Abstract
The availability of an annotated genome sequence for the yeast Saccharomyces cerevisiae has made possible the proteome-scale study of protein function and protein–protein interactions. These studies rely on availability of cloned open reading frame (ORF) collections that can be used for cell-free or cell-based protein expression. Several yeast ORF collections are available, but their use and data interpretation can be hindered by reliance on now out-of-date annotations, the inflexible presence of N- or C-terminal tags, and/or the unknown presence of mutations introduced during the cloning process. High-throughput biochemical and genetic analyses would benefit from a “gold standard” (fully sequence-verified, high-quality) ORF collection, which allows for high confidence in and reproducibility of experimental results. Here, we describe Yeast FLEXGene, a S. cerevisiae protein-coding clone collection that covers over 5000 predicted protein-coding sequences. The clone set covers 87% of the current S. cerevisiae genome annotation and includes full sequencing of each ORF insert. Availability of this collection makes possible a wide variety of studies from purified proteins to mutation suppression analysis, which should contribute to a global understanding of yeast protein function.
The budding yeast Saccharomyces cerevisiae is one of the most studied eukaryotes at the genetic, molecular, and cellular levels. Many of the mechanisms that control molecular and cell biology of the yeast are conserved in other eukaryotes, including mechanisms of such basic functions as DNA replication, progression through the cell cycle, and transcriptional regulation. Together with rapid growth and genetic tractability, this feature makes yeast particularly valuable for biological research.
Sequencing of the S. cerevisiae genome began as a worldwide collaboration and was completed in 1996, providing the first example of a fully sequenced eukaryotic genome. The 12,068 kilobase-pair sequence defined 5885 potential protein-encoding genes on 16 chromosomes (Goffeau et al. 1996). The average size of genomic sequence for protein-coding genes (exons plus introns) is 1.48 kb, with a range of 51 bp to 14,733 bp. About 25% of all ORFs are larger than 2 kb and the average GC content is 40%.
Annotation of protein-coding genes in the S. cerevisiae genome has changed over time as new experimental data and advanced sequence analyses led to improved annotation. In 2003, a comparative analysis of S. cerevisiae with three related species led to the proposed elimination of about 500 previously annotated ORFs and redefinition of start and/or stop codons for at least 300 ORFs (Kellis et al. 2003). This led to the release of a major revision of the genome sequence annotation in 2004, in addition to subsequent, less comprehensive revisions. As of September 2006, the SGD full-genome annotation includes 6604 known or putative genes, of which 5780 are known or putative protein-coding ORFs, with ∼77% of the protein-coding genes partially characterized.
The knowledge gained from extensive annotation of the S. cerevisiae genome over the past decade has made it possible for researchers to take a genome- and proteome-wide view of yeast gene function. The earliest genome-scale ORF collections for S. cerevisiae were constructed using a gap-repair cloning approach (Hudson et al. 1997; Uetz et al. 2000; Zhu et al. 2000, 2001; Ito et al. 2001). Progress has been made studying different aspects of protein activities in global scale, such as protein post-translational modification, mapping pathways, and determining phenotypes that result from systematic gene overexpression, and measuring the interaction of proteins with other proteins, small molecules, or nucleic acids by parallel screening of the whole yeast proteome using these collections (Ito et al. 2001; Zhu et al. 2001; Hall et al. 2004; Huang et al. 2004; Ptacek et al. 2005; Sopko et al. 2006).
Although these ORF collections have proved useful for specific proteomic studies, the ORF inserts are basically locked into the original vector and cannot be moved to another vector without a PCR amplification step (Marsischky and LaBaer 2004). In addition, the fixed presence of an N-terminal tag may affect the function of some proteins and/or the results of subcellular localization studies (Kumar et al. 2002). Recently, a movable ORF collection (MORF) for yeast was generated by Grayhack and colleagues that included 5854 yeast ORFs in the Invitrogen Gateway entry vector pDONR221, allowing for high-fidelity, in-frame, cost-efficient transfer of inserts into a wide variety of expression vectors (Gelperin et al. 2005). The ORFs in this collection were cloned without their natural stop codons, both allowing and requiring the addition of a C-terminal tag. As in most previous collections, the clones in this collection were verified by end-read sequences.
Among the limitations of end-read sequencing is that many clones do not end up with full sequence coverage and are effectively unfinished. Here, we describe a new collection of yeast ORF clones, Yeast FLEXGene (Full Length EXpresssion-ready), in which all of the clones were full-length sequence verified and contain minimal differences between the clone and reference sequences at the amino acid level. This collection is based on the best available gene annotation, constructed in a recombinational cloning vector that enables high-throughput transfer into a wide variety of vectors, and produced with a stop codon at its native location, allowing for the production of either native or N-terminally tagged protein. The majority of clones (68%) have a normalized stop codon potentially enabling some suppression strategies. We set as a goal to obtain at least 5000 completed clones. The current collection includes clones for 5003 genes and covers 87% of the predicted protein-coding sequences for S. cerevisiae, and preliminary evidence suggests that the collection will be useful for a variety of genomic and proteomic-based approaches.
Results
Identification of an ORF target set from the annotated S. cerevisiae genome sequence
To create an initial reference set of target ORFs, the genomic sequence of the 6277 predicted S. cerevisiae ORFs annotated at the time we initiated our study (2000) were downloaded from the Saccharomyces Genome Database (SGD). In addition, the first phase of our cloning effort (Phase One) relied on a pre-existing set of gene-specific primers from Research Genetics that were based on an earlier annotation of the S. cerevisiae genome. Our target set of reference ORFs was not static, however. We adapted to major revisions and the analysis presented here is based on the major revision released in 2004. Thus, our final target set comprises 5774 ORFs (215 additional ORFs and 252 modified ORFs relative to the 1999 set). About 500 initially targeted ORFs were dubious ORFs, pseudogenes, or Ty elements, and were not attempted at later stages.
