coli DH5α and purified (Wizard Maxi-Purification Kit; Promega). After ethanol precipitation, purified plasmid DNA was resuspended in sterile water or TE buffer (10 mM Tris/1 mM EDTA, pH 8.0). These experiments were approved by the Institutional Biosafety Committee at New York Medical College.
Antibiotics and Minimal Inhibitory Concentrations (MIC).
Antibiotic stock solutions (10 mg/ml) of erythromycin and chloramphenicol were made in ethanol, of kanamycin, neomycin, novobiocin, and cefotaxime, in distilled water, of coumermycin A1, in DMSO. Solid BSK-H medium (P-BSK-H) with 0.65% high strength analytical grade agarose (Bio-Rad) in both top and bottom layers was used for plating B31 cells to determine MIC. P-BSK-H without antibiotic was used as control in these experiments. Approximately 2 × 108 bacteria in 2 ml of P-BSK-H were overlaid onto 60-mm Falcon tissue culture dishes (Becton Dickinson) containing a 6-ml bottom layer of solid P-BSK-H. B31 cell concentrations were determined spectrophotometrically by multiplying optical density at 600 nm by 1.4 × 109, or by direct counting under phase-contrast microscopy in a Petroff Hausser cell-counting chamber (Hausser Scientific Partnership, Horsham, PA). These two methods were equivalent to each other and to viable cell counts determined by direct plating (18). Plates seeded with B31 were incubated at 32°C in a humidified 3% CO2 atmosphere for 2–3 wk. MIC of antibiotics was the lowest concentration of antibiotic that prevented growth of colonies on the plates (19).
Electroporation of B31 with Plasmid DNA.
B31 cells were grown in BSK-H medium until a density of approximately 8 × 107 cells per ml. Bacteria were washed twice in Dulbecco's PBS and three times in electroporation solution (EPS) [270 mM sucrose/15% (vol/vol) glycerol]. After the final wash, the bacterial pellet was resuspended in cold EPS to a final cell density of about 1010 cells per ml (20), and 5–8 μg of plasmid DNA was mixed with 40 μl of B31 cell suspensions. Electroporation was carried out in a Gene Pulser II apparatus (Bio-Rad) with 0.1-cm or 0.2-cm gap cuvettes at 1.25 kV for a 0.1-cm gap or 2.5 kV for a 0.2-cm gap, 25 μF capacitance, and 200 Ω producing a time constant of 4–5 ms. A sample containing B31 cells without DNA was electroporated as a control. Immediately after electroporation, B31 cells were placed in 4 ml of BSK-H medium containing 7% rabbit serum without antibiotics and incubated at 32°C overnight. After this recovery period (18–20 h), aliquots of electroporated B31 cultures and controls containing 2 × 108 B31 cells and selective antibiotics at a concentration corresponding to the MIC were plated in 2 ml of melted P-BSK-H medium onto 60-mm tissue culture dishes containing 6 ml of solidified P-BSK with a concentration of selective antibiotic similar to that in the top layer. Colonies of electroporant Borrelia usually appeared 2–4 wk after plating.
Cloning of the egfp Gene into pGK12 to Generate pMS1.
To isolate the B31 flaB putative promoter, the DNA sequence of the flaB gene (GenBank accession no. BB0147) was analyzed. A forward primer F1, 5′-CCATCGATTTTACCGTTAAGCGC-3′, was designed with a ClaI restriction site (underlined) to insert the amplicon into pGK12. The reverse primer F2, 5′-GATAATCATATAGAATTCCTCC-3′, contained an EcoRI restriction site (underlined) for junction with the egfp gene. The egfp gene was obtained from pBI-EGFP (CLONTECH). To amplify the egfp gene, the forward primer G1, 5′-GGGAATTCTATATGGTGAGCAAGGGCG-3′, containing an EcoRI restriction site (underlined) was used. The reverse primer G2, 5′-CCATCGATTTCTTACTTGTACAG-3′, contained a ClaI restriction site (underlined). Primers were synthesized by GenoSys Biotechnology (The Woodlands, TX). PCR amplifications were performed in a Rapid Thermal Cycler (Idaho Technology, Idaho Falls, ID) in the buffer supplied by the manufacturer in the presence of 2 mM MgCl2, 0.25 units of Taq DNA polymerase (GIBCO/BRL), and 0.5 μM each primer in volume of 10 μl. A total of 37 cycles were done under the following machine settings: denaturation at 95°C for 0 s, annealing at 57°C for 0 s, and extension at 72°C for 27 s. After amplification, PCR products were digested with EcoRI, purified by electrophoresis in 1% agarose (SeaKem; FMC), and purified (QIAquick Gel Extraction Kit; Qiagen, Santa Clara, CA). Purified products were ligated by using T4 DNA ligase and reamplified with F1 and G2 primers. The resulting PCR product was a DNA fragment of 925 bp that contained the flaB promoter and the egfp gene. It was digested by ClaI and inserted into the HpaII site of pGK12. The DNA sequence of the junction between the flaB promoter and the egfp gene was sequenced (T7 Sequenase Quick-Denature Plasmid Sequencing Kit and deoxyadenosine 5′-[α-35S]thiotriphosphate, triethylammonium salt; Amersham Pharmacia). All restriction endonucleases, T4 DNA ligase, and appropriate buffers were obtained from New England Biolabs.
