Abstract
The RNA-catalyzed splicing of group I and group II introns is facilitated by proteins that stabilize the active RNA structure or act as RNA chaperones to disrupt stable inactive structures that are kinetic traps in RNA folding. In Neurospora crassa and Saccharomyces cerevisiae, the latter function is fulfilled by specific DEAD-box proteins, denoted CYT-19 and Mss116p, respectively. Previous studies showed that purified CYT-19 stimulates the in vitro splicing of structurally diverse group I and group II introns and uses the energy of ATP binding or hydrolysis to resolve kinetic traps. Here, we purified Mss116p and show that it has RNA-dependent ATPase activity, unwinds RNA duplexes in a non-polar fashion, and promotes ATP-independent strand-annealing. Further, we show that Mss116p binds RNA non-specifically and promotes in vitro splicing of both group I and group II intron RNAs, as well as RNA cleavage by the aI5γ-derived D135 ribozyme. However, Mss116p also has ATP-hydrolysis-independent effects on some of these reactions, which are not shared by CYT-19 and may reflect differences in its RNA-binding properties. We also show that a non-mitochondrial DEAD-box protein, yeast Ded1p, can function almost as efficiently as CYT-19 and Mss116p in splicing the yeast aI5γ group II intron and less efficiently in splicing the bI1 group II intron. Together, our results show that Mss116p, like CYT-19, can act broadly as an RNA chaperone to stimulate the splicing of diverse group I and group II introns and that Ded1p also has an RNA chaperone activity that can be assayed by its effect on splicing mitochondrial introns. Nevertheless, these DEAD-box protein RNA chaperones are not completely interchangeable and appear to function in somewhat different ways using biochemical activities that have likely been tuned by coevolution to function optimally on specific RNA substrates.
Keywords: catalytic RNA, ribozyme, RNA chaperone, RNA-protein interaction, RNA structure
Introduction
Group I and group II introns splice via RNA-catalyzed transesterification reactions that are facilitated by proteins.1 Some of these proteins are RNA splicing factors that bind specifically to the intron RNA and stabilize the catalytically active RNA structure, while others are RNA chaperones that bind RNAs without apparent specificity and disrupt stable inactive structures that are “kinetic traps” during RNA folding. RNA splicing factors for group I and group II introns include both intron-encoded maturases and a variety of host-encoded proteins that act on different introns.1–3 The intron-encoded maturases are related to proteins that function in intron mobility, DNA endonucleases for group I introns and reverse transcriptases for group II introns, and the host proteins include aminoacyl-tRNA synthetases and other dually functional cellular proteins. The host-encoded splicing factors for group I and group II introns differ among organisms, and many if not all appear to be cellular proteins that adapted secondarily to function in RNA splicing. Studies using protein-dependent in vitro splicing systems have shown that splicing factors promote formation of a catalytically active RNA structure by tertiary-structure nucleation, tertiary-structure capture, or some combination of these mechanisms.1,4,5
Although a variety of proteins that bind RNA non-specifically have been shown to function as RNA chaperones in vitro or when over-expressed in vivo,6–8 recent findings show RNA chaperone function for splicing mitochondrial (mt) group I and group II introns in Neurospora crassa and Saccharomyces cerevisiae is fulfilled by specific DEAD-box proteins, denoted CYT-19 and Mss116p, respectively.9,10 DExH/D-box proteins, also referred to as RNA helicases, are a large and ubiquitous protein family, members of which use the energy of ATP binding and hydrolysis to mediate RNA structural rearrangements in a variety of cellular processes, including translation, ribosome assembly, RNA degradation, and RNA splicing.11–13 All DExH/D-box proteins contain a core RNA helicase region, which consists of two RecA-like domains with a series conserved sequence motifs involved in RNA binding and ATP hydrolysis. Three subfamilies, denoted DEAD, DEAH, and DExH, are named according to the amino acid sequence of a conserved motif II involved in ATP binding.13 In many DExH/D-box proteins, the core helicase region is linked to different N- and/or C-terminal domains, and in some cases, these appended domains have been shown to target the proteins to their sites of action via protein-protein or protein-RNA interactions.11–13
In N. crassa, the efficient splicing of a subset of mt group I introns requires both the mt tyrosyl-tRNA synthetase (mt TyrRS; CYT-18 protein), which stabilizes the catalytically active RNA structure,14,15 and the DEAD-box protein CYT-19, which functions to resolve kinetic traps.9 The requirement for CYT-19 in group I intron splicing was demonstrated both genetically and biochemically. A mutation in the cyt-19 gene (cyt-19-1) inhibits the splicing of all CYT-18-dependent group I introns, and purified recombinant CYT-19 functions in conjunction with CYT-18 to promote group I intron splicing in vitro.9 As expected for its RNA chaperone function, the requirement for CYT-19 is pronounced at 25°C, the normal physiological growth temperature of N. crassa, but progressively less pronounced at higher temperatures, which provide an alternate means of overcoming the high activation enthalpy for disruption of stable inactive RNA structures.9 RNA-structure mapping experiments using the Tetrahymena thermophila LSU-ΔP5abc intron, whose splicing at low Mg2+ concentrations is dependent upon CYT-18, showed directly that CYT-19 plus ATP resolves a predominant misfolded intermediate that is rate-limiting for splicing in that RNA.9 Recent studies using a ribozyme derivative of the T. thermophila intron reinforce the conclusion that CYT-19 functions by resolving non-native RNA structures and provide additional mechanistic insight into how this is accomplished.16 In addition to its group I intron splicing defects, the cyt-19-1 mutant is also defective in some RNA processing reactions and likely in mt translation. 9,17,18 Thus, CYT-19 appears to have multiple RNA-related functions.
In S. cerevisiae, the mt TyrRS does not function in group I intron splicing, and instead a variety of other proteins have evolved to stabilize the active structure of group I and group II intron RNAs, including intron-encoded maturases, the mt LeuRS, and the well-studied protein Cbp2p, which functions in splicing the group I intron bI5 (reviewed in Lambowitz et al.1). Yeast mitochondria also contain a DEAD-box protein, Mss116p, whose core helicase region has 52% similarity to that of CYT-19 but more divergent N- and C-terminal domains.10,19 The disruption of the MSS116 gene inhibits splicing of all S. cerevisiae mt group I and group II introns, some RNA processing reactions, and translation of a subset of mt mRNAs, and all of these defects can be suppressed by the expression of CYT-19.10 Experiments using the yeast group II intron aI2, which requires an intron-encoded maturase for structural stabilization, indicated that DEAD-box protein activity accelerates a step after maturase binding, likely the resolution of stable folding intermediates or non-native structures that constitute kinetic traps.10 Together, these findings showed that in S. cerevisiae, both Mss116p and CYT-19 can function broadly in splicing diverse group I and II introns, other mt RNA processing reactions, and mt translation.
The yeast mt group I and group II introns, whose splicing is affected by Mss116p and CYT-19, include those known to require intron-encoded maturases and other splicing factors for structural stabilization, as well as two small subgroup IIB introns aI5γ and bI1, which do not encode maturases.10 Because of their relatively small size, aI5γ and bI1 have been studied extensively as model systems for group II intron self-splicing and RNA folding.3 Recently, we showed that purified recombinant CYT-19 could by itself promote the splicing of the aI5γ and bI1 group II introns in an ATP-hydrolysis dependent manner under near physiological conditions.20 Further, by using reverse-branching and reverse-splicing reactions of bI1, which could be initiated by adding RNA oligomer substrates, we showed that CYT-19 acts as an archetypical RNA chaperone, whose continued presence is not required after RNA folding has occurred.20 This in vitro splicing system opened the possibility of detailed analysis of how DEAD-box proteins affect the folding and splicing of their natural group II intron RNA substrates.