Amplification of ORFs by PCR from a normalized genomic template
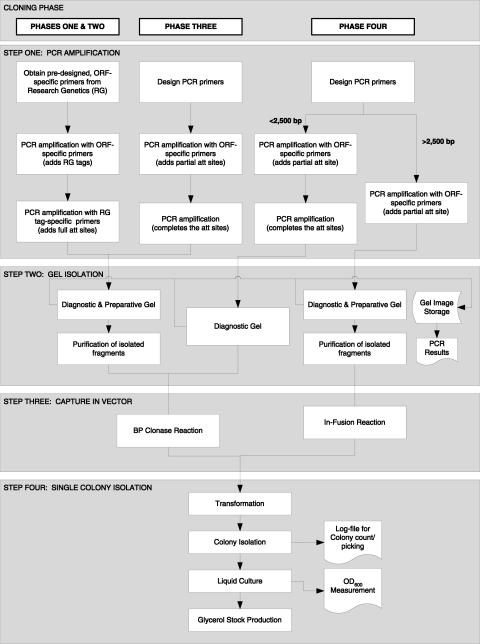
S. cerevisiae served as a test case for genome-scale ORF cloning in our group, which has educated our production of other genome-scale ORF collections. The overall strategy was to use gene-specific PCR to capture the ORFs from a genomic DNA template, which provided a single, quality controlled, and normalized template. This was feasible because only 4% of the predicted ORFs contain introns. These ORFs were maintained in the target list as the presence of introns was not considered problematic for expression in yeast by scientists in this field and because we plan to return to these using cDNA methods. The cloning effort was carried out in four distinct phases, each characterized by production of a group of successfully cloned and verified ORFs, and a set of cloning and/or sequence failures that were passed forward to the next phase. For all four phases, ORFs were amplified by PCR and cloned using nonrestriction enzyme-mediated strategies into a Gateway recombination-based cloning vector (Fig. 1).
Figure 1.
Workflow diagram of clone production. The entire process from the design of primers to production of clone stocks is shown for the four production phases. The process started by designing primers for every ORF in the genome. The primers were used to amplify the ORFs from the genome. Subsequent amplifications with universal primers generated ORF sequences flagged by recombinational cloning sites at either end and were monitored by a diagnostic gel. The product was cloned into a recombinational cloning vector via a BP clonase reaction or In-Fusion reaction. Competent bacterial strains were transformed with the reaction mix to yield colonies that were isolated robotically, cultured in liquid medium, and stored as 15% glycerol stocks.
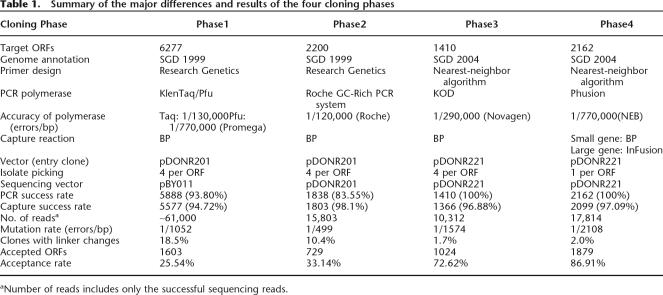
The overall failure rate during the first two phases was ∼70% and failures were primarily due to quality and design issues pertaining to the RG primers and polymerase choice. For Phases Three and Four, we used higher fidelity polymerases than those used for previous phases. Together with the inclusion of newly designed ORF-specific primers, both polymerases improved overall cloning success (Table 1).
Table 1.
Summary of the major differences and results of the four cloning phases
aNumber of reads includes only the successful sequencing reads.
Capture of PCR products in a vector compatible with cloning via enzyme-mediated, site-specific recombination
PCR products were initially captured in the Gateway entry vector pDONR201 and later in pDONR221. Using this system, ORFs are captured in the correct orientation via subtle but noncompatible differences between the 5′ and 3′-flanking att site sequences. Capture was initially done using the BP reaction, a method well suited to high-throughput cloning. However, we found that approach did not efficiently capture fragments 2.5 kb or longer. For this reason, in our last cloning phase, a linearized derivate of pDONR221 was used in conjunction with Clontech’s In-Fusion method for ORFs longer than 2.5 kb. Capture of the PCR amplified fragment in the vector was defined as positive when colonies were detected after transformation, thus allowing single-colony isolation on solid agar (Fig. 1).
Initially, we selected four colonies per ORF and maintained them separately, as we expected that this would increase the likelihood of obtaining at least one mutation-free clone. With experience, our methods have improved such that the benefits of choosing multiple isolates no longer outweigh the costs, as 80% of ORFs can be accepted based on a single isolate. Thus, by Phase Four, we revised our strategy to isolate one colony per ORF. After capture into the vector, half of the capture reaction was plated on solid agar and the remaining transformation mix was stored at −80°C, allowing us to return to the frozen transformation mix without the need to repeat the entire cloning procedure.
DNA sequencing reveals high-fidelity capture of 87% of known and predicted yeast ORFs
To provide a well-annotated collection, we sequenced the complete insert (including the 5′ and 3′ attB sequences), assembled sequence contigs, and compared the assemblies to corresponding reference sequences. Our strategy was to first perform end-read sequencing of all clones using vector-specific (universal) primers followed by insert-specific (internal) primers designed to cover the gaps. Moreover, sequence data were managed, quality-checked, assembled, and analyzed using the Automated Clone Evaluation (ACE) suite of software tools. An assembled sequence contig was considered to be full length if it covered the entire ORF and the flanking sequences relevant for site-specific recombination. After contig assembly, the sequences were compared with their corresponding reference sequences in ACE at both the nucleotide and amino acid levels. For this cloning project, insertion, deletion, and nonsense mutations were defined as nonacceptable “discrepancies” where sequence confidence was high (Phred quality score >25). Low confidence regions were resequenced to obtain better quality. Clones were accepted if the assembled experimental sequences matched the corresponding reference sequences perfectly or contained discrepancies deemed acceptable (any number of silent changes and up to two amino acid changes). The majority of clones had no differences with the reference peptide sequence (82%), and all differences at both the nucleotide and amino acid levels are carefully documented in our distribution database (http://plasmid.hms.harvard.edu) and in Supplemental Table 1. Full-length sequences for all clones are also available at GenBank.
