Abstract
Cellular functions are determined by interactions among proteins in the cells. Recognition of these interactions forms an important step in understanding biology at the systems level. Here, we report an interaction network of Escherichia coli, obtained by training a Support Vector Machine on the high quality of interactions in the EcoCyc database, and with the assumption that the periplasmic and cytoplasmic proteins may not interact with each other. The data features included correlation coefficient between bit score phylogenetic profiles, frequency of their co-occurrence in predicted operons, and a new measure—the distance between translational start sites of the genes. The combined genome context methods show a high accuracy of prediction on the test data and predict a total of 78,122 binary interactions. The majority of the interactions identified by high-throughput experimental methods correspond to indirect interaction (interactions through neighbors) in the predicted network. Correlation of the predicted network with the gene essentiality data shows that the essential genes in E. coli exhibit a high linking number, whereas the nonessential genes exhibit a low linking number. Furthermore, our predicted protein–protein interaction network shows that the proteins involved in replication, DNA repair, transcription, translation, and cell wall synthesis are highly connected. We therefore believe that our predicted network will serve as a useful resource in understanding prokaryotic biology.
Living systems are made up of molecular entities, interactions among which give rise to the complex properties of life that are not apparent in the individual molecules. The complex properties of life can now be addressed at the systems level because of the availability of complete genome sequences of several organisms. Proteins being the dominant molecules of life, much focus has been on understanding protein–protein interactions in the recent past (Barabasi and Oltvai 2004). A wide range of experiments has been carried out in order to map genome-wide protein–protein interactions in different organisms (Rain et al. 2001; Li et al. 2004; Suthram et al. 2005; Krogan et al. 2006; Stelzl et al. 2006). The high-throughput experiments typically limit the observations to only a fraction of all possible interactions since the experiments are carried out under unique conditions. This has led to the development of many algorithms to predict genome-wide protein–protein interactions (Bork et al. 2004). By understanding the network of these complex interactions, it is hoped that our understanding of the living systems will be enhanced considerably.
The interactions between proteins can be classified into two major categories: physical and functional. The former refer to physical association between two proteins, whereas the latter refer to the proteins in a biochemical or a signaling pathway. Inferences of functional interactions can be obtained from methods such as coexpression data from microarray analysis. Physical interactions, on the other hand, can be detected by direct experimental techniques such as pull-down assays, coimmunoprecipitation, or tandem affinity purification coupled to mass spectrometry. Such techniques are also amenable to high-throughput experimentation.
Experimental detection of genome-wide protein–protein interactions is costly, time-consuming, and difficult to implement in a modestly equipped laboratory. Moreover, there is little overlap between the results obtained from different experimental methods (von Mering et al. 2002; Arifuzzaman et al. 2006; Krogan et al. 2006). There thus exists a substantial scope for computational analysis in the areas of evaluation of the quality of experimental data, and more importantly in predicting genome-wide protein–protein interactions.
Algorithms that identify genome-wide interactions between proteins mainly focus on coregulation of interacting proteins. These methods assume that the genes that are coregulated often occur close to each other on the genomes and show conserved gene order. Thus the genes, which are part of an operon, could be functionally linked (Dandekar et al. 1998; Overbeek et al. 1999). These methods, however, fail to identify functional links between the proteins that are distantly located on a genome. A modified method has been used to infer functional links between genes that are distantly located in one genome but whose orthologs may be a part of an operon in another genome (Janga et al. 2005).
The coevolution method attempts to calculate evolutionary rates of substitutions in different proteins and assumes that the proteins that evolve at similar rates are parts of an interacting pair (Fraser et al. 2002). Alternatively, the phylogenetics profile method detects protein interactions by identifying correlations between the presence and the absence of genes across the genomes (Pellegrini et al. 1999).
In this study, we propose a computational method that identifies protein–protein interactions on the basis of distance between translational start sites of two genes and the frequency of co-occurrence of the genes in predicted operons. These two features are combined with phylogenetics profiles using a Support Vector Machine to infer the genome-wide functional linkages in Escherichia coli. The Support Vector Machine when trained on high-quality experimental data performs well in blind tests, with the accuracies of prediction often exceeding 85%. The network of interactions constructed using our predictions exhibits characteristic scale-free topology and an excellent correlation with the experimental gene essentiality data. We therefore believe that the network of interactions predicted by our method will be a useful tool in understanding the biology of E. coli.
Results and Discussion
Data for prediction of protein–protein interactions
Interactions between pairs of proteins were derived using a Support Vector Machine (SVM) trained on the interactions reported in the EcoCyc database (Keseler et al. 2005). These linkages represent hand-curated data obtained essentially through low-throughput experimental approaches. Moreover, these interactions are deduced from multiple experiments and are therefore likely to be free of the false positives or bias that is often associated with high-throughput experiments. Supplemental Tables I and II list the data sets used for supervised learning. The learning set contained 1082 proteins pairs, which were further divided into operonic (654) (Supplemental Table I) and non-operonic data sets (428) (Supplemental Table II) as defined in the EcoCyc database.
Defining a reliable negative data set for predicting functional linkages using machine-learning techniques has been acknowledged to be a difficult problem (Jansen et al. 2003; Ben-Hur and Noble 2006). The negative data for predicting functional linkages in eukaryotes were assumed to be those proteins that are not colocalized in the same subcellular compartment (Jansen et al. 2003). Similarly, in prokaryotes it might seem reasonable that the proteins required for physiological functions in the extracellular or the periplasmic milieu would not physically interact with the proteins of the cytoplasmic space. We therefore formulated a negative data set, in which each protein pair comprises one secreted protein and the other localized in the cytoplasm. Thus, proteins with a known secretory signal were first identified, and were considered separately from the proteins without any signal peptide. The top-scoring 40 proteins with the predicted signal sequence at the N terminus were considered to be periplasmic, while 346 proteins that do not possess any of the known signal sequences along the entire length of the polypeptide were considered to be cytoplasmic (Supplemental Table III). Combining one periplasmic and one cytoplasmic protein therefore generated 13,840 pairs. The pairs in the negative data set did not belong to any known complex, or metabolic pathway, and therefore were assumed to lack functional linkages.