Although CYT-19 can promote the splicing of yeast mt group II introns in vivo and in vitro, for detailed analysis of the aI5γ and bI1 splicing reactions, as well as other yeast mt RNA splicing reactions, it was desirable to use the native yeast protein Mss116p, which normally promotes the splicing of these introns in vivo. Here, we expressed Mss116p in Escherichia coli, purified the active recombinant protein, and characterized its biochemical activities. We show that like CYT-19, Mss116p can promote group I and group II intron splicing reactions at near physiological conditions in an ATP-hydrolysis dependent manner, but that Mss116p also has ATP-hydrolysis independent effects on these reactions that are not shared by CYT-19. These effects can be correlated with Mss116p’s relatively high, nucleotide-independent RNA-binding affinity, which may impede cycling on and off RNA substrates. We also show that a non-mitochondrial DEAD-box protein, yeast Ded1p, functions almost as efficiently as CYT-19 and Mss116p in splicing the yeast aI5γ group II intron and somewhat less efficiently in splicing the yeast bI1 group II intron. Our results indicate that small biochemical differences between related DEAD-box proteins can correlate with larger and more diverse effects on their reactions with natural substrates.
Results
Expression and purification of Mss116p
At the outset, we tested several E. coli expression systems for Mss116p. The highest yields of active protein were obtained with plasmid pMAL-Mss116p, which expresses Mss116p with an N-terminal MalE fusion that is cleavable by TEV protease. The fusion protein was purified via PEI-precipitation and amylose-affinity chromatography, then cleaved with TEV protease, and further purified through additional hydroxyapatite and amylose-affinity chromatography steps to remove the cleaved MalE tag (see Materials and Methods). After protease cleavage, the expressed protein corresponds to the mature, intramitochondrial form of Mss116p lacking the mt targeting peptide (amino acid residues 1–36), but with two extra N-terminal amino acid residues (GlySer). The yield of purified Mss116p protein was 0.5 to 1.5 mg per liter culture, and the purity was ≥95%, as judged by SDS-polyacrylamide gels stained with Coomassie blue. An Mss116p mutant, with a single amino acid substitution (K158E) in the Walker A motif of the ATPase active site, was expressed and purified by the same methods used for the wild-type protein.
Mss116p has RNA-dependent ATPase activity
Table 1 shows that purified Mss116p, like CYT-19 and other DExH/D-box proteins involved in RNA transactions, has RNA-dependent ATPase activity. At saturating concentrations of ATP and a variety of different RNAs (the T. thermophila LSU-ΔP5abc group I intron, the S. cerevisiae aI5γ and bI1 group II introns, poly(A), poly(C), or poly(U)), kcat values for ATP hydrolysis ranged from 54 to 112 min−1 . These kcat values as well as KmATP values determined for several RNAs (see Table 1 legend) are comparable those found previously for CYT-19 and other DExH/D-box proteins.9,21 Only low ATPase activity was found with poly(dC), and no significant activity above background was detected in the absence of RNA (-RNA) or with poly(G) and poly(dA), unlike CYT-19, which has low background ATPase activity in the absence of RNA.9 Mss116p’s very low ATPase activity in the absence of RNA could reflect very tight regulation by RNA binding, or that its extensive purification more completely removes contaminating RNA and ATPases. Mss116p’s ATPase activity in the presence of aI5γ, bI1, or poly(C) was relatively constant over a wide range of Mg2+ concentrations, with the activities for aI5γ and bI1 (but not poly(C)) declining somewhat only at Mg2+ concentrations ≥ 35 mM (Table 2). The Mss116p-K158E protein, which has a mutation in the ATPase active site, had no detectable ATPase activity with any of the above RNAs that maximally stimulated the ATPase activity of the wild-type protein (data not shown).
Table 1.
Mss116p has RNA-dependent ATPase activity
| Nucleic acid | kcat (min−1) |
|---|---|
| Tt LSU- P5abc | 72 ± 8 |
| Sc aI5γ | 112 ± 6 |
| Sc bI1 | 110 ± 14 |
| Poly(A) | 90 ± 26 |
| Poly(C) | 62 ± 12 |
| Poly(G) | n.d. |
| Poly(U) | 54 ± 16 |
| Poly(dA) | n.d. |
| Poly(dC) | 10 ± 2 |
| -RNA | n.d. |
ATPase activity was assayed in reaction medium containing 100 mM KCl/8 mM MgCl2 at 30°C with 50 nM Mss116p in the presence of saturating unlabeled ATP (3 mM) and RNA (300 nM for intron RNAs and 0.25 μg/μl for oligonucleotides). The table summarizes kcat values determined by measuring the initial rate (r0), corresponding to <1% of final product in a 100-min time course at saturating ATP and RNA concentrations; all reactions were linear for >15 min. Saturation was confirmed by measuring r0 at multiple RNA concentrations. KmATP values for a subset of RNAs were determined by measuring r0 at a series of saturating and subsaturating ATP concentrations: Tt LSU-ΔP5abc, KmATP = 88 ± 30 μM; Sc aI5γ, KmATP = 171 ± 25 μM; Sc bI1, KmATP = 222 ± 40 μM; poly(C), KmATP = 633 ± 98 μM. Based on these results, 3 mM ATP was assumed to be saturating for all RNAs. All values are the mean ± the standard deviation for triplicate assays. n.d., not detectable above background.
Table 2.
Effect of Mg2+ concentration on RNA-dependent ATPase activity of Mss116p
|
kobs(min−1)
|
|||
|---|---|---|---|
| Mg2+ (mM) | aI5γ | bI1 | poly (C) |
| 1 | 122 ± 18 | 64 ± 16 | 10 ± 10 |
| 3 | 90 ± 22 | 58 ± 12 | 60 ± 8 |
| 5 | 88 ± 16 | 78 ± 12 | 60 ± 12 |
| 8 | 112 ± 6 | 110 ± 14 | 62 ± 12 |
| 10 | 84 ± 18 | 80 ± 14 | 58 ± 4 |
| 15 | 68 ± 22 | 62 ± 18 | 36 ± 12 |
| 25 | 82 ± 12 | 56 ± 6 | 44 ± 12 |
| 35 | 34 ± 3 | 50 ± 12 | 48 ± 14 |
| 50 | 32 ± 4 | 28 ± 6 | 46 ± 6 |
ATPase activity was assayed with 50 nM Mss116p in reaction medium containing 100 mM KCl and different Mg2+ concentrations at 30°C, in the presence of 3 mM ATP and 300 nM intron RNAs or 0.25 μg/μl poly(C). The RNA concentrations are saturating in reaction medium containing 100 mM KCl and 8 mM Mg2+. kobs values were calculated from initial rates in time courses (see Table 1). In the absence of RNA, ATPase activity was not detectable at any of the Mg2+ concentrations (not shown). Values are the mean ± the standard deviation for triplicate assays.
Duplex-unwinding and strand-annealing by Mss116p
The duplex-unwinding and strand-annealing activities of Mss116p were assayed first using RNA oligonucleotide substrates that form a 16-bp duplex (Figure 1, Table 3). Mss116p readily unwound this 16-bp RNA duplex with either a 5′- or 3′-single-stranded region in an ATP-dependent manner (Figure 1(a) and (b), left panels; Table 3). No reaction was observed in the absence of ATP or in the presence of the non-hydrolyzable ATP analog AMP-PNP (data not shown). Notably, Mss116p also unwound blunt-end duplexes with rate constants similar to that for the tailed substrates (Figure 1(c), left panel; Table 3). As all of these reactions were performed at saturating Mss116p concentrations relative to substrate (data not shown), the similar rate constants suggest that single-stranded regions do not significantly affect the rate-limiting step for duplex unwinding.
Figure 1.