ORF size, PCR primer attributes, and GC content contribute to cloning failure
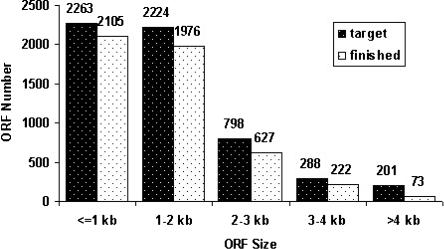
Despite repeated attempts, there were a number of clones for which we were never successful at creating acceptable clones. We analyzed these recalcitrant ORFs and identified several factors that may have contributed to cloning failure. Clearly, large genes were more difficult to clone: the yeast ORF collection we describe here covers more than 93% of ORFs <1 kb, 85% of ORFs 1–4 kb, but 36% of ORFs >4 kb (Fig. 2). Among the 128 large ORFs (>4 kb) for which we failed to obtain a qualified clone, 24 failed at the capture step of clone production, and 104 failed at sequence validation. In general, GC content did not seem to be a contributing factor, except in the extreme cases where ORFs with very low GC content were more likely to fail. Because there are only 39 ORFs in the yeast genome with GC content <30%, this factor did not have a big impact on our overall cloning efforts.
Figure 2.
Size distribution of the yeast gene clone collection as compared with the sizes of the predicted protein-coding sequences as defined by the current annotation at SGD. The cloning success rate was more than 90% for genes smaller than 1 kb and about 85% for medium sized ORFs (1–4 kb). For large ORFs (>4kb), only 36% were cloned and accepted after sequence analysis.
In our amplification strategy, primers target gene-specific regions of ∼20–30 nucleotides in length that correspond to the extreme 5′ and 3′ ends of the coding sequence. We were surprised to find that many different genes share identical 5′ and 3′ ends, making it difficult to amplify all desired ORFs. To determine the extent to which primer specificity contributed to cloning failure, we compared primer sequences for all ORFs with one another. Matching primer pairs typically causes favored amplification of the shortest gene sharing the primer sequences. We also examined whether primer sequences could bind elsewhere in the genome (not necessarily at the ends of other genes). This situation leads to failed amplification or amplified junk sequence. We found that high primer sequence similarity with other ORFs, as well as high primer stickiness to genomic DNA, reduced the cloning success rate from 87% to 70%. Details are listed in Supplemental Table 2 and Supplemental Figure 1.
Failure to clone some sequences could reflect errors in the target ORF sequences
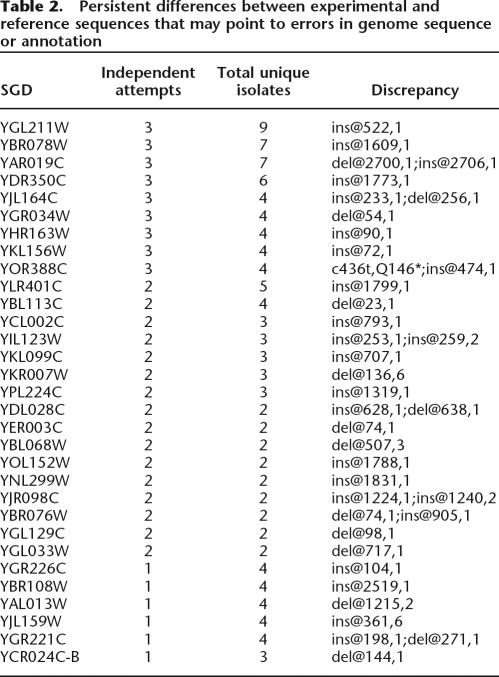
For 31 ORFs, we observed that multiple independent clones had the same frameshifting nucleotide discrepancy compared with the genome reference. In these cases, failure to obtain an isolate matching the genome sequence may be due to an error in the genome sequence, rather than an error in the amplification and cloning process. ORF YBR078W provides an instructive example. For this ORF, we analyzed a total of seven isolates from three cloning efforts with different PCR primers and enzymes. All clones carry the same single base-pair insertion at position 1609 of the coding sequence, which is unlikely to be an artifact of cloning. This and similar cases are summarized in Table 2 and may be helpful in annotation of the yeast genome.
Table 2.
Persistent differences between experimental and reference sequences that may point to errors in genome sequence or annotation
Yeast ORF clones are useful for protein expression and analysis
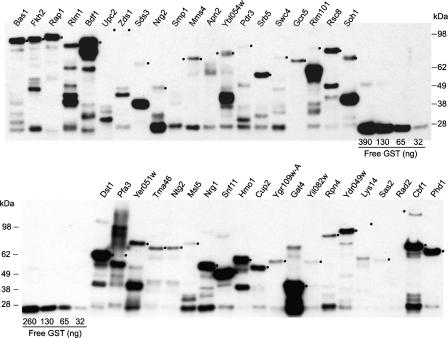
Our rationale for sequence verification of all clones in the yeast collection was to ensure that the clones are useful for protein expression-based assays. To further test the utility of the clones in protein-based assays, we transferred a functionally related set of clones from the entry vector to a bacterial expression vector, induced expression, and purified the proteins. In total, we selected 257 clones that encode known and predicted transcription factors for transfer into the protein expression vector pDEST-GST (LaBaer et al. 2004). Immunoblot analysis of the 257 purified proteins revealed that 58% (148/257) yielded at least 300 ng of protein of the expected size. For 68% of the well-expressed proteins (101/148), a band of the expected size was also the most prominent one (Fig. 3).