Agarose Gel Electrophoresis and Southern Blot Hybridization.
Plasmid DNA from wild-type B31 and derivatives containing plasmids pGK12 and pMS1 were isolated (Plasmid Mini Kit; Qiagen), electrophoresed on 0.7% agarose gel at 30 V for 6 h, transferred to Magna Graph nylon membranes (Micron Separations, Westboro, MA), and hybridized with a DNA probe generated by PCR. The probe was a 720-bp PCR product corresponding to the chloramphenicol-resistance gene of pGK12 (11). It was amplified with the forward primer Cm1, 5′-GATAAAGATCTAGGAGGCATAT-3′, and the reverse primer Cm2, 5′-TAAAAAGCTTCTTGAACTAACG-3′ (both primers synthesized by Genosys Biotechnology), and purified (QIAquick Gel Extraction Kit). The DNA probe was labeled with digoxigenin (DIG DNA Labeling and Detection Kit; Roche Molecular Biochemicals), according to the manufacturer's instructions.
Characterization of pGK12 and pMS1 Extracted from B. burgdorferi B31.
pGK12 and pMS1 DNA extracted from B. burgdorferi electroporants were transformed into competent E. coli DH5α cells (21). Transformants were selected on LB agar plates containing 20 μg/ml chloramphenicol. Plasmid DNA of five transformants was purified (Qiagen Plasmid Mini Kit), their DNA restriction patterns were analyzed with NdeI, HpaII, and BpmI and compared with those obtained with pGK12 and pMS1 originally introduced into B31.
Estimation of pMS1 Copy Number.
Competitive PCR was used to determine pMS1 plasmid copy number in a B31 cell. Two competitors containing internal deletions were constructed, one for the egfp gene and a second for the flaB gene. The competitor for egfp had a 240-bp deletion in the region amplified with primers G1 and G2. This deletion was constructed by digesting the egfp gene with endonuclease BpmI, an enzyme with two restriction sites in the egfp gene sequence. The competitor for flaB was constructed by amplifying an 880-bp fragment of flaB gene from total B31 DNA by using flaB specific primers (forward primer F3, 5′-CTAGTGGGTACAGAATTAATCGAGC-3′, reverse primer F4, 5′-GCCTGCGCAATCATTGCCATTGC-3′). This amplicon was cloned by blunt-end ligation methods into EcoRV site of pBlueScript II SK+ vector (Stratagene), and an internal deletion of 189 bp was introduced by using SmlI. Doses of competitors from 3.3–1,000 fg (six 3-fold dilutions) were mixed with fixed amounts of total B31 DNA to perform quantitative PCR for egfp and flaB. Target DNA and competitor DNA were assumed to be present at equimolar concentrations in those reactions when the competitor and target PCR products were at similar intensities in agarose gel electropherograms (22).
Immunological Detection of EGFP in B31 Containing pMS1.
Proteins of wild-type B31 and variants containing pGK12 and pMS1 were extracted from lysates of 5 × 108 cells (17), and analyzed by 12% SDS/PAGE. Resolved polypeptides were transferred to Hybond ECL nitrocellulose membranes (Amersham Pharmacia), and were probed with EGFP-specific polyclonal Living Colors antibodies (CLONTECH), developed with horseradish peroxidase-conjugated goat anti-rabbit Ig (Bio-Rad), and visualized with Hyperfilm MP Autoradiography Film (Amersham Pharmacia) for 3 s and 10 s.
RNA Isolation and Reverse Transcription (RT)-PCR to Detect EGFP mRNA.
B31 cells were grown in 10 ml until culture density was 4 × 108 cells per ml. One-half of this culture was incubated at 25°C in the presence of oxygen for 6 h, while the other half was incubated at 32°C in 3% CO2. Total RNA from each culture was isolated by extraction with guanidine thiocyanate/phenol/chloroform (23), treated with RNase-free DNase (Promega) for 3 h at 37°C to eliminate any contaminating DNA, extracted with phenol/chloroform, and precipitated with ethanol. The amount of total RNA was determined spectrophotometrically by absorbance at 260 nm. Primers G1 and G2 specific for egfp gene were used to detect egfp mRNA in total RNA preparations by RT-PCR (Access RT-PCR System; Promega), according to the manufacturer's instructions.