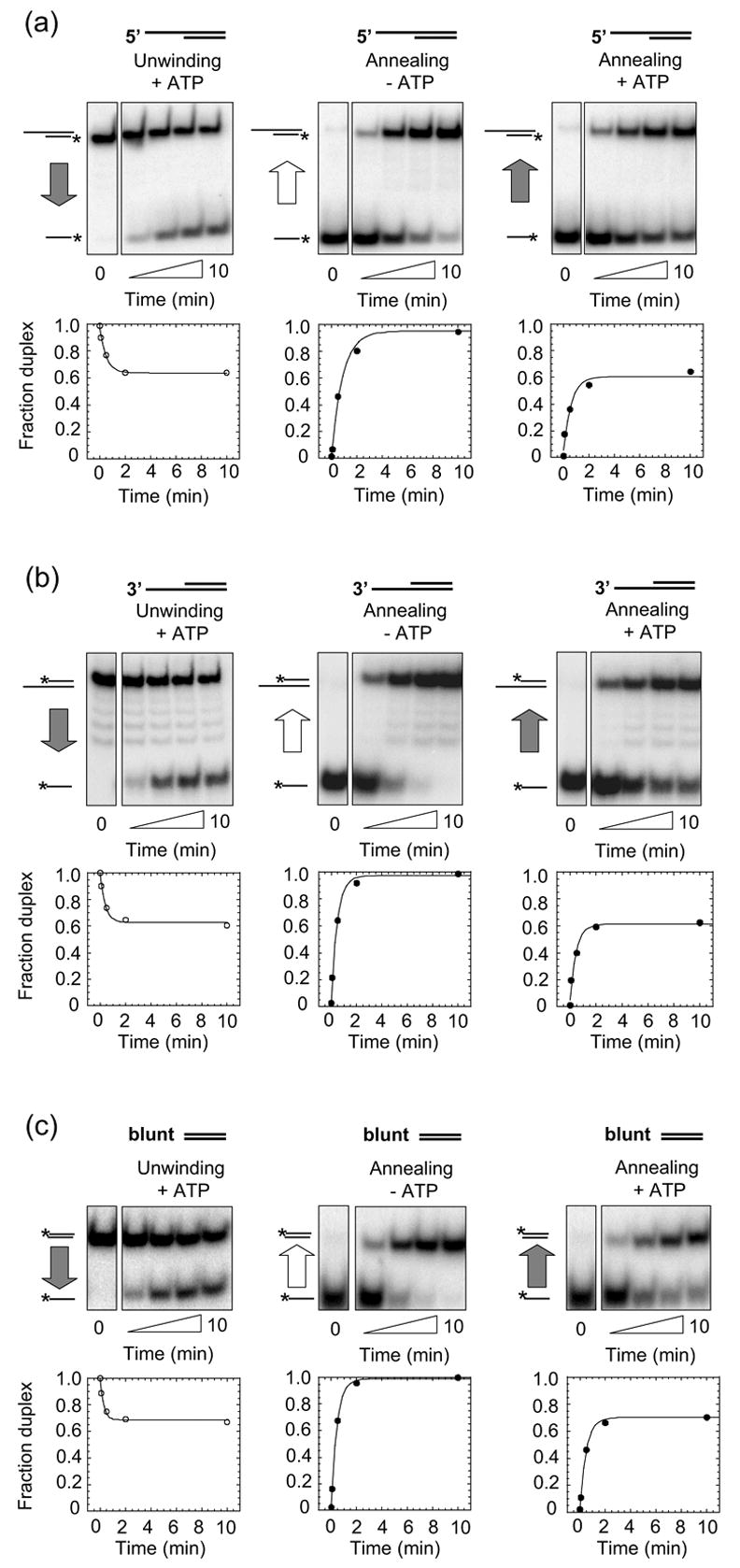
Mss116p duplex-unwinding and strand-annealing activities. Reactions were done with RNA oligonucleotides that form a 16-bp duplex with (a) a 25-nt 5′ overhang; (b) a 25-nt 3′ overhang; or (c) blunt-ends. For each substrate, the figure shows representative duplex- unwinding reactions with 2 mM ATP (left panels), strand-annealing reactions without ATP (middle panels), and strand-annealing reactions with 2 mM ATP (right panels). Plots beneath the gels show fraction duplex versus reaction time fitted to the integrated form of a homogenous first-order rate law. Substrates and products are depicted schematically to the left of the gel, with the asterisk indicating the radiolabel and the vertical arrow indicating the direction of the reaction. Gray arrows indicate presence and white arrows indicate absence of ATP in the reactions. The zero time points for the duplex-unwinding reactions were recorded before ATP addition, and the zero time points for the strand-annealing reactions were recorded before Mss116p addition. Strand annealing in the absence of Mss116p was not significant (<5% of the RNA spontaneously annealed within 10 min; data not shown). Rate constants calculated from at least two independent time courses for each reaction are summarized in Table 3.
Table 3.
Rate constants for duplex-unwinding and strand-annealing of RNA oligonucleotide substrates by Mss116p.
| kunw (min−1) | kann[no ATP] (x109 M−1 min−1) | kann[ATP] (x109 M−1 min−1) | |
|---|---|---|---|
| 5′ overhang | 0.91 ± 0.07 | 4.7 ± 0.8 | 5.6 ± 1.7 |
| 3′ overhang | 1.21 ± 0.31 | 5.6 ± 0.5 | 7.8 ± 2.0 |
| Blunt | 1.0 ± 0.2 | 6.2 ± 0.6 | 4.9 ± 1.1 |
Rate constants were calculated from at least two repeats of reactions shown in Figure 1. RNA substrates contained a 16-bp duplex with a 5′- or 3′-overhang or blunt ends (see Materials and Methods).
In addition to this RNA helicase activity, Mss116p promoted the annealing of the oligonucleotides comprising the above substrates in the absence of ATP (Figure 1, middle panels; Table 3). Mss116p enhanced the bimolecular rate constant for duplex formation by several orders of magnitude, close to the diffusion-imposed limit for a bi-molecular reaction22 and comparable to the maximum rate reported for the profound strand-annealing activity of the DEAD-box protein Ded1p.23
Like Ded1p, Mss116p also promoted strand-annealing in the presence of ATP. Under these conditions, annealing did not proceed to completion, but ended at a defined amplitude (Figure 1, right panels). The bimolecular annealing rate constants in the presence of ATP were similar to those in the absence of ATP, and the amplitude was virtually identical to that for the unwinding reaction under identical reaction conditions (Figure 1, compare left and right panels; Table 3). These observations suggest that Mss116p, like Ded1p, promotes a steady state between duplex unwinding and strand annealing when ATP is present.23
Mg2+-concentration dependence of duplex-unwinding and strand-annealing reactions
While the duplex-unwinding and strand-annealing activities of Mss116p were assayed at low Mg2+ concentrations (0.5 mM) to allow comparison of these activities with those of other DEAD-box proteins, a correlation of the unwinding and annealing activities of Mss116p with its ability to promote group I and group II intron splicing required analysis under RNA-splicing conditions. Accordingly, Figure 2 shows experiments in which we determined Mss116p’s unwinding and annealing rate constants in splicing reaction medium containing different Mg2+ concentrations at 30°C. Because increasing Mg2+ concentrations have been shown to decrease the unwinding rate constants for other RNA helicases (e.g., Rogers et al.24), we utilized substrates with a 13-bp duplex region, which was unwound faster than the 16-bp duplex used above and thus facilitated measurements over a greater range of Mg2+ concentrations.
Figure 2.