Having defined the positive and the negative data sets for supervised machine learning, predictions were carried out for assessing functional linkages between all the possible protein pairs in the E. coli K12 genome. Three different data features, namely, frequency of co-occurrence in predicted operons, phylogenetic profile correlation score, and minimum distance between the two genes on any of the 266 genomes, were used for these predictions as described in Methods.
Frequency of co-occurrence in predicted operons
Several different prediction methods exist in the literature for the identification of operons. These are based on features such as intergenic distance, functional correlation, and conserved gene neighborhood (Salgado et al. 2000; Ermolaeva et al. 2001; Zheng et al. 2002). We used a Support Vector Machine to identify operons in all the prokaryotic genomes using intergenic distance between the transcription start sites of the genes as the data feature.
The average accuracy of fivefold cross-validation for operon prediction was 82%. The model with the highest accuracy was used to predict operons in the 124 prokaryotic genomes. This led to the prediction of a total of 786 polycistronic transcription units in E. coli K12.
Phylogenetic profile method
The phylogenetic profile of a gene can be represented in two different ways: a binary profile (Pellegrini et al. 1999) or a normalized bit score profile (Enault et al. 2003). In the former, the presence or absence of a homolog is represented as 1 or 0, respectively, whereas in the latter, the same is represented as the normalized bit score obtained from BLAST. Two genes displaying a similar phylogenetic profile, as assessed by Pearson correlation coefficient, therefore can be assumed to be functionally linked. We constructed phylogenetic profiles for all the genes of E. coli based on both the methods.
Effect of gene conservation on prediction accuracy
Assessment of functional linkage between two genes may not always be possible by inspection of their phylogenetic profiles, especially if these two genes are present in only a few closely related organisms. Moreover, the phylogenetic profiles of the genes that are specific to a lineage are likely to show a high correlation coefficient among them, since these are absent in a large number of genomes, but present only in a few closely related species. This might result in reporting false-positive interactions among these genes. Thus, the choice of training the SVM using these data can critically affect the results of prediction. This is especially true for the negative training data. We therefore assessed the effect of training data on the accuracy of predictions by taking into consideration the conservation of genes in the phylogenetic profiles.
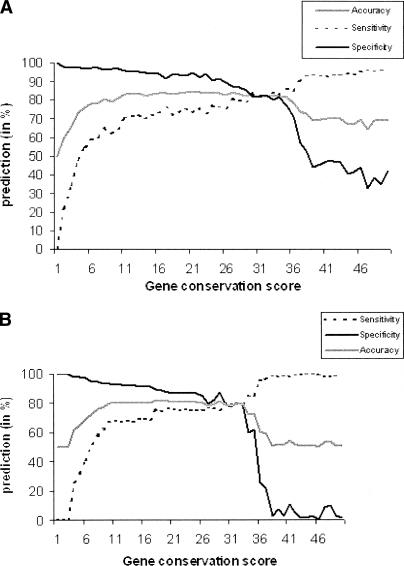
The conservation score for any gene was defined as the total number of genomes that possess homologs of the gene under consideration (Sun et al. 2005). The conservation score of a pair of genes was, in turn, defined as the minimum conservation score of any of the two genes in the pair. To understand the effect of conservation score of a gene pair on prediction accuracy of the phylogenetic profile method using bit score, we used two-thirds of the randomly picked data from the positive and negative data sets for training and the remaining one-third for testing the prediction accuracy. The process was repeated 50 times, each time by incrementing the cutoff conservation score by 1 in the negative data set while retaining the same positive data. Figure 1A shows that the prediction accuracy of protein–protein interactions gradually increases until the conservation score of 20 because of an increase in sensitivity, and then decreases because of a decrease in specificity. The accuracy remains constant beyond the conservation score of 12, and therefore we considered phylogenetic profiles of the genes that are conserved in at least 12 genomes.
Figure 1.
Effect of gene conservation score on the accuracy of protein–protein interaction predictions using phylogenetic profiles. (A) Profiles of the two genes in a pair are compared using the normalized bit scores. (B) Profiles of the two genes in a pair are compared using the presence (1) or absence (0) of the genes. Accuracy is defined as the average of sensitivity and specificity as described in Methods. It is clearly seen that the prediction accuracy is poorer at low and high conservation scores. See text for details.
Similar analysis was performed with the binary phylogenetic profile, where the outcome of the analysis was also similar. These results, as shown in Figure 1B, lead to two major conclusions: (1) the prediction accuracy of both the methods increases by increasing the cutoff value of the conservation score for the genes in the negative data set, and (2) the sensitivity of the predictions is low at low conservation scores, while the specificity is low at high conservation scores. Specificity rapidly decreases for both the methods at high conservation scores. However, for the binary phylogenetic profiles, the specificity drops to zero at high conservation scores.
Gene distance method
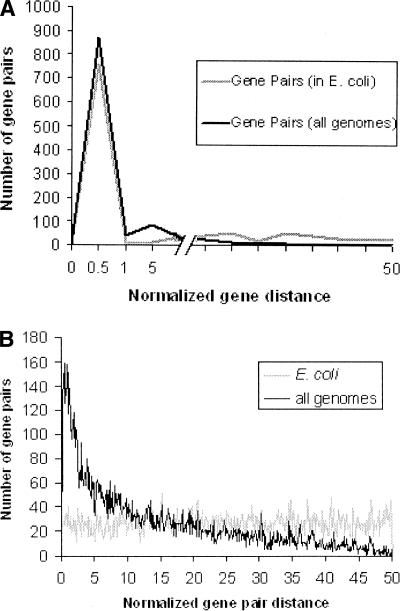
Genes that are closer to each other usually show stronger functional linkages and the possibility of coregulation by an operon or transcriptional coupling (Korbel et al. 2004). Analysis of the 1082 gene pairs that code for components of protein complexes showed that a large number of them (428) are not part of operons. These gene pairs were distantly located in the E. coli K12 genome, but interestingly their orthologs were closer to each other in at least one other genome as shown in Figure 2A. This suggests that functionally related genes tend to occur close to each other in at least one genome.
Figure 2.