Microscopy.
EGFP fluorescence in E. coli and B31 was visualized with an Olympus Bx60 system microscope equipped with a mercury light source and a PM-20 automatic photomicrographic system. For phase-contrast as well as fluorescence observations and photography, a Plan phase-contrast objective (magnification, ×100; numerical aperture, 1.25) was used. Fluorescence was visualized by using an excitation cube (41001, 611 HQ: FITC; Chroma Technology, Brattleboro, VT) with a wide-bandpass (470- to 490-nm) excitation filter.
Results
Antibiotic Susceptibility of B31.
There have been several studies of the susceptibility of B. burgdorferi to antibiotics to identify therapeutic agents for Lyme disease (24–29). The concentration of antibiotics and the numbers of B. burgdorferi cells used in these therapeutically oriented assays are different from those used to select borrelial electroporants containing cloning vectors (2, 19). This is because it is necessary to use antibiotic and Borrelia concentrations in cloning experiments that optimize the selection of electroporants while minimizing selection of spontaneous mutants. MIC of B31 to different antibiotics was therefore determined in solid P-BSK-H medium by using cell numbers (2 × 108 B31) used in electroporation. The following antibiotic concentrations gave no or one to three colonies of B31 after growth at 32°C for at least 21 days: erythromycin, 0.03 μg/ml; chloramphenicol, 2 μg/ml; kanamycin, 160 μg/ml; coumermycin A1, 0.02 μg/ml; novobiocin, 5.0 μg/ml; cefotaxime 0.12 μg/ml. These results indicate that B31 electroporants able to grow in the presence of antibiotics under these conditions would much more likely contain electroporated plasmids than be spontaneous antibiotic-resistant B31 mutants.
Electroporation of Wide-Host-Range Plasmids into B31.
Several considerations were taken into account in choosing plasmids for electroporation into B31. They were selected containing replicons and antibiotic resistance genes from bacteria whose DNA has a G + C content close to that of Borrelia to facilitate their expression. Because previous experiments indicated that the efficiency of electroporation of linear DNA into Borrelia was low (6), small cloning vectors with covalently closed DNA able to replicate in both Gram-positive bacteria and E. coli appeared to be appropriate choices (11). Replication in E. coli was particularly desirable to facilitate isolation of the large amounts of plasmid DNA required for electroporation (10). Finally, because β-lactam antibiotics and tetracyclines are preferred choices in the treatment of Lyme disease, selection of resistance to these antibiotics was undesirable, as it could potentially create Borrelia strains resistant to antibiotic treatment (29). For this reason, plasmids with resistance genes to chloramphenicol and erythromycin were used for these experiments as they encode modifying enzymes whose genes are not present in the Borrelia genome, thus decreasing the opportunities for recombination (1).
Two plasmids consistent with these criteria (pGK12, pHP13) were electroporated into B31. No colonies were obtained with B31 electroporated with pHP13 DNA on multiple occasions. In contrast, B31 electroporated with plasmid pGK12 DNA yielded erythromycin-resistant colonies on solid agar after 21 days of culture; no electroporants were obtained from selective plates containing chloramphenicol in these experiments. B31 originating from erythromycin-resistant colonies grew to concentrations of 5 × 108 cells per ml in liquid media containing 0.03 or 0.06 μg/ml erythromycin, suggesting that pGK12 in these B31 erythromycin-resistant electroporants was stable in the presence of antibiotic for at least 40 generations. Erythromycin-resistant B31 electroporants were unable to grow in liquid medium containing chloramphenicol. Agarose gel electrophoresis of total and plasmid DNA extracted from several B31 erythromycin-resistant clones demonstrated a DNA band of 4.3 kb corresponding to pGK12 (Fig. 1a, lanes 3–5). This band was absent in wild-type B31 (Fig. 1a, lane 2). Moreover, amplification of plasmid DNA extracted from this band with primers amplifying pGK12 erythromycin and chloramphenicol resistance genes generated amplicons of the expected size (data not shown).
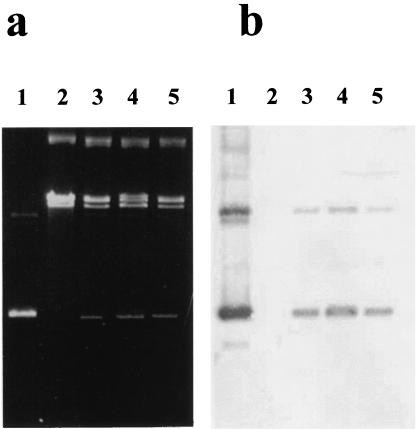
Figure 1.