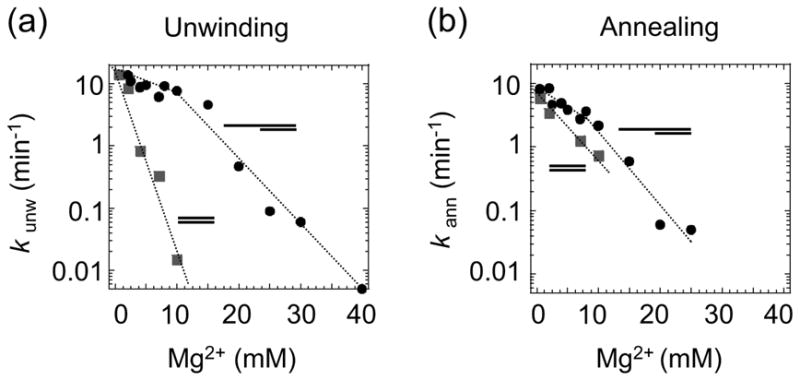
Mg2+-concentration dependence of the Mss116p duplex-unwinding and strand- annealing reactions. The Mg2+-concentration dependence of the rate constants for (a) duplex unwinding (kunw), and (b) strand annealing (kann) of RNA oligonucleotides containing a 13-bp duplex region with a 5′ overhang (filled circles) or blunt-ends (filled squares). Reactions were done in the presence of 1 mM ATP.
For the substrate with the 5′ overhang, the unwinding rate constant (kunw) decreased in a biphasic manner with increasing Mg2+ concentrations (Figure 2(a)). Between 0.5 and 10 mM Mg2+, kunw decreased by a factor of ~2 , whereas between 15 and 25 mM Mg2+, kunw decreased by a factor of ~50. Further analysis showed that the 400 nM Mss116p used in these experiments was saturating at concentrations up to ~15 mM Mg2+, while at higher Mg2+ concentrations, the functional binding was decreased to the extent that the enzyme was no longer saturating (data not shown).
For the blunt-end substrate, the decrease of the rate constant with increasing Mg2+ concentrations was not biphasic and much more pronounced than that seen with the tailed substrate. Between 0.5 and 10 mM Mg2+, the unwinding rate constant for the blunt-end substrate decreased by a factor of ~1,000, even though the unwinding rate constants at 0.5 mM Mg2+ were similar for substrates with and without single-stranded overhangs (Figure 2(a)). This more pronounced effect of increasing Mg2+ concentration reflects a scaling of the functional binding of Mss116p with Mg2+. While at Mg2+ concentrations <1 mM, the 400 nM Mss116p used in these experiments was saturating for the blunt-end substrate, higher Mg2+ concentrations decreased the functional binding of the blunt-end substrate, so that Mss116p was no longer saturating (data not shown). The distinct Mg2+ dependencies of the unwinding rate constants for blunt-end and tailed substrates indicate that Mss116p interacts differently with these substrates.
The rate constants for the strand-annealing reactions of both tailed and blunt-end substrates, also decreased with increasing Mg2+ concentrations (Figure 2(b)). For the 5′-tailed substrate, the biphasic nature of the Mg2+ dependence of the annealing rate constant was less pronounced than the unwinding rate constant, although the decrease also appeared biphasic. In addition, differences in the Mg2+ dependence of the annealing rate constants between blunt-end and tailed substrates were smaller than the differences observed for the scaling of unwinding rate constants with Mg2+. These observations suggest that duplex unwinding and strand annealing are two distinct activities that most likely involve different interactions of Mss116p with the RNA.
Mss116p binds non-specifically to group I and group II intron RNAs
The ability of Mss116p to bind group I and group II intron RNAs was tested by equilibrium- binding assays. The experiments were done in reaction medium containing 100 mM KCl and 5 or 8 mM Mg2+ at 30°C and in the presence or absence of 1 mM ATP, AMP-PNP, and ADP. Figure 3 shows a set of representative binding curves at 8 mM Mg2+, and similar results were obtained at 5 mM Mg2+ (data not shown). Mss116p bound to each of the RNAs tested with a Kd of 1 to 11 nM, with little if any significant difference between the different RNAs or in the presence or absence of ATP, AMP-PNP, and ADP. Similar apparently non-specific binding of the same group I and group II intron RNAs was found previously for CYT-19 in the presence of ATP or without added nucleotide (Kd = 20–40 nM),20 but CYT-19 differed in that the Kd decreased 20- to 40-fold in the presence of AMP-PNP (Kd = 0.5–1 nM).9,20 CYT-19 preparations purified more extensively than those used previously showed qualitatively similar behavior, but with tighter binding in the absence of nucleotide or presence of ATP, leading to a smaller differential decrease upon addition of AMP-PNP (data not shown). The behavior of CYT-19 is similar to that reported for other DExH/D-box proteins25 and is thought to reflect that AMP-PNP locks the two domains in a “closed” conformation that has a higher affinity for RNA.26 The relatively unchanged non-specific binding affinity for Mss116p in the presence of AMP-PNP may reflect that it is more disposed to remain in a closed configuration in the absence of nucleotide or a greater contribution to RNA binding from another region of the protein, such as the C-terminal domain (see Discussion).
Figure 3.
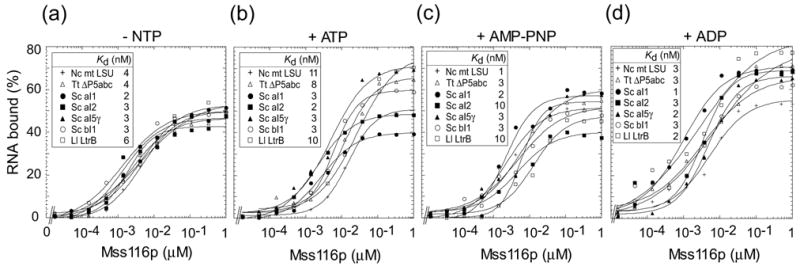
Equilibrium-binding of Mss116p to group I and group II intron RNAs. 32P-labeled RNAs (5 pM) were incubated with increasing concentrations of Mss116p in reaction medium containing 100 mM KCl/8 mM MgCl2 for 90 min at 30°C (a) without nucleotide (-NTP), or with 1 mM (b) ATP, (c) AMP-PNP, or (d) ADP. RNAs were group I introns N. crassa mt LSU-ΔORF and T. thermophila LSU-ΔP5abc, and group II introns S. cerevisiae aI1-ΔORF, aI2-ΔORF, aI5γ, and bI1 and L. lactis Ll.LtrB-ΔORF. The plots show the percent input RNA bound versus Mss116p concentration. Essentially identical binding curves for each condition were obtained when the RNA and protein were incubated for 10 or 120 min before filtration through nitrocellulose, or in reaction medium containing 100 mM KCl/5 mM MgCl2 (data not shown).
Mss116p plus ATP stimulates splicing of the N. crassa mt LSU-ΔORF group I intron
To test whether Mss116p could promote group I intron splicing in vitro, we first used a well-characterized substrate for CYT-19, the N. crassa mt LSU-ΔORF intron.9 This group I intron cannot self-splice under any conditions and requires a splicing factor, the N. crassa CYT-18 protein, for structural stabilization (Paukstelis et al.27 and references therein). We showed previously that at 25°C, the physiological growth temperature of N. crassa, the CYT-18-dependent splicing of the mt LSU-ΔORF intron is inefficient, but the reaction is strongly stimulated by CYT-19 plus ATP.9
Figure 4 shows splicing of the N. crassa mt LSU-ΔORF intron with CYT-18 alone (left) or with CYT-18 plus Mss116p (right). The reactions were done with 20 nM 32P-labeled precursor RNA and 100 nM of each protein in reaction medium containing 100 mM KCl, 5 mM Mg2+, and 1 mM ATP at 25°C. As expected, under these conditions, the splicing reaction with CYT-18 alone was slow (kobs= 1–2 x 10−3 min−1), with only a small proportion of the precursor RNA spliced after 120 min. As found previously for CYT-19,9 the addition of Mss116p in the presence of ATP greatly stimulated the reaction (kobs = 56 x 10−3 min−1 for precursor RNA disappearance), with nearly all the precursor spliced within 60 min.