Figure 3.
Western blots of 40 known or predicted yeast transcription factors. Representative Western blot analysis of 40 known or candidate yeast TFs. N-terminally GST-tagged proteins were overexpressed in and purified from E. coli in high-throughput, as described in Methods. Five microliters out of 60 uL total of each purified protein were analyzed by Western blots using anti-GST antibody. Serial dilutions of recombinant GST (Sigma) were included for estimation of protein concentrations. Bands of the correct size or the positions of the expected size are indicated by a dot on the right side of the band.
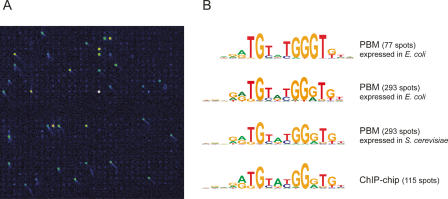
In a pilot study we applied the purified proteins to a protein-binding microarray (PBM) to identify DNA sequence motifs bound by the query protein(s) (Bulyk et al. 1999, 2001; Mukherjee et al. 2004). In a previous study, whole-genome yeast intergenic microarrays were used to test the DNA-binding specificities of the yeast transcription factors Abf1, Rap1, and Mig1 purified from S. cerevisiae (Mukherjee et al. 2004). Here we wished to explore the possibility of using heterologously expressed proteins, which is simpler to obtain, for a similar analysis. To do this, we expressed Rap1 in E. coli, purified the protein, and assayed the protein via PBM (Fig. 4A). A total of 77 distinct intergenic regions were bound in vitro. All of these regions were previously identified as in vitro targets of Rap1 purified from yeast (Mukherjee et al. 2004). As shown in Figure 4B, the DNA sequence motif derived from these 77 sequences closely matches the motif identified using Rap1 purified from yeast (Mukherjee et al. 2004) and the motif derived from regions bound by Rap1 in vivo as determined by genome-wide location analysis (Lee et al. 2002).
Figure 4.
Whole-genome yeast intergenic microarray bound by S. cerevisiae Rap1. (A) Close-up view of a portion of a microarray spotted with all yeast intergenic regions, bound by Rap1 overexpressed in and purified from E. coli in high-throughput. Fluorescence intensities are shown in false color, with white indicating saturated signal intensity, yellow indicating high signal intensity, green indicating moderate signal intensity, and blue indicating low signal intensity. (B) Sequence logos for Rap1 DNA-binding site motifs determined from genomic DNA-binding site identification experiments. We previously performed a set of triplicate PBM experiments using Rap1 expressed in and purified from S. cerevisiae, resulting in 293 intergenic regions bound with a Bonferroni-corrected P value of 0.001 (Mukherjee et al. 2004). Here, as the data on Rap1 overexpressed in and purified from E. coli were generated by a single PBM experiment, fewer spots (77) met our significance threshold for binding. The top two motifs were derived from the 77 and 293 most significantly bound spots in the PBM shown in A. The third motif from the top was derived from our previous set of triplicate PBMs using Rap1 purified from S. cerevisiae (Mukherjee et al. 2004). The motif at the bottom was derived from all intergenic regions bound in vivo in ChIP-chip (Lee et al. 2002). Motifs were generated using BioProspector (Liu et al. 2001) and exhibited the following group specificity scores (top to bottom): 1.3 × 10−97, 2.4 × 10−207, 1.1 × 10−222, and 8.7 × 10−92.
Discussion
Genome-sequencing projects have produced an immense amount of information regarding the organization, evolution, and coding capacity of genomes. Availability of this information has propelled biological research in the direction of genome- or proteome-scaled approaches. The need to develop tools and resources to facilitate this type of research is ever increasing. Large-scale functional proteomics studies, for example, rely on the availability of cloned copies of DNA-encoding the proteins, which make it possible to express proteins in vivo or in vitro and use them in a wide variety of assays (Uetz et al. 2000; Ito et al. 2001; Zhu et al. 2001; Hall et al. 2004; Huang et al. 2004; Gelperin et al. 2005; Ptacek et al. 2005; Sopko et al. 2006).
An ideal collection of protein-encoding clones would embody the virtues of comprehensive coverage of all ORFs, simplified transfer of ORFs to any protein expression vector and full-length sequence validation of all ORFs. In this report, we have described the cloning and verification of yeast FLEXGene ORF clones that meet this “gold standard” for clone quality. In our vector choice, we exploited the availability of recombination-based cloning technology, making it possible for the ORFs in our collection to be easily moved from one vector to another, facilitating the widest possible range of functional experimentation. Importantly, the clones in the collection we describe here were clonally isolated and full-length sequence verified. The collection covers 87% of S. cerevisiae protein-coding sequences (Supplemental Table 1) and 82% of the clones in this collection match perfectly to the reference peptide sequence from current ORF annotation (18% of clones carry one or two amino acid changes). These clones all have GenBank listings and can be searched and are available at http://plasmid.hms.harvard.edu.
The effort to build this ORF collection was carried out in four distinct phases, in which clones that failed in a prior phase were carried forward to the next phase. Despite the fact that failed clones were carried forward, we found that several major factors contributed to a much higher failure rate in earlier phases than in later phases. These included: (1) incorrectly designed and/or synthesized PCR primers; (2) the use of PCR enzymes with low fidelity; (3) difficulties sequencing inserts in the entry vector pDONR201, which made it impossible to achieve full-sequence assemblies for many clones; and (4) erroneous genome annotation. We addressed each of these issues in the subsequent cloning phases and achieved a twofold higher success rate in later versus initial phases in terms of obtaining full-length verified clones (Table 1).