Agarose gel electrophoresis and Southern hybridization of plasmid DNA isolated from B. burgdorferi B31 erythromycin-resistant clones electroporated with plasmid pGK12. (a) Agarose gel electrophoresis. (b) Southern blot of agarose gel in a. Lane 1, pGK12 DNA purified from E. coli MM294; Lane 2, Plasmid DNA from wild-type B. burgdorferi B31; Lanes 3–5, Plasmid DNA of three different B. burgdorferi erythromycin-resistant electroporants containing pGK12.
Plasmid pGK12 Replicates Independently of the B31 Chromosome and Plasmids.
Further confirmation of the extrachromosomal location of pGK12 in B31 was suggested by the ability of plasmid DNA extracted from erythromycin-resistant clones containing pGK12 to transform E. coli DH5α to chloramphenicol resistance (data not shown). Plasmid DNA extracted from these chloramphenicol-resistant E. coli had molecular and restriction enzyme patterns similar to those of the pGK12 originally electroporated into B31, indicating that pGK12 was mainly extrachromosomal and did not undergo recombination that could alter its original DNA structure. Results of DNA hybridization of plasmid DNA from erythromycin-resistant B31 with a plasmid DNA probe containing a 720-bp DNA sequence present in the chloramphenicol resistance gene of pGK12 indicated that most of the hybridizing DNA was in the DNA band corresponding to the circular covalently closed autonomous pGK12 (Fig. 1b, lanes 3–5). There were no hybridization signals in the region of the gel that contained Borrelia plasmid DNA and only a weak signal in the region that corresponded to open circular pGK12 (Fig. 1b, lanes 3–5). The presence of a discrete, constant, and visible plasmid DNA band corresponding to pGK12 in agarose gel electrophoresis of plasmid DNA extracted from erythromycin-resistant Borrelia grown in media with the antibiotic further suggested that replication of pGK12 is stable in B31 in the presence of the antibiotic. Replication stability of pGK12 in B31 was further tested by growing one erythromycin-resistant B31 isolate in liquid media for at least 20 generations without antibiotic and then comparing colony counts of this recombinant B31 on solid media plates with and without antibiotic. The count of colonies growing on solid medium with antibiotic was only 75% of those plated on solid medium without antibiotics, indicating some partial stability of the pGK12 replication in this bacteria in the absence of antibiotic.
Use of pGK12 as a Cloning Vector in B31.
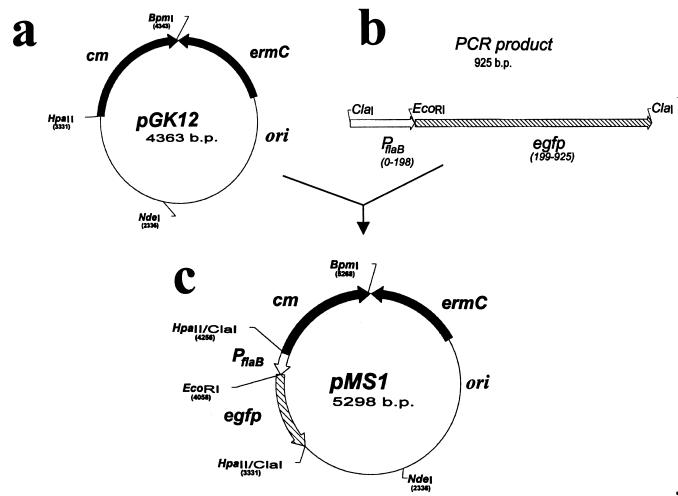
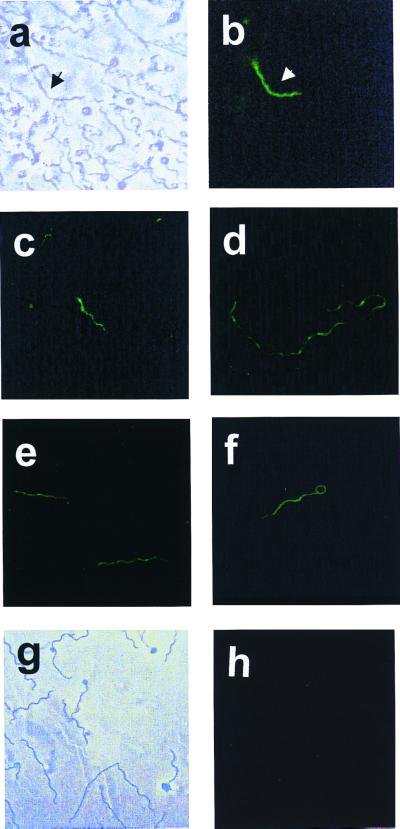
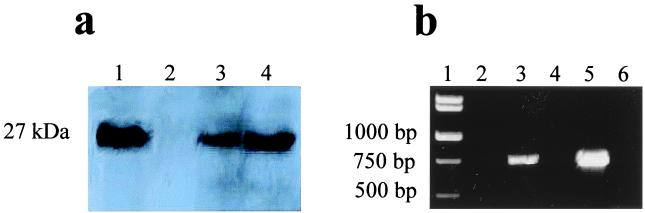
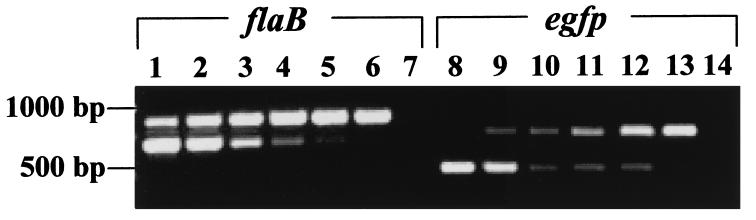
The potential usefulness of pGK12 as cloning vector in Borrelia was examined by developing a recombinant pGK12 plasmid containing the egfp gene (30). To optimize expression of the egfp gene in Borrelia, a segment of DNA of 198 bp containing the flaB (flagellin) gene promoter was added to the 5′ end of the DNA segment of 722 bp containing egfp from pB1-EGFP (Fig. 2f). The final recombinant plasmid (pMS1; 5,298 bp) was constructed by introducing the DNA fragment containing the flaB gene promoter and the egfp gene into the HpaII site of pGK12 (Fig. 2 a and b). The correctness