Figure 4.
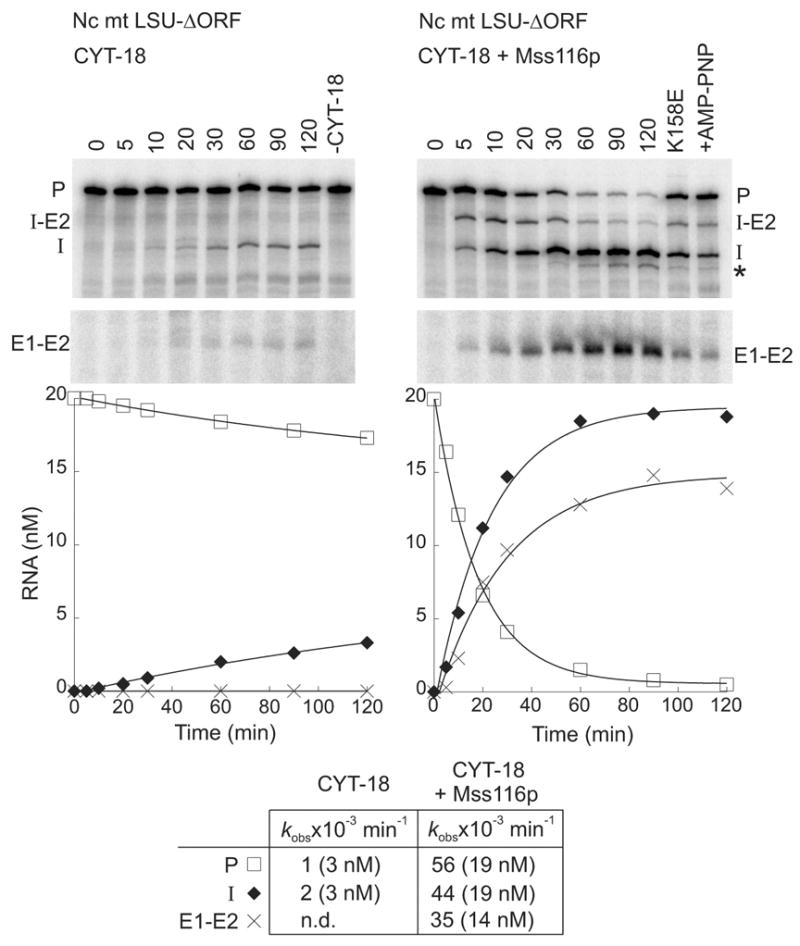
Mss116p promotes splicing of the N. crassa mt LSU-ΔORF group I intron. Splicing time courses for CYT-18 alone (left) and CYT-18 plus Mss116p (right) were done by incubating 32P-RNA substrate (20 nM) with one or both proteins (each at 100 nM) in reaction medium containing 100 mM KCl, 5 mM MgCl2, and 1 mM ATP at 25°C. The splicing products were analyzed in a denaturing 4% polyacrylamide gel, which was dried and quantified with a phosphorimager. In controls (right lanes), RNA was incubated for 120 min with 1 mM ATP without CYT-18 (-CYT-18), with mutant Mss116p-K158E plus 1 mM ATP, or with wild-type Mss116p plus 1 mM AMP-PNP (+AMP-PNP). The plots beneath the gel show the disappearance of precursor RNA and the appearance of products as a function of time, with the data fit to a single exponential. The table at the bottom summarizes kobs values, with the numbers in parentheses indicating the concentration of RNA (nM) reacted or produced after 120 min. Abbreviations: E1–E2, ligated exons; I, intron; I-E2, intermediate after the first step of splicing; P, precursor RNA; *, slightly shorter intron RNA resulting from use of an upstream cryptic 3′-splice site; n.d., not detected.
Surprisingly, while the stimulation of the splicing reaction by Mss116p was decreased in the presence of AMP-PNP or for the Mss116p-K158E mutant, indicating a requirement for ATP hydrolysis (Figure 4, right hand lanes), it was not abolished completely, in contrast to previous results for CYT-19.9 Both the residual Mss116p stimulation in the absence of ATP hydrolysis and the difference in behavior for CYT-19 were confirmed by additional time-course experiments carried out in parallel with the two proteins using the same reagents (Figure 5(a) and (b), respectively). These experiments also showed only minimal stimulation by Mss116p in the absence of ATP, but surprisingly high stimulation by Mss116p in the presence of ADP, which was again not observed in parallel experiments for CYT-19 (Figure 5(a) and (b), and data not shown). For Mss116p, controls showed similar stimulation by ADP incubated with hexokinase plus glucose to remove any residual ATP,28 and for ATP quantitatively converted to ADP by incubation with hexokinase plus glucose; in both cases, ATP hydrolysis was confirmed to be complete by thin-layer-chromatography with trace [γ-32P]-ATP added just prior to the hexokinase plus glucose (data not shown). Together, these findings show that Mss116p stimulates the CYT-18-dependent splicing of the N. crassa mt LSU intron both by an ATP-hydrolysis-dependent mechanism, like CYT-19, and by one or more ATP-hydrolysis-independent mechanisms, which are not observed for CYT-19. Further, the ATP-hydrolysis-independent stimulation appears to be favored by the binding of specific nucleotides, particularly ADP.
Figure 5.
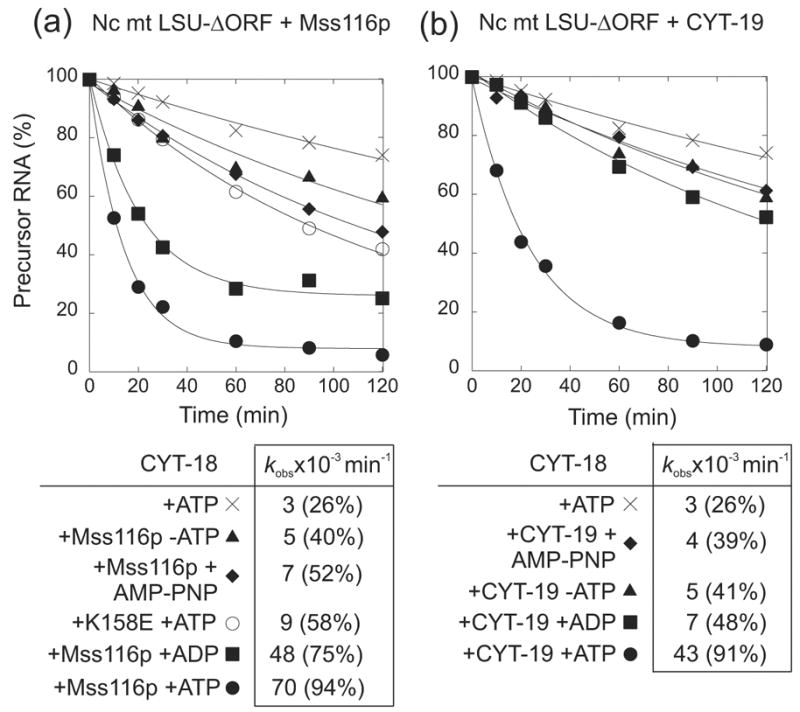
Mss116p promotes CYT-18-dependent splicing of the N. crassa mt LSU-ΔORF group I intron by both ATP-hydrolysis-dependent and ATP-hydrolysis independent mechanisms. Splicing time courses were done with 20 nM 32P-labeled precursor RNA, 100 nM CYT-18 dimer, and (a) 100 nM Mss116p or (b) 100 nM CYT-19 in reaction medium containing 100 mM KCl and 5 mM MgCl2 at 25°C in the presence or absence of 1 mM ATP, AMP-PNP, and ADP. The time course for Mss116p-K158E in (a) was done in the presence of 1 mM ATP. The tables below the plots summarize kobs values for the disappearance of precursor RNA, with the numbers in parentheses indicating the percent of precursor RNA reacted after 120 min.