Normalized gene distance distributions among the data sets used for prediction of protein–protein interactions. The normalized gene pair distance is defined as the number of base pairs that separate the translation start sites of two genes in a pair, divided by the total genome length and then scaled to 100. (A) (Gray) Normalized distances between the known interacting protein pairs in E. coli; (black) the minimum normalized distance between the same proteins on any of the other genomes. (B) Normalized distances between the protein pairs, which are hypothesized to be noninteracting. (Gray line) The normalized distance distribution in E. coli; (black line) the minimum of this between the same pair of proteins in any other genome.
Intergenic distance was analyzed for all the hypothesized noninteracting gene pairs in the negative data set (Fig. 2B). Interestingly, the normalized distance between the translational start sites in these pairs of proteins is evenly distributed between 0 (minimum possible normalized distance) and 50 (maximum possible normalized distance) in E. coli. The same distribution is skewed toward lower values when a minimum normalized distance between the same pairs across all the genomes was calculated. Comparison of the normalized intergenic distances across all the genomes showed that 80% of the interacting proteins are spaced closely (normalized gene distance <1) in at least one genome, whereas only 10% of the hypothesized noninteracting pairs possess a normalized gene distance <1.
Cross-validation accuracy of prediction of protein–protein interactions
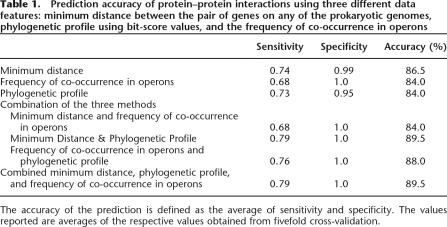
To compare the accuracy of prediction of different methods for protein–protein interactions, we calculated the average sensitivity and specificity of each prediction by fivefold cross-validation. Table 1 shows the comparison of fivefold cross-validation for the three different data features and their combinations. The combination of all the three data features appears to be the best choice for prediction of protein–protein interactions. The average accuracy of predictions reaches as high as 89.5% in the best prediction. The network predicted using two features, namely, minimum distance and phylogenetic profile, showed similar sensitivity and specificity values as those predicted using all three data features. Nonetheless, the number of false positives predicted during cross-validation using only two features was higher than that using three features. Therefore, we used all three data features to predict genome-wide functional linkages.
Table 1.
Prediction accuracy of protein–protein interactions using three different data features: minimum distance between the pair of genes on any of the prokaryotic genomes, phylogenetic profile using bit-score values, and the frequency of co-occurrence in operons
The accuracy of the prediction is defined as the average of sensitivity and specificity. The values reported are averages of the respective values obtained from fivefold cross-validation.
Genome-wide prediction of protein interactions in E. coli
The prediction of genome-wide interactions in E. coli resulted in a total of 78,122 binary interactions (Table 1). The high specificity values clearly suggest that the number of false-positive interactions in the predicted network would be minimal. On the other hand, sensitivity being determined by the total number of identified interactions among all the theoretically possible interactions, lower sensitivity values suggest that the total number of interactions reported by us might represent a lower estimate. Thus, we believe that the number of all pairwise protein–protein interactions in E. coli will be in excess of 78,122.
Comparison of the predicted network with high-throughput experimental data
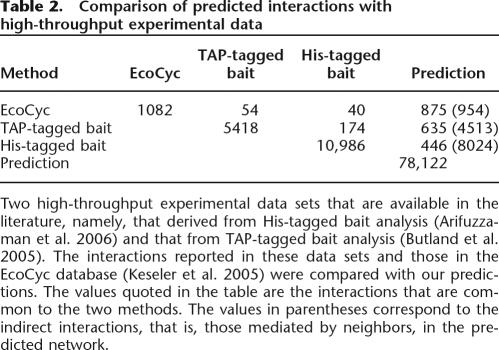
The predicted protein–protein interactions were compared with the results of two high-throughput experimental data sets (Butland et al. 2005; Arifuzzaman et al. 2006). It was observed that among the 5418 interactions identified by the pull-down assays by the TAP-tagged bait protein method, our method predicted 635 direct interactions and 4513 indirect (binary, ternary, or quaternary) interactions (Butland et al. 2005). Interestingly, 254 protein pairs among the 635 direct interactions are reportedly of high confidence (Butland et al. 2005). On the other hand, our method identified 446 direct and 8024 indirect interactions out of the total 10,986 unique interactions determined by pull-down assays using the His-tagged bait method (Arifuzzaman et al. 2006; Table 2).
Table 2.
Comparison of predicted interactions with high-throughput experimental data
Two high-throughput experimental data sets that are available in the literature, namely, that derived from His-tagged bait analysis (Arifuzzaman et al. 2006) and that from TAP-tagged bait analysis (Butland et al. 2005). The interactions reported in these data sets and those in the EcoCyc database (Keseler et al. 2005) were compared with our predictions. The values quoted in the table are the interactions that are common to the two methods. The values in parentheses correspond to the indirect interactions, that is, those mediated by neighbors, in the predicted network.
The overlap between our predictions and the experimental studies is higher when we consider indirect interactions. This might be because one may identify proteins that interact with bait directly, or indirectly while using pull-down assays. It is therefore likely that the experimental techniques not only identify direct physical interactions, but also complexes involving several proteins. Taking this into consideration, we find that the overlap between our predicted set and the two experimental data sets is quite high (∼80%).
A large number of new interactions are predicted by our method. These interactions could be physical, or might represent functional linkages not necessarily of physical character. For example, it is known that LacI regulates expression of the lac operon. However, LacI does not interact physically with any of the three structural genes of the lac operon. Such interactions between LacI and the genes of lac operon cannot be identified using pull-down methods. Similarly, our predictions identified interactions between RpoA and Sec A, B, D. These interactions might also represent functional linkages between these proteins. As is well known, transcription and translation in prokaryotes are coupled processes, and the Sec-dependent pathway is also coupled to the translation process. Thus, proteins of the transcription machinery might be functionally linked to the Sec A, B, and D proteins. These examples suggest that the present method not only predicts physical interactions but also provides insights into regulatory and functional linkages.
Analysis of predicted protein–protein interaction network
Most of the real world networks are known to possess scale-free topology (Barabasi and Albert 1999). The most important characteristic of the scale-free networks is that the degree distribution (distribution of number of links) follows the power law, that is, p(k) ∼ ak−γ, where γ < 3. The scale-free networks possess a small number of nodes with a large number of links, whereas a large number of nodes possess very few links (Barabasi and Bonabeau 2003). We analyzed the scale-free behavior of our predicted interaction network, which is described in the following discussion.