of the DNA sequence of the flaB promoter and egfp junction in the pMS1 plasmid was confirmed by DNA sequencing (data not shown). pMS1 DNA isolated from E. coli was introduced by electroporation into B31, and the electroporants were plated on solid medium containing erythromycin. Several erythromycin-resistant clones were obtained and further propagated in liquid medium with erythromycin. PCR amplification of plasmid DNA extracted from these clones using primers specific for the egfp gene generated amplicons of the expected size, indicating the presence of the egfp gene in these clones. Similarly, RT-PCR with primers specific for egfp cDNA using RNA extracted from B31 containing pMS1 generated amplicons of the expected size. Expression of egfp in these B31 electroporants was further confirmed by fluorescence microscopy. Fig. 3 shows several micrographs of B31 cells containing the pMS1 plasmid and expressing EGFP. Direct microscopic observation of B31 cells containing pMS1 indicated that EGFP was expressed at low levels in all cells. However, comparison of phase-contrast and fluorescent micrographs (Fig. 3 a and b) shows that the expression of EGFP is only sufficiently intense to be registered by photography in a fraction of B31 electroporants. Exposure of the cells to atmospheric air with gentle shaking increased expression of EGFP (data not shown) (31). Western blots of cell lysates of these B31 electroporants confirmed the presence of a 27-kDa band that reacted with rabbit antibodies against EGFP (Fig. 4a). Additional immunoblot protein bands were also present in B31 cells lacking and containing pMS1; the origin of these bands is unclear. B31 cells containing pMS1 also contained specific mRNA for EGFP (Fig. 4b). Competitive PCR experiments (Fig. 5) indicated that there were equivalent amounts (330 fg/0.5 ng total DNA) of egfp and flaB in B. burgdorferi containing pMS1, suggesting that there is a similar number of copies of pMS1 per chromosome(s) in the recombinant B31 cell.
Figure 2.
Construction of plasmid pMS1 containing the egfp gene. (a) Plasmid pGK12. (b) Ligated PCR amplification products containing the flagellin B promoter-Pfla and egfp gene. (c) Recombinant plasmid pMS1.
Figure 3.
Micrographs of B. burgdorferi B31 containing plasmid pMS1. (a) Phase contrast microphotograph of B. burgdorferi B31 with pMS1; (b–f) Fluorescent microphotographs of B. burgdorferi with pMS1. (g) Phase contrast microphotograph of wild-type B. burgdorferi B31; (h) Fluorescent microphotograph of wild-type B. burgdorferi B31.
Figure 4.
Detection of EGFP and egfp mRNA in B. burgdorferi B31 containing plasmid pMS1. (a) Immunodetection of EGFP. Lane 1, protein lysate of E. coli containing pMS1. Lane 2, protein lysate of wild-type B. burgdorferi B31. Lanes 3 and 4, B. burgdorferi isolates containing pMS1. (b) RT-PCR of egfp RNA. Lane 1, molecular weight standards. Lane 2, amplification of RNA from B. burgdorferi containing pMS1 without reverse transcriptase. Lane 3, amplification of RNA from B. burgdorferi containing pMS1 with reverse transcriptase. Lanes 4 and 5, the same as 2 and 3, but with cultures of B. burgdorferi shaken in the presence of oxygen. Lane 6, RT-PCR performed without RNA.