Mss116p inhibits splicing of the Tetrahymena thermophila LSU-ΔP5abc group intron
Another surprising result was obtained for the T. thermophila LSU-ΔP5abc intron. This intron is unable to self-splice at low (5 mM) Mg2+ concentrations, due to deletion of the large peripheral structure P5abc,29 but can splice if CYT-18 is provided for structural stabilization.30 We showed previously that the CYT-18-dependent splicing of the T. thermophila LSU-ΔP5abc intron is stimulated by CYT-19 plus ATP, and we used this system to test the ability of CYT-19 to resolve different types of non-native secondary and tertiary structures (Mohr et al.,9 and unpublished data). Surprisingly, the addition of Mss116p plus ATP inhibited the CYT-18-dependent splicing of the T. thermophila LSU-ΔP5abc intron, and this inhibition was proportional to the amount of Mss116p protein added (Figure 6(a)). Additional experiments showed similar inhibition by wild-type Mss116p or Mss116p-K158E in the absence of nucleotide or presence of ATP, AMP-PNP, ADP, but no inhibition when Mss116p was denatured by boiling (Figure 6(b)). By contrast, in parallel reactions with the same reagents, CYT-19 plus ATP stimulated the CYT-18-dependent splicing of the T. thermophilia LSU-ΔP5abc intron (Figure 6(a) and (b)), in agreement with previous results.9 These findings suggest that Mss116p is more discriminatory than CYT-19, and in one case, actually impedes splicing. This inhibition of splicing by Mss116p may reflect that its “non-specific” binding stabilizes an inactive structure of the intron RNA or impedes the binding of CYT-18.
Figure 6.
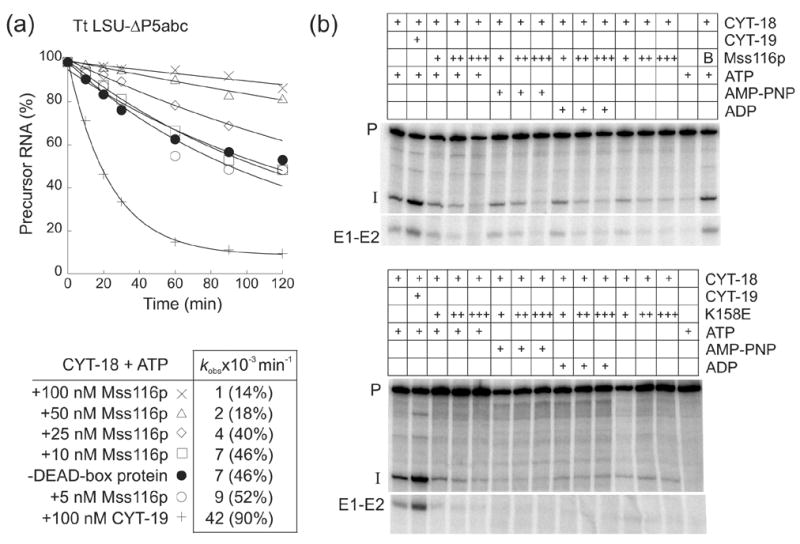
Mss116p inhibits CYT-18-dependent splicing of the T. thermophila LSU-ΔP5abc group I intron. (a) Splicing time courses were done with 50 nM 32P-labeled precursor RNA, 50 nM CYT-18 dimer, and 5 to 100 nM Mss116p or 100 nM CYT-19 in reaction medium containing 100 mM KCl and 5 mM MgCl2 at 30°C. The products were analyzed in a denaturing 4% polyacrylamide gel, which was dried and scanned with a phosphorimager. The table below the plots summarizes kobs values for the disappearance of precursor RNA, with the numbers in parentheses indicating the percent of precursor RNA reacted after 120 min. (b) Splicing reactions were carried out as above for 30 min with different concentrations of wild-type Mss116p or Mss116p-K158E in the presence or absence of 1 mM ATP, AMP-PNP, or ADP. In panel (a) reactions were initiated by adding CYT-18; in panel (b), precursor RNA was preincubated with 50 nM CYT-18 for 10 min at 30°C, and the reaction was initiated by adding the DEAD-box protein plus 1 mM GTP. Concentrations of wild-type Mss116p and Mss116p-K158E were 25 nM (+), 50 nM (++), and 100 nM (+++). In the top gel, the far right lane shows a control reaction in which 50 nM wild-type Mss116p was boiled (B) for 5 min prior to the start of the splicing reaction. Abbreviations are as in Figure 4.
Mss116p plus ATP promotes splicing of the S. cerevisiae aI5γ and bI1 group II introns
We next tested whether Mss116p plus ATP alone could promote the splicing of the S. cerevisiae aI5γ and bI1 group II introns, which had been shown previously to be promoted by CYT-19 plus ATP under near physiological conditions (100 mM KCl, 5 mM MgCl2 and 30°C).20 Figure 7 shows splicing time courses for both group II introns carried out with 100 nM Mss116p and 20 nM 32P-labeled precursor RNA in reaction medium containing 100 mM KCl and 8 mM Mg2+ at 30°C. The Mg2+ concentration was increased from that used previously for the CYT-19-promoted splicing reactions to alleviate problems due to the very sharp cutoff of Mss116p- or CYT-19-promoted splicing of aI5γ and bI1 around 5 mM Mg2+ (see Mohr et al.,20 and below).
Figure 7.
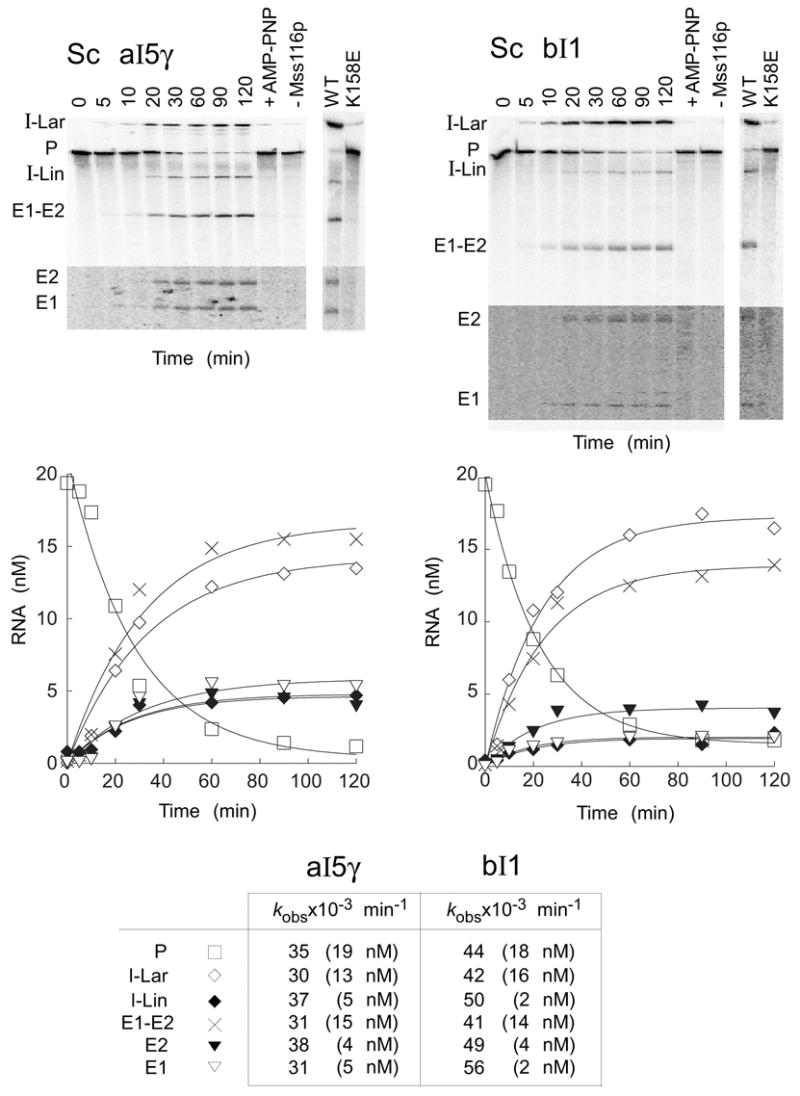
Mss116p-promoted splicing of group II introns aI5γ and bI1. Splicing time courses for aI5γ (left) and bI1 (right) were done by incubating 20 nM 32P-labeled RNA substrate with 100 nM Mss116p protein and 1 mM ATP in reaction medium containing 100 mM KCl and 8 mM MgCl2 at 30°C. Splicing products were analyzed in a denaturing 4% polyacrylamide gel, which was dried and quantified with a phosphorimager. The bottom segments of the gels are darker exposures to show E1 and E2, which result form SER. Control lanes show RNA substrate incubated for 120 min with Mss116p plus 1 mM AMP-PNP (+AMP-PNP) or without Mss116p (-Mss116p). The additional lanes (far right) show precursor RNAs incubated with wild-type (WT) Mss116p and mutant Mss116p-K158E under the same conditions for 120 min. The plots beneath the gels show disappearance of precursor RNA and appearance of products as a function of time, with the data fit to a single exponential. The table at the bottom summarizes kobs values, with the numbers in parentheses indicating the concentration of RNA (nM) reacted or produced after 120 min. The I-Lin band may contain a mixture of linear intron and broken lariat RNA. Abbreviations: E1, 5′ exon; E2, 3′ exon; E1–E2, ligated exons; I-Lar, intron lariat; I-Lin, linear intron; P, precursor RNA.