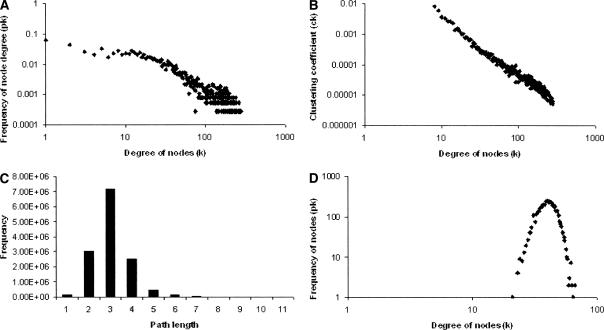
The degree distribution of the predicted network with 3798 nodes and 78,122 edges follows the power law with an average number of links being 41 and the degree exponent of 1.26 (Fig. 3A). This indicates that the network possesses a high diversity of node degree. Moreover, the node degree is similar to that obtained by the two experimental methods: TAP-tagged bait-associated proteins (1.24), and His-tagged bait-associated proteins (1.41). A degree <2 implies that the total number of links grows faster than the total number of nodes (Seyed-Allaei et al. 2005). Further analysis of the clustering coefficient, C(k), revealed that the distribution of clustering coefficient also follows the power law [C(k) ∼ k−β], with modularity exponent β = −1.94, suggesting that the low-degree nodes are more cohesive than the high-degree nodes (Fig. 3B). This, in turn, illustrates the disassortive nature of the E. coli interaction network, indicating that the high-degree nodes in general are not linked to the other high-degree nodes.
Figure 3.
Topological features of the predicted protein–protein interaction network in E. coli K12. (A) The degree distribution clearly shows that the network follows scale-free properties. (B) The distribution of the clustering coefficient shows that the network is not hierarchical. Both the axes represent the respective logarithmic values. (C) Distribution of the shortest path between pairs of proteins in the predicted network. (D) The null network possesses a Gaussian degree distribution as observed in the log–log plot.
The number of links (path length) required to connect each node to every other node in the predicted network was determined, which is also referred to as the “small world property” (Fig. 3C). The diameter of the predicted network, that is, the longest graph distance between any two nodes, was found to be 11. This implies that the nodes are densely linked and thus possess small world properties. In other words, small metabolic perturbations might not affect the overall functioning of the organism.
Hubs in protein interaction networks have been postulated to represent essential genes in a genome (Jeong et al. 2001; Albert 2005). Indeed, we observed that the proteins like DNA polymerase subunits (HolA and HolC) and the RNA polymerase subunits (RpoA, RpoB, RpoC, and RpoZ) that are essential for survival possess a degree >100. Further analysis revealed that >80% of the known essential genes possess a degree >41 (Baba et al. 2006). This is in accordance with the earlier observation that essential genes are densely connected (Yu et al. 2004). The average number of interactions for essential genes in our predicted network was 115 (Supplemental Fig. 1a).
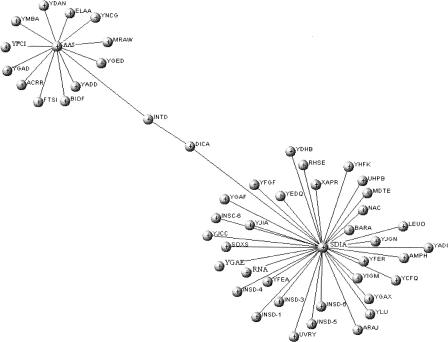
Degree distribution of the essential genes showed that certain essential genes possess degrees as low as 2–3. As suggested by Pržulj et al. (2004), lethal nodes are not always the highly connected nodes in the network. Certain nodes are lethal because their deletion disconnects the network into two subgraphs and thereby disrupts the network structure. Such nodes have been referred to as “articulation points.” Analysis of one of the low-degree essential genes, dicA, supported this argument. DicA is a prophage-based transcriptional regulator and regulates the expression of cell division inhibitor DicB. DicA has been predicted to interact with the prophage integrase IntD and a DNA-binding transcriptional activator, SdiA (Fig. 4). In our predicted network, SdiA interacts with 35 proteins, whereas IntD interacts with Aas (2-acyl-glycerophospho-ethanolamine acyltransferase). Aas, in turn, interacts with 13 other proteins. The topology of the subnetwork clearly shows that although DicA possesses a low degree, its elimination would disrupt the connection between SdiA and Aas subgraphs. Intriguingly, the topology of the subnetwork suggests that IntD would also behave similarly to DicA, yet IntD is not known to be essential. The nonessentiality of IntD could be attributed to the occurrence of several prophage integrase genes in the E. coli genome, which might function analogously.
Figure 4.
DicA subnetwork. dicA is known to be an essential gene in E. coli. Intriguingly, we observed that DicA has a degree of only 2. However, as is clear from the subgraph, deletion of DicA leads to disconnected islands in the subnetworks, thereby offering an explanation to its essentiality.
Another important correlation of our predicted network with the available experimental data relates to the list of nonessential genes. Analysis of the degree distribution of 741 genes identified to be nonessential by Posfai et al. (2006) interestingly showed that the average number of interactions (〈k〉) for this set of genes is only 18 (Supplemental Fig. 1b). The 〈k〉 for nonessential genes is very low compared to the same for the complete network (〈k〉 = 41) or for the essential genes (〈k〉 = 115). This observation further supports the good agreement between the predicted interaction network and independently obtained experimental data.
To further enhance confidence in our predicted network, a null network was constructed by randomizing the edges in the network to generate 78,122 interactions using the same number of nodes. The degree distribution was analyzed for this network. The random network possessed a Gaussian degree distribution unlike the scale-free predicted network (Fig. 3D). Moreover, in the random network, the average number of interactions (〈k〉) for the essential genes was 40.8, and that for the nonessential genes was 30.8. This suggests that the predicted network is biologically relevant and provides useful information.