Figure 5.
Competitive PCR of egfp and flaB DNAs in B. burgdorferi containing pMS1. All reactions except negative controls contained 0.5 ng of total DNA from B. burgdorferi cells with pMS1. In flaB (lanes 1–7) and egfp (lanes 8–14), the competitive amplifications were performed with primers specific to the flaB and egfp genes, respectively. The PCR reactions contained 1 pg of correspondent competitor (lanes 1 and 8), 300 fg (lanes 2 and 9), 100 fg (lanes 3 and 10), 33 fg (lanes 4 and 11), 10 fg (lanes 5 and 12), and 3.3 fg (lanes 6 and 13). Lanes 7 and 14 correspond to reactions carried out without DNA.
Discussion
We have been able to electroporate the Lactococcus lactis-derived, wide-host-range plasmid, pGK12 (11), into B31 and propagate it extrachromosomally for several generations, using the erythromycin resistance gene ermC harbored by pGK12 as a selective marker for genetic experiments in this pathogen. We were also able to use pGK12 as a cloning vector to obtain expression of EGFP in this bacterium. The advantages of pGK12 as a cloning vector in Borrelia over previous systems used in this pathogen to introduce foreign DNA are manyfold. They include its small size, the augmented efficiency of selection of electroporants with erythromycin, the easy isolation of pGK12 DNA from Borrelia because of its sustained extrachromosomal replication, the lack of recombination and integration of pGK12 into the borrelial chromosome, and the increased possibility of manipulation in vitro as pGK12 DNA can be readily isolated in large amounts from E. coli. The improved electroporation frequency we achieved with pGK12 may be because of our use of high concentrations of covalently closed, supercoiled pGK12 DNA, a DNA structure that, as has been shown in other bacterial systems, favors DNA introduction into bacteria because of its resistance to exonucleases (32).
Our success in using erythromycin to select true electroporants may be because of the lack of spontaneous mutants and a sustained postantibiotic effect on the growth of susceptible Borrelia even in the face of the antibiotic inactivation that can occur with the long incubation periods needed to detect putative B31 electroporants (33). Moreover, erythromycin resistance mutations mediated by alternative mechanisms to methylation of the 23S RNA (the mechanism encoded by ermC) will otherwise be very low, as the genetic information for these other functions is not present in the B. burgdorferi genome (1). Analysis of the B. burgdorferi genome indicates that it contains two adenosyl methylase genes with low DNA homology to the staphylococcal adenosyl methylase gene in pGK12 responsible for erythromycin resistance (1, 11). This lack of DNA homology probably explains the inability of the staphylococcal gene to recombine with homologous DNA regions of Borrelia and to insert the vector into the borrelial chromosome (1). Although we avoid generating Borrelia strains that might be resistant to antibiotics in common clinical use for treating Lyme disease, it might be argued that erythromycin itself is occasionally used to treat Lyme disease (34). However, B. burgdorferi strains naturally resistant to erythromycin exist, and we have found high levels of resistance to this antibiotic in the virulent B. burgdorferi N40 strain and in fresh clinical isolates (M.S. and F.C.C., unpublished results). Our use of a noninfectious, high passage B. burgdorferi strain and adequate terminal sterilization of all cultures containing erythromycin-resistant Borrelia minimizes any potential for their escape from the laboratory. Added levels of containment are given by the nonconjugative nature of pGK12, the inability of B. burgdorferi to survive as free bacteria in the environment, and its lack of competence to uptake foreign DNA naturally (11, 35). Interestingly, erythromycin-resistant B31 electroporants did not express resistance to chloramphenicol, whereas E. coli transformed with pGK12 isolated from their DNA were resistant to this antibiotic, indicating that the lack of expression of chloramphenicol resistance in Borrelia was not because of changes in the cm DNA sequence. The lack of expression of chloramphenicol resistance in B31 may be because of the inability of B31 to recognize the transcription and translation signals present in staphylococcal cm, posttranslational modifications of the gene product, and/or the recently described interference of components of reagent rabbit sera with the activity of chloramphenicol acetyl transferase (36).