As found previously for CYT-19, Mss116p plus ATP alone promoted the robust splicing of both group II introns, and the reactions were entirely dependent upon ATP hydrolysis, with only a very light intron lariat band due to slow self-splicing in the absence of Mss116p (-Mss116p), with Mss116p plus AMP-PNP, or with the Mss116p-K158E mutant protein (Figure 7). Other experiments showed a similar lack of stimulation by Mss116p plus 1 mM ADP (data not shown). In the splicing reactions promoted by Mss116p plus ATP, the disappearance of precursor RNA (P) and appearance of ligated exons (E1–E2) and excised intron lariat RNA (I-Lar) were monophasic for both introns with kobs = 30–35 x 10−3 min−1 for aI5γ and 41–44 x 10−3 min−1 for bI1, similar to the rate constants for CYT-19 plus ATP,20 and nearly all of the precursor RNA spliced within 120 min.
In addition to excised intron lariat and ligated-exon products, the Mss116p-promoted splicing reactions also produced smaller amounts of linear intron RNA (I-Lin) and free 5′ and 3′ exons (E1 and E2). The former could result from splicing via 5′-splice site hydrolysis or breakage of intron lariat,31 while the latter results from cleavage of ligated exons by the excised intron RNA, a reaction known as spliced-exon reopening (SER).32 Both the I-Lin and SER products are present in somewhat higher amounts than seen previously for the CYT-19-promoted splicing reactions of the same introns,20 but these differences reflect at least in part the higher Mg2+ concentration used in the Mss116p reactions (see below).
The splicing rates for aI5γ and bI1 in the presence of Mss116p and ATP (kobs = 30–44 x 10−3 min−1) are similar to those for CYT-1920 and comparable to rates under optimal self-splicing conditions (kobs = 28–120 x 10−3 min−1),31,33 even though the latter reactions are carried out at substantially higher temperature. Nevertheless, the splicing rates remain much slower than in vivo, possibly reflecting that the aI5γ and bI1 introns require additional as yet unidentified proteins for structural stabilization.
Mg2+-concentration dependence of Mss116p-promoted splicing of aI5γ and bI1
As found previously for CYT-19, the Mg2+ concentration is a critical parameter for Mss116p-promoted splicing of the aI5γ and bI1 group II introns.20 Figure 8 shows that in reaction medium containing 100 mM KCl at 30°C, the Mss116p-promoted splicing of aI5γ and bI1 becomes detectable at 5 and 6 mM Mg2+, respectively, with a very sharp cutoff such that no reaction is observed at lower Mg2+ concentrations. Above these concentrations, the reactions have a broad rate optimum between 6 and 15 mM Mg2+ for aI5γ (kobs = 22–41 x 10−3 min−1) and between 7 and 25 mM Mg2+ for bI1 (kobs = 21–51 x 10−3 min−1) (shaded in Figure 8, bottom). Within this range of Mg2+ concentrations, the time courses for precursor RNA disappearance were very similar, with splicing complete after 2 h. The reactions were inhibited at higher Mg2+ concentrations, becoming undetectable at 50 mM Mg2+ for both introns. The very sharp cutoff at 5–6 mM Mg2+ may reflect highly cooperative Mg2+-binding by group II intron RNAs. The inhibition of splicing at Mg2+ concentrations >25–35 mM could be due to impaired binding, decreased duplex-unwinding and strand-annealing activities (see Figure 2), or increased stabilization of inactive RNA structures at high Mg2+ concentrations. Higher Mg2+ concentrations also increased the production of I-Lin and SER products (Figure 8), presumably reflecting the formation of somewhat different active RNA structures that favor these products. For these reasons, we selected 8 mM Mg2+, the lowest Mg2+ concentration that gives close to the optimal rate and amplitude, as a standard condition for the Mss116p-promoted aI5γ and bI1 splicing reactions.
Figure 8.