We further observed that ∼2120 proteins interact directly or indirectly with the essential genes. The shortest path-length distribution of the essential gene network showed that a large number of nodes are connected to the essential genes by a path length of 3 (Supplemental Fig. 1c). Similarly, the path-length distribution of the nonessential gene network showed that most proteins interact with the nonessential genes through at least four nodes (Supplemental Fig. 1d). The diameter for the essential gene network was found to be 8, whereas for the nonessential gene network it was 14. Thus, not only the degree distribution but also the small world properties are able to distinguish between the experimentally characterized essential and nonessential genes.
Functional analysis of predicted network
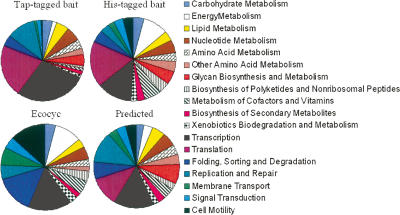
It is of great interest to analyze the protein–protein interactions in the context of their functional categories. KEGG classifies all the E. coli proteins in 19 different functional categories. We observed that the important cellular pathways—transcription and translation—possess the highest average degree of interaction in all the networks (Fig. 5). Our predicted network also showed a high average degree for replication and repair pathways. Interestingly, all the metabolic pathways possessed smaller average degrees. This appears to suggest that perturbations in transcription or translation are more likely to affect the physiology of the cells than perturbations in the metabolic pathways. Interestingly, Pržulj et al. (2004) have earlier observed that in the yeast protein–protein interaction network, stress and defense and transport pathways are less connected than transcription and translation. It was also observed that proteins involved in cellular organization possess a low degree but consist of the highest articulation points. Encouragingly, a high average degree was observed for the cell wall synthesis pathway in the predicted network. Thus, the overall physiology of a prokaryotic cell might be determined by the processes involved in the maintenance and expression of genetic information and those in cell wall biosynthesis, rather than those in the metabolic processes.
Figure 5.
Comparative analysis of average degree of interactions between proteins belonging to different functional pathways in the experimental and the predicted interactomes. It is clear from the distributions that transcription and translation processes exhibit the largest average degree.
An interesting analysis of the predicted protein–protein interaction network pertains to the well-known thioredoxin system. The thioredoxin system plays an important role in maintaining the cellular environment in reduced conditions and thereby enabling proper functioning of several enzymes. Thioredoxin, although overexpressed during oxidative stress, is essential for proper functioning of a large number of proteins involved in the light-activated Calvin cycle and transcription regulation (Arner and Holmgren 2000). E. coli K12 possesses two thioredoxins, TrxA and TrxC, that are reduced by thioredoxin reductase, TrxB. In our predicted network, TrxA was observed to interact with 33 proteins, TrxB with 47 proteins, and TrxC with 12 proteins (Supplemental Fig. 2). Interestingly, out of the total 85 proteins, 80 possess at least one cysteine residue. Since the principal mechanism of the thioredoxin-mediated redox reaction is via dithiol exchange, it appears that the 80 proteins might indeed be natural substrates of the thioredoxins. Occurrence of the well-known physiological partners of thioredoxins such as ribonucleotide reductase (Rnr), alkyl hydroperoxide reductase (AhpC), and chaperone protein (DnaK) suggest that our predicted network of thioredoxins might be of high confidence (Kumar et al. 2004). Apart from the known partners, we also found proteins such as the predicted thioredoxin-domain-containing protein YbbN, the predicted transferase with NAD(P)-binding domain YbhK, and the predicted oxidoreductase YjjX to interact with the thioredoxins. Thus, the predicted thioredoxin subnetwork not only substantiates the known interactions but also enables identification of possible additional pathways regulated by the thioredoxin system.
Yet another observation from our predictions pertains to the toxin–antitoxin system that helps bacteria maintain segregational stability and fight stress conditions. Several toxin–antitoxin pairs have been identified in E. coli (Hayes 1998). In our predicted network, all the toxin–antitoxin pairs (TA) have been predicted as interacting partners. RelBE, one of the well-characterized TA pairs, is known to alter gene expression levels during amino acid and carbon source starvation. We observe that the identification of the glutamine transporter (GlnQ) and ribose-5-phosphate isomerase (RpiB) as RelE-interacting partners is not surprising (Supplemental Fig. 3a). Interestingly the Nickel transporter subunit NikE and the transcription regulator of Manganese transport protein MntH, MntR, were also predicted to interact with RelE. This suggests that in addition to nutrient stress, RelBE might play a role in overcoming heavy metal stress. Furthermore, a few hypothetical proteins such as YnjB and YnjC, which have been predicted to act as membrane transporters, also occur in the RelBE subnetwork. In other words, the RelBE system might enable cells to recover from a variety of different stresses. This is in accordance with the fact that the RelBE locus tends to protect cells from the detrimental effects of stress, rather than being suicidal (Pedersen et al. 2002).
E. coli possesses two paralogs of the RelBE system, RelBEK12 and RelBESOS. RelBESOS, also known as YafQ/DinJ, possesses a LexA-binding site in its promoter region and therefore gets activated during the SOS response elicited by DNA damage (Lewis et al. 1994). The physiological role of YafQ/DinJ is not well characterized. However, on the basis of predicted interacting partners, we are able to propose that YafQ/DinJ might inhibit HsdR, an endonuclease, thereby allowing perpetuation of the modified DNA (Supplemental Fig. 3b). In addition to allowing DNA modification to combat the SOS response, YafQ is also predicted to interact with the enzymes of tryptophan and glutamine biosynthesis. Although the functional implications of this observation are not clear, we propose that YafQ is involved in repression of amino acid biosynthesis. This implies that YafQ/DinJ protect the cell from DNA damage by allowing DNA modification and inhibiting protein synthesis.
The excellent correlation between our predicted protein interaction network in E. coli K12 and a wide variety of experimental data suggests that this interaction network will be a useful resource to understand the biology of E. coli. The network can be used, for example, to identify functions of the genes that have been annotated as hypothetical, or of function unknown. We suggest that the cliques in the network, which include genes with characterized functions and a few genes with unknown function, will be able to assign the pathway in which such genes reside. Another interesting feature that can be addressed using the interaction network is to identify large multiprotein complexes by the identification of cliques. Analysis of the modular nature of our predicted E. coli interactome can help in deciphering unknown functions and pathways.