The ability to introduce foreign plasmid DNA able to replicate extrachromosomally into B31 has not been reported before. Most currently available genetic systems developed for B. burgdorferi have been directed at recombining introduced, nonreplicating DNA into the bacterial chromosome or obtaining transient expression of nonreplicating genes introduced into the bacterial cytoplasm (5, 9). The ability of pGK12 to replicate extrachromosomally suggests that DNA replication signals present in pGK12 are recognized by gene products involved in the replication of borrelial DNA, and underlines the conservation of DNA replication functions across different bacterial species (37). The role played by plasmid genes and their gene products in replication of plasmid DNA in Borrelia vis-à-vis replication of Borrelia DNA gene and gene products will need further study (37). For example, it has been shown that host-encoded RNA polymerase is involved in the conversion of pGK12 ssDNA to dsDNA in B. subtilis but not in L. lactis, indicating that the plasmid may make use of different enzymes in different hosts to achieve replication (37).
The ability of pGK12 to serve as a cloning vector for the expression of foreign genes in Borrelia, shown by the expression of both staphylococcal ermC and jellyfish egfp genes, confirms that the transcriptional and translational apparatuses of Borrelia can recognize transcriptional and translational signals of other organisms (38). Competitive PCR copy number studies suggested that there is a similar number of copies of pMS1 and chromosomes in each B31 cell. This low copy number may explain the relatively high frequency of segregation of the plasmid while growing in media without erythromycin, the low levels of expression of the egfp gene, and the lack of deleterious metabolic effects in Borrelia expressing this foreign gene. These findings also suggest that pMS1 can be an adequate vehicle to study gene regulation in this bacteria as alternative Borrelia promoters can be cloned upstream the egfp gene (39). The facts that pGK12 can propagate in B. burgdorferi, that 25% of the cells are able to segregate pGK12 in medium without antibiotic, and that the plasmid does not recombine with the borrelial chromosome suggest that pGK12 can be potentially used as a platform to introduce unique in vitro-inactivated genes and transposons into the chromosome of Borrelia. Electroporation of Borrelia with pGK12 containing the wild-type allele complementing these mutants will open the way for the efficient functional genetic analysis of the putative virulence genes of this pathogen. There is also little doubt that a series of pGK12-derived and improved cloning vectors for Borrelia can be constructed with multiple cloning sites and strong Borrelia promoters and translational signals, as well as with the ability to generate gene fusions and to insert in the chromosome. However, our experience suggests that the improvement of the cloning vectors will be restricted by the fact that one of the limiting factors in developing a borrelial cloning vector may be the size of the DNA that can be introduced by electroporation in this pathogen.
Acknowledgments
We thank Gerard Venema for kindly providing us with pGK12 and Daniel R. Zeigler for pHP13. Victoria Gorbacheva provided us with essential and critical advice at the beginning of this research. Marina Nechaeva gave us advice and reagents for experiments with EGFP. Stuart A. Newman generously gave us advice about and allowed us the use of his fluorescent microscopes. Henry P. Godfrey advised us on computer analysis of DNA sequences, immunological methods, and in the preparation of the manuscript. Harriett V. Harrison contributed to the preparation of the manuscript. This work was supported by Public Health Service Grant R01 AI 43063 (to F.C.C.).
Abbreviations
- B31
Borrelia burgdorferi strain B31
- GFP, green fluorescence protein
EGFP, enhanced GFP
- MIC
minimal inhibitory concentration
- RT-PCR
reverse transcription–PCR
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080068797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080068797
References
- 1.Fraser C M, Casjens S, Huang W M, Sutton G G, Clayton R, Lathigra R, White O, Ketchum K A, Dodson R, Hickey E K, et al. Nature (London) 1997;390:580–586. doi: 10.1038/37551. [DOI] [PubMed] [Google Scholar]
- 2.Samuels D S, Garon C F. Antimicrob Agents Chemother. 1993;37:46–50. doi: 10.1128/aac.37.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lafay B, Lloyd A T, McLean M J, Devine K M, Sharp P M, Wolfe K H. Nucleic Acids Res. 1999;27:1642–1649. doi: 10.1093/nar/27.7.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Picardeau M, Lobry J R, Hinnebusch B J. Mol Microbiol. 1999;32:437–445. doi: 10.1046/j.1365-2958.1999.01368.x. [DOI] [PubMed] [Google Scholar]