Despite multiple attempts using different primers and cloning strategies, however, the collection still lacks a qualified clone for some ORFs. Examining the factors contributing to lost ORFs will inform future projects, particularly those involving eukaryotic genes. Factors such as ORF size (Fig. 2), GC content, primer similarity, and primer stickiness to other genes or to genomic DNA (Supplemental Table 2; Supplemental Fig. 1) make some ORFs more difficult to clone from a genomic template than others. In total, 427 ORFs in the target list were more difficult to clone due to one or more of these factors (i.e., ORFs size ≥4000; GC ≤30%; two or more other ORFs share the same 5′ or 3′ primer region; and/or more than two genome binding sites for either 5′ or 3′ primer). Our collection covers 59% of these difficult ORFs, in contrast to 90% coverage for all other ORFs. In addition, we have found that for some genes, failure to obtain an isolate that matches the reference sequence appears to be due to an error in the genome sequence or annotation, rather than an error in the amplification and cloning process (Table 2). Annotation of the yeast genome is an ongoing effort that relies on experimental data, and our results may be useful in precisely defining the ORF sequences of these genes. Even though our template DNA came from the same yeast strain as that used to generate the genomic sequence, some of the differences we detected could be the result of subtle differences between our isolate of genomic template and that used for genome sequencing.
Our aim was to create a high-quality clone collection useful for the broadest possible variety of functional studies of yeast proteins. We used the protein-binding microarray (PBM) approach to identify the DNA sequence motif of Rap1 to demonstrate the use of the clones described here in protein-based assays (Fig. 4). The usefulness of the resource was also demonstrated in a high-throughput screen to identify the cellular targets of a small-molecule inhibitor of the TOR pathway (Butcher et al. 2006). This high-quality and comprehensive clone collection makes possible a wide variety of genome-scale studies, including protein expression, purification, and analysis, which should contribute to our understanding of protein functions of S. cerevisiae.
Methods
Preparation of genomic DNA
Genomic DNA was purified from S. cerevisiae strain S288C identical to the strain used for the initial published genome sequence (Goffeau et al. 1996) as described elsewhere (LaBaer et al. 2004).
Primer design, synthesis, and PCR amplification
For phases one and two, first-step PCR primers were designed and synthesized by Research Genetics as Yeast Gene Pair Primers (SGD website). Gene-specific primer sequences were designed based on the initial S. cerevisiae ORF annotation. For Phases One and Two, second-step PCR was performed with universal primers as follows: forward primer, 5′-GGGGACAAGTTTGTACAAAAAAG CAGGCTTCCAGCTGACCACCATG; reverse primer (under-line, attB sequence; bold, RG tag sequence), 5′-GGGGAC CACTTTGTACAAGAAAGCTGGGTATC CCCGGGAATTGCCA.
For Phases Three and Four, first-step PCR, optimal gene-specific primers were designed using a modified nearest-neighbor algorithm (Sugimoto et al. 1995). The Tm was set to 60°C. attB sequences were appended to the gene-specific region as follows: forward primer, 5′-TACAAAAAAGCAG GCTCCACC-atg then gene specific sequences; reverse primer, 5′-GTACAAGAAAGCTGGGTC-normalized STOP codon then gene-specific sequences (underline, partial attB sequences). For second-step PCR, universal primers were as follows: forward primer, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCC; reverse primer, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTC (underline, partial attB sequences). For all phases, a Kozak consensus sequence was included. PCR amplification conditions were set up according to product specification from manufacture (Table 1).
Capture of amplified ORFs
For capture of amplified ORFs in Phase One, PCR fragments were gel purified prior to BP, followed by transformation into DH5α, plated into 6-well plates. Four single colonies were picked by hand for each ORF. After growing in liquid culture and making glycerol stocks, DNA was prepared for LR transfers into pBY011 (Butcher et al. 2006) for sequencing. For capture of amplified ORFs in Phase Two, the size of PCR fragments after step one PCR was checked via agarose gel electrophoresis and PCR fragments from step two PCR were gel purified prior to BP capture into pDONR201, followed by transformation into DH5α (T1-resistant) and plating into 48-well plates for robotic colony isolation. Four individual colonies per ORF were picked. End-read sequencing was done on entry clones (pDONR201) at Agencourt. For capture of amplified ORFs in Phase Three, capture was done as described for Phase Two, except that the vector was pDONR221. End-read sequencing was done on entry clones (pDONR221) at the Dana Farber/Harvard Cancer Center DNA Resource Core (Harvard Medical School). Internal-read sequencing was done at Ludwig Institute for Cancer Research (University of California San Diego). For Phase Four, there were two possible workflows for PCR amplification and ORF capture. For ORFs <2500 bp, second-step PCR was used to generate att sites. Next, ORFs were captured by direct BP reaction. For ORFs >2500 bp, after the first-step PCR, products were separated by agarose gel electrophoresis, isolated, and purified. Next, we performed In-Fusion cloning into linearized pDONR221 and 50% of the transformation mix was plated to LB/agar (the remainder was frozen at −80°C with 15% glycerol). One clone per ORF was isolated for glycerol stock production and sequencing. End-read and internal-read sequencing were both done at Harvard Medical School.
To obtain a linear pDONR221 version that would allow for directional cloning, we introduced unique restriction sites in the 5′ (NcoI) and 3′ (XhoI) att sequences by site-directed mutagenesis, and sequence verified the correct insertion after In-Fusion reactions.