Methods
Completely sequenced genomes of 295 bacteria were downloaded from the NCBI ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/). Bacteria with linear genomes were not considered for the analysis. A total of 266 bacteria genomes remained after excluding the bacteria with linear genomes (Supplemental Table VII). The homologous sequences of all the known open reading frames (ORFs) of E. coli K-12 were searched using BLASTp against the 266 genomes with e−04 as the cutoff value. Orthologs of the E. coli genes were identified by bidirectional best BLAST hit (Hirsh and Fraser 2001). Wherever genome sequences of different bacterial strains of the same species were available, we selected the one that shares the maximum number of orthologs with E. coli K12 to reduce the bias in phylogenetic profiles. Therefore, 124 genome sequences belonging to different bacterial species were used for generating the phylogenetic profiles and for determining the frequency of co-occurrence of a gene pair (Supplemental Table VIII).
Phylogenetic profiles
Bit scores for all the open reading frames of E. coli were obtained by BLAST against 124 genomes, and these were then used to generate a profile across 124 genomes, which was doubly normalized: (1) each bit score of a profile was divided by the maximum value of the bit score over all the genes; (2) the minimum bit score of the E. coli ORFs among all its homologs in other genomes was considered. In order to assess if a pair of genes were coevolving, the Pearson correlation coefficient of their respective phylogenetic profiles was calculated. The E. coli genome encodes 4237 proteins. A 4237 × 4237 symmetric matrix was constructed, where each of the matrix elements was represented by the correlation coefficient thus calculated.
Frequency of co-occurrence in predicted operons
A total of 1113 operon sequences were downloaded from the EcoCyc database (Keseler et al. 2005). Partially identified operons and those with the alternative transcription termination were excluded. Among the remaining 699 operons, there were 119 operons with at least one gene whose name was not present in the gene coordinate file (NC_000913.ptt) of E. coli K12. After mapping the remaining 580 operons to the E. coli K12 gene coordinate file, there are 497 gene pairs in which the genes in a pair belong to the same operon and the two are adjacent to each other. Furthermore, there are 616 gene pairs in which the genes in a pair were codirectionally transcribed, that is, were adjacent to each other but belong to different transcription units. Intergenic distances between the genes in the former gene pairs were taken as the positive data set (Supplemental Table IV), whereas those in the latter were taken as the negative data set (Supplemental Table V). These data sets were then used to train a Support Vector Machine for prediction of operons in all the 124 genomes.
The frequency of co-occurrence of all the pairs of proteins (4237 × 4237) in the predicted operons across all the genomes was assigned as a score to each of the protein pairs. Thus, each element in the symmetric matrix represents the frequency of co-occurrence of genes i and j, within the predicted operons across all the genomes.
Gene distance method
With 4237 genes in the E. coli genome, the distance between the transcriptional start sites of each gene against the rest of the genes (4236) was calculated by dividing the absolute value of the distance (in nucleotide bases) by the total genome length. All the genomes under consideration being circular, we calculated the absolute value of distance in both clockwise and counterclockwise directions, and then considered the minimum of the two values. These values were then scaled to 100.
Similarly, we calculated the distances between orthologs of the genes in all the 266 genomes. The interaction score between any two genes was taken as the minimum of the distances between the genes and their orthologs across all the 266 genomes. The final matrix is once again 4237 × 4237 symmetric, and each element in the matrix is the minimum of the distances between the genes i and j in any of the 266 genomes.
Prediction of protein–protein interactions using the Support Vector Machine
The 4237 × 4237 symmetric matrix generated by each of the above methods was considered to represent all the possible pairwise interactions between the proteins. Each element of the matrix represented the score of the interaction. These interaction scores were used as different data features for the training of the Support Vector Machine with the positive data and negative data as described below.
Preparation of the positive and negative data set
The data on E. coli protein complexes and pathways were downloaded from the EcoCyc database (Keseler et al. 2005). Any two proteins that are part of the same complex were considered to be functionally interacting. The self-interactions between homodimers in complexes were excluded.
The negative reference data set comprised those pairs of proteins that are not colocalized in the same compartment in E. coli. Thus, the periplasmic location of all the proteins was predicted using SIGCLEAVE in EMBOSS (Sarachu and Colet 2005). The protein was considered as periplasmic if it contained at least one predicted signal sequence within the 50 residues from the N terminus. The top-scoring 40 proteins were considered to be periplasmic. In all, 346 proteins did not possess any signal sequence throughout the entire polypeptide stretch, and therefore were considered to be cytoplasmic. The accuracy of identifying the signal sequence by SIGCLEAVE has been reported to be 75%–80%, and therefore the sets of proteins chosen to be periplasmic and cytoplasmic are likely to be of correct subcellular localization (http://bioweb.pasteur.fr/docs/EMBOSS/sigcleave.html). Each pair was unique to the two data sets.
Cross-validation for model selection and prediction accuracy
We used LibSvm to predict the protein–protein interactions (Chih-Chung and Chih-Jen 2001). The software enables users to define several parameters and allows a choice of inbuilt kernel function including linear, Radial Basis Function (RBF), polynomial, and sigmoid.
In fivefold cross-validation, the data set consisting of positives and negatives was randomly divided into five equal size sets. Training and testing was carried out five times, using the “svm-train” and “svm-predict” utility of the LibSvm software package (Chih-Chung and Chih-Jen 2001). In each round of cross-validation, four sets were used for training, and the remaining set was used for testing. The RBF function was used for training, and the best cost and γ were selected using the grid search algorithm grid.py in the LibSvm software package. In each step of testing, sensitivity and specificity values were calculated. The accuracy of prediction was taken to be the average of sensitivity and specificity, which were defined as follows:
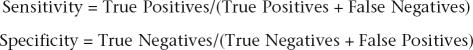 |
Among the five models that were generated, the model with the highest accuracy was retained as the best model and was used for further predictions on a genome-wide scale.
Analysis of network attributes
A variety of graph-theoretic statistics, such as the degree distribution, clustering coefficient, and characteristic path length and diameter were analyzed. Locally written Perl scripts were used to analyze the degree distribution and clustering coefficient. Path length was analyzed using Cytoscape2.2 (Shannon et al. 2003). The subnetwork graphs for illustrations were generated using VISANT (Hu et al. 2005).