- 5.Sohaskey C D, Arnold C, Barbour A G. J Bacteriol. 1997;179:6837–6842. doi: 10.1128/jb.179.21.6837-6842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuels D S, Mach K E, Garon C F. J Bacteriol. 1994;176:6045–6049. doi: 10.1128/jb.176.19.6045-6049.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Samuels D S, Marconi R T, Huang W M, Garon C F. J Bacteriol. 1994;176:3072–3075. doi: 10.1128/jb.176.10.3072-3075.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samuels D S, Garon C F. Microbiology. 1997;143:519–522. doi: 10.1099/00221287-143-2-519. [DOI] [PubMed] [Google Scholar]
- 9.Stevenson B, Bono J L, Elias A, Tilly E, Rosa P. J Bacteriol. 1998;180:4850–4855. doi: 10.1128/jb.180.18.4850-4855.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie T D, Tsong T Y. Biophys J. 1992;63:28–34. doi: 10.1016/S0006-3495(92)81580-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kok J, van der Vossen J M B M, Venema G. Appl Environ Microbiol. 1984;48:726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haima P, Bron S, Venema G. Mol Gen Genet. 1987;209:335–342. doi: 10.1007/BF00329663. [DOI] [PubMed] [Google Scholar]
- 13.Horinouchi S, Weisblum B. J Bacteriol. 1982;150:804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horinouchi S, Weisblum B. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luchansky J B, Muriana P M, Klaenhammer T R. Mol Microbiol. 1988;2:637–646. doi: 10.1111/j.1365-2958.1988.tb00072.x. [DOI] [PubMed] [Google Scholar]
- 16.Cabello F, Timmis K, Cohen S N. Nature (London) 1976;259:285–290. doi: 10.1038/259285a0. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Stoenner H G. Appl Microbiol. 1974;28:540–543. doi: 10.1128/am.28.4.540-543.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Waterworth P M. In: Laboratory Methods in Antimicrobial Chemotherapy. Reeves D S, Philips I, Williams J D, Wise R, editors. Edinburgh: Churchill Livingstone; 1978. pp. 31–40. [Google Scholar]
- 20.Samuels D S. Methods Mol Biol. 1995;47:253–259. doi: 10.1385/0-89603-310-4:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chung G T, Niemela S L, Miller R H. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zimmermann K, Mannhalter J W. In: The PCR Technique: Quantitative PCR. Larrick J W, editor. Natick, MA: Eaton; 1997. pp. 3–17. [Google Scholar]
- 23.Chomczynski P, Sacchi N. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 24.Dever L L, Jorgensen J H, Barbour A G. J Clin Microbiol. 1992;30:2692–2697. doi: 10.1128/jcm.30.10.2692-2697.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agger W A, Callister S M, Jobe D A. Antimicrob Agents Chemother. 1992;36:1788–1790. doi: 10.1128/aac.36.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dever L L, Jorgensen J H, Barbour A G. Antimicrob Agents Chemother. 1993;37:1115–1121. doi: 10.1128/aac.37.5.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gasser R, Wendelin I, Reisinger E, Bergloff J, Feigl B, Schafhalter I, Eber B, Grisold M, Klein W. Infection. 1995;23:S39–S43. doi: 10.1007/BF02464959. [DOI] [PubMed] [Google Scholar]
- 28.Kazragis R J, Dever L L, Jorgensen J H, Barbour A G. Antimicrob Agents Chemother. 1996;40:2632–2636. doi: 10.1128/aac.40.11.2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Millner M M, Thalhammer G H, Dittrich P, Spork K D, Brunner M, Georgopoulos A. Infection. 1996;24:174–177. doi: 10.1007/BF01713334. [DOI] [PubMed] [Google Scholar]
- 30.Chris D, Resnekov O. Methods Enzymol. 1999;302:136–153. doi: 10.1016/s0076-6879(99)02015-7. [DOI] [PubMed] [Google Scholar]
- 31.Heim R, Prasher D C, Tsien R Y. Proc Natl Acad Sci USA. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joseph J W, Kolodner R. J Biol Chem. 1983;258:10411–10417. [PubMed] [Google Scholar]
- 33.Champney W S, Tober C L. Antimicrob Agents Chemother. 1999;43:1324–1328. doi: 10.1128/aac.43.6.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadelman R B, Wormser G P. Lancet. 1998;352:557–565. doi: 10.1016/S0140-6736(98)01146-5. [DOI] [PubMed] [Google Scholar]
- 35.Barbour A G. Yale J Biol Med. 1984;57:521–525. [PMC free article] [PubMed] [Google Scholar]
- 36.Sohaskey C D, Barbour A G. Antimicrob Agents Chemother. 1999;43:655–660. doi: 10.1128/aac.43.3.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leenhouts K J, Tolner B, Bron S, Kok J, Venema G, Seegers J F M L. Plasmid. 1991;26:55–66. doi: 10.1016/0147-619x(91)90036-v. [DOI] [PubMed] [Google Scholar]
- 38.Aron L, Toth C, Godfrey H P, Cabello F C. FEMS Microbiol Lett. 1996;145:309–314. doi: 10.1111/j.1574-6968.1996.tb08594.x. [DOI] [PubMed] [Google Scholar]
- 39.Valdivia R H, Falkow S. Mol Microbiol. 1996;22:367–368. doi: 10.1046/j.1365-2958.1996.00120.x. [DOI] [PubMed] [Google Scholar]