Identification of known or predicted transcription factor genes
A total of 421 S. cerevisiae ORFs were identified as candidate transcription factors (TFs) according to their annotations in several curated databases. A total of 329 ORFs were identified as candidate TFs based on at least one of the following three criteria: (1) annotated in Gene Ontology as “DNA-binding” and one of “Transcription Factor,” “Transcriptional Activator,” or “Transcriptional Repressor” in the Yeast Proteome Database (Costanzo et al. 2000); (2) annotated as “Transcription Factor” in the Munich Information for Protein Sequences (MIPS) Database (Mewes et al. 2002); (3) selected for genome-wide location analysis (ChIP-chip) in a prior global study (Harbison et al. 2004). An additional 92 ORFs were added to this list, as lower confidence TFs candidates, based on their curation in the Pfam database as containing at least one of 44 known DNA-binding domains (Bateman et al. 2004).
Subcloning of ORFs into expression vector, bacterial expression, and purification of proteins
All of the 96-well purifications were performed robotically with optimized protocols on the Biomek Fx (Beckman Coulter) from cell lysis to elution of the bound protein. For GST affinity purification, the robotic deck was setup using 25 mL of lysis buffer (100 mM NaH2PO4, 10 mM Tris at pH 8.0), 25 mL of wash buffer (100 mM Tris, 500 mM NaCl, 2 mM EDTA, 0.1% Triton X-100, 10% glycerol), and 15 mL of elution buffer (wash buffer plus 1% glutathione). All buffers are maintained in chilled conditions on ice and contain complete protease inhibitors (Roche). For purification, cell pellets were thawed for 15 min at room temperature and placed on the deck of the robot at the appropriate location. The cells were lysed in 200 μL of lysis buffer and resuspended thoroughly by repeated pipetting with the help of the 96-well pod of the robot. Ten microliters of DNase mix (10 mg/mL DNase [Sigma] in 900 mM MnCl2, 100 mM MgCl2) was added to the lysate and incubated with shaking (900 rpm) for 10 min. Next, the lysate was allowed to bind to 30 μL of MagneGST (Promega) with shaking at 900 rpm for 10 min in the clockwise direction and 10 min in the anticlockwise direction. The beads were separated with the help of a magnabot, and the remaining lysate was removed and discarded with the help of the 96-well head of the robot. The MagneGST beads with bound protein were washed three times with wash buffer by shaking at 900 rpm for a total of 5 min each time, 2.5 min in the clockwise direction and 2.5 min in the anticlockwise direction. The bound protein was eluted in 50 μL of elution buffer.
Immunoblotting
Proteins were analyzed on precast 4%–12% XT Criterion gradient gels (BioRad) according to the manufacturer’s protocols. Immunoblots were probed with HRP-conjugated rabbit anti-GST antibody (Sigma) at 20 ng/mL final concentration and developed using SuperSignal West Femto Maximum Sensitivity Substrate (Pierce) according to the manufacturer’s protocols.
Protein-binding microarray experiments and data analysis
Whole-genome yeast intergenic microarrays were synthesized essentially as described previously (Ren et al. 2000). PBM experiments and data analysis were performed essentially as described previously, using Rap1 at a final concentration of 20 nM in the protein-binding reaction mixture (Mukherjee et al. 2004; Berger and Bulyk 2006). After quality control filters were applied to the data from the single PBM experiment, 5987 spots (92%) remained for subsequent analysis. A total of 77 features were significantly bound at a Bonferroni-corrected P value significance threshold of 0.001. To search these sequences for over-represented DNA sequence motifs, we used BioProspector (Liu et al. 2001) at widths ranging from 6 to 18 bp (Huber and Bulyk 2006). Motifs were then evaluated according to their group specificity scores (Hughes et al. 2000).
Informatics
Each step of the high-throughput clone production process (from oligo design to final glycerol stock) was tracked in FLEXGene, which is a production laboratory information management system (LIMS) developed in our lab (LaBaer et al. 2004).
Sequence analysis and verification was performed using ACE, a web-based automatic sequence validation package developed in our group for high-throughput clone sequence validation. The features and implementation of this system will be described elsewhere (E. Taycher, A. Rolfs, Y. Hu, D. Zuo, S. Mohr, J. Williamson, and J. LaBaer, in prep).
Accepted clones were imported into and can be publicly searched and requested via the Plasmid Information Database (PlasmID; http://plasmid.hms.harvard.edu). The features and implementation of this system is described elsewhere (Zuo et al. 2007).
Acknowledgments
We thank all past and present members of the Harvard Institute of Proteomics who have contributed to the development of the techniques that made this work possible. Special thanks to Dr. Leonardo Brizuela, Dr. Joeseph Pearlberg for helpful discussions and advice, and Stephanie Ness (LICR, San Diego) for her efforts in performing DNA sequencing. We thank Katrina Saulrieta and Zachary Smith for technical assistance. This work has been supported by grants R01 HG002923 (J.L.) and R01 HG003420 (M.L.B.) from the National Institutes of Health, and funds from the Ludwig Institute for Cancer Research.