Acknowledgments
We thank the Bioinformatics center of CDFD and the SUN Centre of Excellence in Medical Bioinformatics for access to their computational facilities. We also thank M. Vidyasagar for discussions on SVMs and graph theory, and Biju Issac for several stimulating discussions. Financial support from the Department of Biotechnology is gratefully acknowledged. K.G. is a CSIR Senior Research Fellow, and S.C.M. is a Wellcome Trust International Senior Research Fellow supported by grant WT070006.
Footnotes
[Supplemental material is available online at www.genome.org.]
Article published online before print. Article and publication date are online at http://www.genome.org/cgi/doi/10.1101/gr.5900607
References
- Albert R. Scale-free networks in cell biology. J. Cell Sci. 2005;118:4947–4957. doi: 10.1242/jcs.02714. [DOI] [PubMed] [Google Scholar]
- Arifuzzaman M., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Maeda M., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Itoh A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Nishikata K., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Takita C., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Saito R., Ara T., Nakahigashi K., Huang H.C., Hirai A., Ara T., Nakahigashi K., Huang H.C., Hirai A., Nakahigashi K., Huang H.C., Hirai A., Huang H.C., Hirai A., Hirai A., et al. Large-scale identification of protein–protein interactions of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E.S., Holmgren A., Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur. J. Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Tomita M., Wanner B.L., Mori H., Wanner B.L., Mori H., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2:E1–E11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi A.L., Albert R., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Bonabeau E., Bonabeau E. Scale-free networks. Sci. Am. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- Barabasi A.L., Oltvai Z.N., Oltvai Z.N. Network biology: Understanding the cell’s functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Ben-Hur A., Noble W.S., Noble W.S. Choosing negative examples for the prediction of protein–protein interactions. BMC Bioinformatics. 2006;7:1–6. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork P., Jensen L.J., von Mering C., Ramani A.K., Lee I., Marcotte E.M., Jensen L.J., von Mering C., Ramani A.K., Lee I., Marcotte E.M., von Mering C., Ramani A.K., Lee I., Marcotte E.M., Ramani A.K., Lee I., Marcotte E.M., Lee I., Marcotte E.M., Marcotte E.M. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Butland G., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Peregrin-Alvarez J.M., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Li J., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Yang W., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Yang X., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Canadien V., Starostine A., Richards D., Beattie B., Krogan N., Starostine A., Richards D., Beattie B., Krogan N., Richards D., Beattie B., Krogan N., Beattie B., Krogan N., Krogan N., et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature. 2005;433:531–537. doi: 10.1038/nature03239. [DOI] [PubMed] [Google Scholar]
- Chih-Chung C., Chih-Jen L., Chih-Jen L. LIBSVM: A library for support vector machines. 2001. [Google Scholar]
- Dandekar T., Snel B., Huynen M., Bork P., Snel B., Huynen M., Bork P., Huynen M., Bork P., Bork P. Conservation of gene order: A fingerprint of proteins that physically interact. Trends Biochem. Sci. 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- Enault F., Suhre K., Abergel C., Poirot O., Claverie J.M., Suhre K., Abergel C., Poirot O., Claverie J.M., Abergel C., Poirot O., Claverie J.M., Poirot O., Claverie J.M., Claverie J.M. Annotation of bacterial genomes using improved phylogenomic profiles. Bioinformatics. 2003;19:i105–i107. doi: 10.1093/bioinformatics/btg1013. [DOI] [PubMed] [Google Scholar]
- Ermolaeva M.D., White O., Salzberg S.L., White O., Salzberg S.L., Salzberg S.L. Prediction of operons in microbial genomes. Nucleic Acids Res. 2001;29:1216–1221. doi: 10.1093/nar/29.5.1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser H.B., Hirsh A.E., Steinmetz L.M., Scharfe C., Feldman M.W., Hirsh A.E., Steinmetz L.M., Scharfe C., Feldman M.W., Steinmetz L.M., Scharfe C., Feldman M.W., Scharfe C., Feldman M.W., Feldman M.W. Evolutionary rate in the protein interaction network. Science. 2002;296:750–752. doi: 10.1126/science.1068696. [DOI] [PubMed] [Google Scholar]
- Hayes F. A family of stability determinants in pathogenic bacteria. J. Bacteriol. 1998;180:6415–6418. doi: 10.1128/jb.180.23.6415-6418.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsh A.E., Fraser H.B., Fraser H.B. Protein dispensability and rate of evolution. Nature. 2001;411:1046–1049. doi: 10.1038/35082561. [DOI] [PubMed] [Google Scholar]
- Hu Z., Mellor J., Wu J., DeLisi C., Mellor J., Wu J., DeLisi C., Wu J., DeLisi C., DeLisi C. VisANT: Data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 2005;33:W352–W357. doi: 10.1093/nar/gki431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janga S.C., Collado-Vides J., Moreno-Hagelsieb G., Collado-Vides J., Moreno-Hagelsieb G., Moreno-Hagelsieb G. Nebulon: A system for the inference of functional relationships of gene products from the rearrangement of predicted operons. Nucleic Acids Res. 2005;33:2521–2530. doi: 10.1093/nar/gki545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Yu H., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Greenbaum D., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Kluger Y., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Krogan N.J., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Chung S., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Emili A., Snyder M., Greenblatt J.F., Gerstein M., Snyder M., Greenblatt J.F., Gerstein M., Greenblatt J.F., Gerstein M., Gerstein M. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Jeong H., Mason S.P., Barabasi A.L., Oltvai Z.N., Mason S.P., Barabasi A.L., Oltvai Z.N., Barabasi A.L., Oltvai Z.N., Oltvai Z.N. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- Keseler I.M., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D., Collado-Vides J., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D., Gama-Castro S., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D., Ingraham J., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D., Paley S., Paulsen I.T., Peralta-Gil M., Karp P.D., Paulsen I.T., Peralta-Gil M., Karp P.D., Peralta-Gil M., Karp P.D., Karp P.D. EcoCyc: A comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel J.O., Jensen L.J., von Mering C., Bork P., Jensen L.J., von Mering C., Bork P., von Mering C., Bork P., Bork P. Analysis of genomic context: Prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 2004;22:911–917. doi: 10.1038/nbt988. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., Li J., Pu S., Datta N., Tikuisis A.P., Pu S., Datta N., Tikuisis A.P., Datta N., Tikuisis A.P., Tikuisis A.P., et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- Kumar J.K., Tabor S., Richardson C.C., Tabor S., Richardson C.C., Richardson C.C. Proteomic analysis of thioredoxin-targeted proteins in Escherichia coli. Proc. Natl. Acad. Sci. 2004;101:3759–3764. doi: 10.1073/pnas.0308701101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L.K., Harlow G.R., Gregg-Jolly L.A., Mount D.W., Harlow G.R., Gregg-Jolly L.A., Mount D.W., Gregg-Jolly L.A., Mount D.W., Mount D.W. Identification of high affinity binding sites for LexA which define new DNA damage-inducible genes in Escherichia coli. J. Mol. Biol. 1994;241:507–523. doi: 10.1006/jmbi.1994.1528. [DOI] [PubMed] [Google Scholar]
- Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., Vidalain P.O., Han J.D., Chesneau A., Hao T., Han J.D., Chesneau A., Hao T., Chesneau A., Hao T., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Fonstein M., D’Souza M., Pusch G.D., Maltsev N., Fonstein M., D’Souza M., Pusch G.D., Maltsev N., D’Souza M., Pusch G.D., Maltsev N., Pusch G.D., Maltsev N., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen K., Christensen S.K., Gerdes K., Christensen S.K., Gerdes K., Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- Pellegrini M., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O., Marcotte E.M., Thompson M.J., Eisenberg D., Yeates T.O., Thompson M.J., Eisenberg D., Yeates T.O., Eisenberg D., Yeates T.O., Yeates T.O. Assigning protein functions by comparative genome analysis: Protein phylogenetic profiles. Proc. Natl. Acad. Sci. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai G., Plunkett G., III, Feher T., Frisch D., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Plunkett G., III, Feher T., Frisch D., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Feher T., Frisch D., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Frisch D., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Keil G.M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Umenhoffer K., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Kolisnychenko V., Stahl B., Sharma S.S., de Arruda M., Stahl B., Sharma S.S., de Arruda M., Sharma S.S., de Arruda M., de Arruda M., et al. Emergent properties of reduced genome Escherichia coli. Science. 2006;312:1044–1046. doi: 10.1126/science.1126439. [DOI] [PubMed] [Google Scholar]
- Pržulj N., Wigle D.A., Jurisica I., Wigle D.A., Jurisica I., Jurisica I. Functional topology in a network of protein interactions. Bioinformatics. 2004;20:340–348. doi: 10.1093/bioinformatics/btg415. [DOI] [PubMed] [Google Scholar]
- Rain J.C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Simon S., Lenzen G., Petel F., Wojcik J., Schachter V., Lenzen G., Petel F., Wojcik J., Schachter V., Petel F., Wojcik J., Schachter V., Wojcik J., Schachter V., Schachter V., et al. The protein–protein interaction map of Helicobacter pylori. Nature. 2001;409:211–215. doi: 10.1038/35051615. [DOI] [PubMed] [Google Scholar]
- Salgado H., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Moreno-Hagelsieb G., Smith T.F., Collado-Vides J., Smith T.F., Collado-Vides J., Collado-Vides J. Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 2000;97:6652–6657. doi: 10.1073/pnas.110147297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarachu M., Colet M., Colet M. wEMBOSS: A web interface for EMBOSS. Bioinformatics. 2005;21:540–541. doi: 10.1093/bioinformatics/bti031. [DOI] [PubMed] [Google Scholar]
- Seyed-Allaei H., Bianconi G., Marsili M., Bianconi G., Marsili M., Marsili M. Scale-free networks with an exponent less than two. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2005;73:046113. doi: 10.1103/PhysRevE.73.046113. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T., Ramage D., Amin N., Schwikowski B., Ideker T., Amin N., Schwikowski B., Ideker T., Schwikowski B., Ideker T., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Zenkner M., Schoenherr A., Koeppen S., Schoenherr A., Koeppen S., Koeppen S., et al. A human protein–protein interaction network: A resource for annotating the proteome. Cell. 2006;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- Sun J., Xu J., Liu Z., Liu Q., Zhao A., Shi T., Li Y., Xu J., Liu Z., Liu Q., Zhao A., Shi T., Li Y., Liu Z., Liu Q., Zhao A., Shi T., Li Y., Liu Q., Zhao A., Shi T., Li Y., Zhao A., Shi T., Li Y., Shi T., Li Y., Li Y. Refined phylogenetic profiles method for predicting protein–protein interactions. Bioinformatics. 2005;21:3409–3415. doi: 10.1093/bioinformatics/bti532. [DOI] [PubMed] [Google Scholar]
- Suthram S., Sittler T., Ideker T., Sittler T., Ideker T., Ideker T. The Plasmodiumprotein network diverges from those of other eukaryotes. Nature. 2005;438:108–112. doi: 10.1038/nature04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P., Snel B., Cornell M., Oliver S.G., Fields S., Bork P., Cornell M., Oliver S.G., Fields S., Bork P., Oliver S.G., Fields S., Bork P., Fields S., Bork P., Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- Yu H., Greenbaum D., Xin L.H., Zhu X., Gerstein M., Greenbaum D., Xin L.H., Zhu X., Gerstein M., Xin L.H., Zhu X., Gerstein M., Zhu X., Gerstein M., Gerstein M. Genomic analysis of essentiality within protein networks. Trends Genet. 2004;6:227–231. doi: 10.1016/j.tig.2004.04.008. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Szustakowski J.D., Fortnow L., Roberts R.J., Kasif S., Szustakowski J.D., Fortnow L., Roberts R.J., Kasif S., Fortnow L., Roberts R.J., Kasif S., Roberts R.J., Kasif S., Kasif S. Computational identification of operons in microbial genomes. Genome Res. 2002;12:1221–1230. doi: 10.1101/gr.200602. [DOI] [PMC free article] [PubMed] [Google Scholar]