Footnotes
[Supplemental material is available online at www.genome.org]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.6037607
References
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Khanna A., Marshall M., Moxon S., Sonnhammer E.L., Marshall M., Moxon S., Sonnhammer E.L., Moxon S., Sonnhammer E.L., Sonnhammer E.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger M.F., Bulyk M.L., Bulyk M.L. Protein binding microarrays (PBMs) for the rapid, high-throughput characterization of the sequence specificities of DNA binding proteins. In: Bina M., editor. Gene mapping, discovery, and expression. The Humana Press, Inc.; Totowa, NJ: 2006. pp. 245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulyk M.L., Gentalen E., Lockhart D.J., Church G.M., Gentalen E., Lockhart D.J., Church G.M., Lockhart D.J., Church G.M., Church G.M. Quantifying DNA-protein interactions by double-stranded DNA arrays. Nat. Biotechnol. 1999;17:573. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- Bulyk M.L., Huang X., Choo Y., Church G.M., Huang X., Choo Y., Church G.M., Choo Y., Church G.M., Church G.M. Exploring the DNA-binding specificities of zinc fingers with DNA microarrays. Proc. Natl. Acad. Sci. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher R.A., Bhullar B.S., Perlstein E.O., Marsischky G., LaBaer J., Schreiber S.L., Bhullar B.S., Perlstein E.O., Marsischky G., LaBaer J., Schreiber S.L., Perlstein E.O., Marsischky G., LaBaer J., Schreiber S.L., Marsischky G., LaBaer J., Schreiber S.L., LaBaer J., Schreiber S.L., Schreiber S.L. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat. Chem. Biol. 2006;2:103–109. doi: 10.1038/nchembio762. [DOI] [PubMed] [Google Scholar]
- Costanzo M.C., Hogan J.D., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Hogan J.D., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Cusick M.E., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Davis B.P., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Fancher A.M., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Hodges P.E., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Kondu P., Lengieza C., Lew-Smith J.E., Lingner C., Lengieza C., Lew-Smith J.E., Lingner C., Lew-Smith J.E., Lingner C., Lingner C., et al. The yeast proteome database (YPD) and Caenorhabditis elegans proteome database (WormPD): Comprehensive resources for the organization and comparison of model organism protein information. Nucleic Acids Res. 2000;28:73–76. doi: 10.1093/nar/28.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelperin D.M., White M.A., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., White M.A., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Wilkinson M.L., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Kon Y., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Kung L.A., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Wise K.J., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Lopez-Hoyo N., Jiang L., Piccirillo S., Yu H., Jiang L., Piccirillo S., Yu H., Piccirillo S., Yu H., Yu H., et al. Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes & Dev. 2005;19:2816–2826. 546. doi: 10.1101/gad.1362105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Barrell B.G., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Bussey H., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Davis R.W., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Dujon B., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Feldmann H., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Galibert F., Hoheisel J.D., Jacq C., Johnston M., Hoheisel J.D., Jacq C., Johnston M., Jacq C., Johnston M., Johnston M., et al. Life with 6000 genes. Science. 1996;274:563–567. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- Hall D.A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M., Zhu X., Royce T., Gerstein M., Snyder M., Royce T., Gerstein M., Snyder M., Gerstein M., Snyder M., Snyder M. Regulation of gene expression by a metabolic enzyme. Science. 2004;306:482–484. doi: 10.1126/science.1096773. [DOI] [PubMed] [Google Scholar]
- Harbison C.T., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Gordon D.B., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Lee T.I., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Rinaldi N.J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Macisaac K.D., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Danford T.W., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Hannett N.M., Tagne J.B., Reynolds D.B., Yoo J., Tagne J.B., Reynolds D.B., Yoo J., Reynolds D.B., Yoo J., Yoo J., et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Zhu H., Haggarty S.J., Spring D.R., Hwang H., Jin F., Snyder M., Schreiber S.L., Zhu H., Haggarty S.J., Spring D.R., Hwang H., Jin F., Snyder M., Schreiber S.L., Haggarty S.J., Spring D.R., Hwang H., Jin F., Snyder M., Schreiber S.L., Spring D.R., Hwang H., Jin F., Snyder M., Schreiber S.L., Hwang H., Jin F., Snyder M., Schreiber S.L., Jin F., Snyder M., Schreiber S.L., Snyder M., Schreiber S.L., Schreiber S.L. Finding new components of the target of rapamycin (TOR) signaling network through chemical genetics and proteome chips. Proc. Natl. Acad. Sci. 2004;101:16594–16599. doi: 10.1073/pnas.0407117101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber B., Bulyk M., Bulyk M. Meta-analysis discovery of tissue-specific DNA sequence motifs from mammalian gene expression data. BMC Bioinformatics. 2006;7:7. doi: 10.1186/1471-2105-7-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson J.R., Jr., Dawson E.P., Rushing K.L., Jackson C.H., Lockshon D., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Dawson E.P., Rushing K.L., Jackson C.H., Lockshon D., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Rushing K.L., Jackson C.H., Lockshon D., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Jackson C.H., Lockshon D., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Lockshon D., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Conover D., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Lanciault C., Harris J.R., Simmons S.J., Rothstein R., Harris J.R., Simmons S.J., Rothstein R., Simmons S.J., Rothstein R., Rothstein R., et al. The complete set of predicted genes from Saccharomyces cerevisiae in a readily usable form. Genome Res. 1997;7:1169–1173. doi: 10.1101/gr.7.12.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J.D., Estep P.W., Tavazoie S., Church G.M., Estep P.W., Tavazoie S., Church G.M., Tavazoie S., Church G.M., Church G.M. Computational identification of cis-regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J. Mol. Biol. 2000;296:1205–1214. doi: 10.1006/jmbi.2000.3519. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Ozawa R., Yoshida M., Hattori M., Sakaki Y., Yoshida M., Hattori M., Sakaki Y., Hattori M., Sakaki Y., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E., Patterson N., Endrizzi M., Birren B., Lander E., Endrizzi M., Birren B., Lander E., Birren B., Lander E., Lander E. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Kumar A., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Agarwal S., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Heyman J.A., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Matson S., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Heidtman M., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Piccirillo S., Umansky L., Drawid A., Jansen R., Liu Y., Umansky L., Drawid A., Jansen R., Liu Y., Drawid A., Jansen R., Liu Y., Jansen R., Liu Y., Liu Y., et al. Subcellular localization of the yeast proteome. Genes & Dev. 2002;16:707–719. doi: 10.1101/gad.970902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBaer J., Qiu Q., Anumanthan A., Mar W., Zuo D., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Qiu Q., Anumanthan A., Mar W., Zuo D., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Anumanthan A., Mar W., Zuo D., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Mar W., Zuo D., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Zuo D., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Murthy T.V., Taycher H., Halleck A., Hainsworth E., Lory S., Taycher H., Halleck A., Hainsworth E., Lory S., Halleck A., Hainsworth E., Lory S., Hainsworth E., Lory S., Lory S., et al. The Pseudomonas aeruginosa PA01 gene collection. Genome Res. 2004;14:2190–2200. doi: 10.1101/gr.2482804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T., Rinaldi N., Robert R., Odom D., Bar-Joseph Z., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Rinaldi N., Robert R., Odom D., Bar-Joseph Z., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Robert R., Odom D., Bar-Joseph Z., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Odom D., Bar-Joseph Z., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Bar-Joseph Z., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Gerber G., Hannett N., Harbison C., Thompson C., Simon I., Hannett N., Harbison C., Thompson C., Simon I., Harbison C., Thompson C., Simon I., Thompson C., Simon I., Simon I., et al. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- Liu X., Brutlag D., Liu J., Brutlag D., Liu J., Liu J. BioProspector: Discovering conserved DNA motifs in upstream regulatory regions of co-expressed genes. Pac. Symp. Biocomput. 2001;6:127–138. [PubMed] [Google Scholar]
- Marsischky G., LaBaer J., LaBaer J. Many paths to many clones: A comparative look at high-throughput cloning methods. Genome Res. 2004;14:2020–2028. doi: 10.1101/gr.2528804. [DOI] [PubMed] [Google Scholar]
- Mewes H., Frishman D., Guldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Frishman D., Guldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Guldener U., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Mannhaupt G., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Mayer K., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Mokrejs M., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Morgenstern B., Munsterkotter M., Rudd S., Weil B., Munsterkotter M., Rudd S., Weil B., Rudd S., Weil B., Weil B. MIPS: A database for genomes and protein sequences. Nucleic Acids Res. 2002;30:31–34. doi: 10.1093/nar/30.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Berger M.F., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Jona G., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Wang X.S., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Muzzey D., Snyder M., Young R.A., Bulyk M.L., Snyder M., Young R.A., Bulyk M.L., Young R.A., Bulyk M.L., Bulyk M.L. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat. Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Devgan G., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Michaud G., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Zhu H., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Zhu X., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Fasolo J., Guo H., Jona G., Breitkreutz A., Sopko R., Guo H., Jona G., Breitkreutz A., Sopko R., Jona G., Breitkreutz A., Sopko R., Breitkreutz A., Sopko R., Sopko R., et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–684. doi: 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- Ren B., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Robert F., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Wyrick J.J., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Aparicio O., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Jennings E.G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Schreiber J., Hannett N., Kanin E., Hannett N., Kanin E., Kanin E., et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Snyder M., Oliver S.G., Cyert M., Hughes T.R., Oliver S.G., Cyert M., Hughes T.R., Cyert M., Hughes T.R., Hughes T.R., et al. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 2006;21:319–330. doi: 10.1016/j.molcel.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Sugimoto N., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Nakano S., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Katoh M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Matsumura A., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Nakamuta H., Ohmichi T., Yoneyama M., Sasaki M., Ohmichi T., Yoneyama M., Sasaki M., Yoneyama M., Sasaki M., Sasaki M. Thermodynamic parameters to predict stability of RNA/DNA hybrid duplexes. Biochemistry. 1995;34:11211–11216. doi: 10.1021/bi00035a029. [DOI] [PubMed] [Google Scholar]
- Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., Lockshon D., Narayan V., Srinivasan M., Pochart P., Narayan V., Srinivasan M., Pochart P., Srinivasan M., Pochart P., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- Zhu H., Klemic J.F., Chang S., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Klemic J.F., Chang S., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Chang S., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Bertone P., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Casamayor A., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Klemic K.G., Smith D., Gerstein M., Reed M.A., Snyder M., Smith D., Gerstein M., Reed M.A., Snyder M., Gerstein M., Reed M.A., Snyder M., Reed M.A., Snyder M., Snyder M. Analysis of yeast protein kinases using protein chips. Nat. Genet. 2000;26:283–289. doi: 10.1038/81576. [DOI] [PubMed] [Google Scholar]
- Zhu H., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bilgin M., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bangham R., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Hall D., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Casamayor A., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Bertone P., Lan N., Jansen R., Bidlingmaier S., Houfek T., Lan N., Jansen R., Bidlingmaier S., Houfek T., Jansen R., Bidlingmaier S., Houfek T., Bidlingmaier S., Houfek T., Houfek T., et al. Global analysis of protein activities using proteome chips. Science. 2001;293:2101–2105. doi: 10.1126/science.1062191. [DOI] [PubMed] [Google Scholar]
- Zuo D., Mohr S.E., Hu Y., Taycher E., Rolfs A., Kramer J., Williamson J., LaBaer J., Mohr S.E., Hu Y., Taycher E., Rolfs A., Kramer J., Williamson J., LaBaer J., Hu Y., Taycher E., Rolfs A., Kramer J., Williamson J., LaBaer J., Taycher E., Rolfs A., Kramer J., Williamson J., LaBaer J., Rolfs A., Kramer J., Williamson J., LaBaer J., Kramer J., Williamson J., LaBaer J., Williamson J., LaBaer J., LaBaer J. PlasmID: A centralized repository for plasmid clone information and distribution. Nucleic Acids Res. 2007;35:D680–D684. doi: 10.1093/nar/gkl898. [DOI] [PMC free article] [PubMed] [Google Scholar]