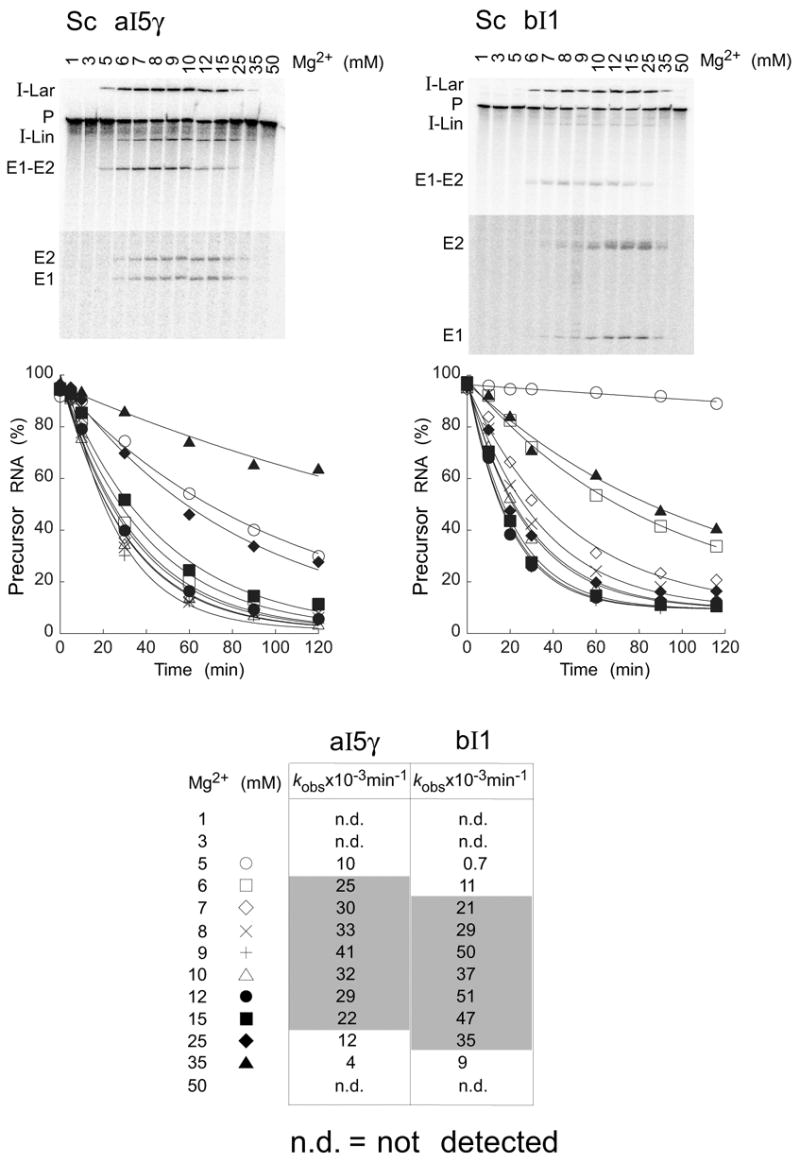
Mg2+-concentration dependence of Mss116p-promoted splicing of aI5γ and bI1. Splicing time courses for aI5γ (left) and bI1 (right) were done by incubating 20 nM 32P-labeled precursor RNAs with 100 nM Mss116p plus 1 mM ATP in reaction media containing 100 mM KCl and the indicated Mg2+ concentrations at 30°C. Products were analyzed and quantified, as in Figure 7. The gels show the extent of splicing after 20 min at different Mg2+ concentrations, and the plots show disappearance of precursor RNA as a function of time, with the data fit to a single exponential. The bottom segments of the gels are darker exposures to show E1 and E2, which result form SER. The table at the bottom summarizes kobs values for aI5γ and bI1 precursor RNA disappearance at different Mg2+ concentrations. The shaded area highlights the broad Mg2+ optimum. Abbreviations are as in Figure 7.
Mss116p-concentration dependence of aI5γ and bI1 splicing
Figure 9 shows splicing time courses done with 20 nM 32P-labeled precursor RNA and different concentrations of Mss116p in reaction medium containing 100 mM KCl and 8 mM Mg2+ at 30°C. For both introns, the results showed that the rate constant increases with increasing protein concentration, as expected if the reaction is limited by the binding of Mss116p. At low protein concentrations, the molar amount of precursor RNA spliced exceeds the amount of Mss116p present in the reaction, indicating turnover of Mss116p. However, as found for CYT-19,20 turnover was limited; little additional splicing occurred after 2 h at any protein concentration and was difficult to quantify because of RNA instability. For both group II introns, the amount of Mss116p required for maximal stimulation of splicing (50–100 nM) was substantially lower than that for CYT-19 (500 nM-1 μM; Mohr et al.,20 and data not shown).
Figure 9.
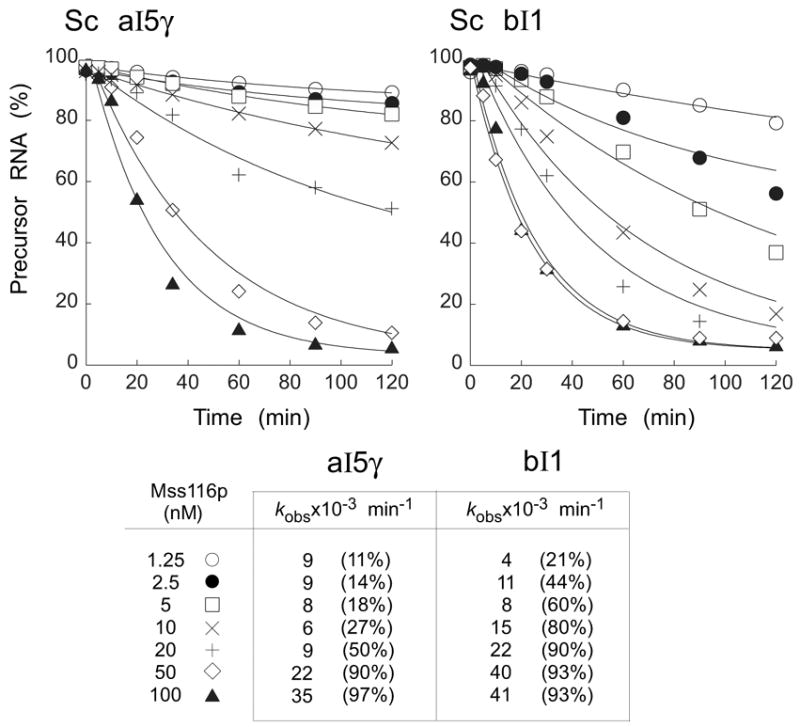
Mss116p-concentration dependence of aI5γ and bI1 group II intron splicing. Splicing time courses for aI5γ (left) and bI1 (right), respectively, were done with 20 nM 32P-labeled precursor RNA and 1.25 to 100 nM Mss116p in reaction media containing 100 mM KCl and 8 mM MgCl2 at 30°C. The plots show disappearance of precursor RNA as a function of time, with data fit to a single exponential. The table at the bottom summarizes kobs values and amplitudes (%) at 120 min. Abbreviations are as in Figure 7.
Behavior of Mss116p in RNA chaperone assays
The RNA chaperone activity of CYT-19 was demonstrated previously by showing that CYT-19 plus ATP could act on bI1 lariat RNA to promote its reverse-branching and reverse-splicing into RNA-oligomer substrates and that these reactions occurred with undiminished efficiency when CYT-19 was removed by protease digestion prior to adding the RNA oligomer substrate.20 However, we were unable to find conditions where Mss116p plus ATP promotes the reverse branching or reverse splicing of bI1. 100 mM Mss116p, which maximally stimulates bI1 splicing, had no effect on its reverse branching or reverse splicing into the RNA-oligomer substrates, while 500 nM Mss116p inhibited even the slow background reaction that was observed in the absence of added protein, presumably due to non-specific binding (data not shown). Similar results were obtained with an assay system based on the refolding of misfolded forms of the T. thermophila LSU ribozyme (data not shown).16
Mss116p plus ATP stimulates RNA-oligomer cleavage by the D135 ribozyme
To investigate the mechanism of action of Mss116p and dissect individual steps at which it acts, we turned to a simplified ribozyme derivative of aI5γ, which consists of aI5γ domains 1, 3, and 5.34 Under conditions similar to those required for self-splicing of aI5γ (500 mM KCl, 100 mM MgCl2 at 42°C), D135 RNA has been shown to cleave an RNA-oligomer substrate containing the 5′ exon-intron junction in an efficient multi-turnover reaction (kobs ~ 0.6 min−1).35,36 First, we looked for milder conditions under which Mss116p plus ATP stimulates RNA-oligomer cleavage by the D135 ribozyme relative to the RNA-only reaction. Figure 10(a) shows reactions in which 50 nM Mss116p plus 1 mM ATP was incubated with 50 nM D135 ribozyme and 5 nM RNA substrate at 25°C in reaction media containing 100 mM KCl and different Mg2+ concentrations. The results show that Mss116p plus ATP stimulated RNA cleavage by the D135 ribozyme under these conditions at Mg2+ concentrations between 12.5 and 25 mM, and like group II intron splicing, the reaction was completely dependent upon ATP hydrolysis, with no detectable stimulation in the absence of Mss116p or in the presence of Mss116p plus 1 mM AMP-PNP (Figure 10(a)) or 1 mM ADP (data not shown). The higher Mg2+ concentration required for activity of the D135 RNA compared to intact intron (see Figure 8) presumably reflects that D135 RNA requires additional structural stabilization due to the lack of intron RNA domains 2, 4, and 6.
Figure 10.
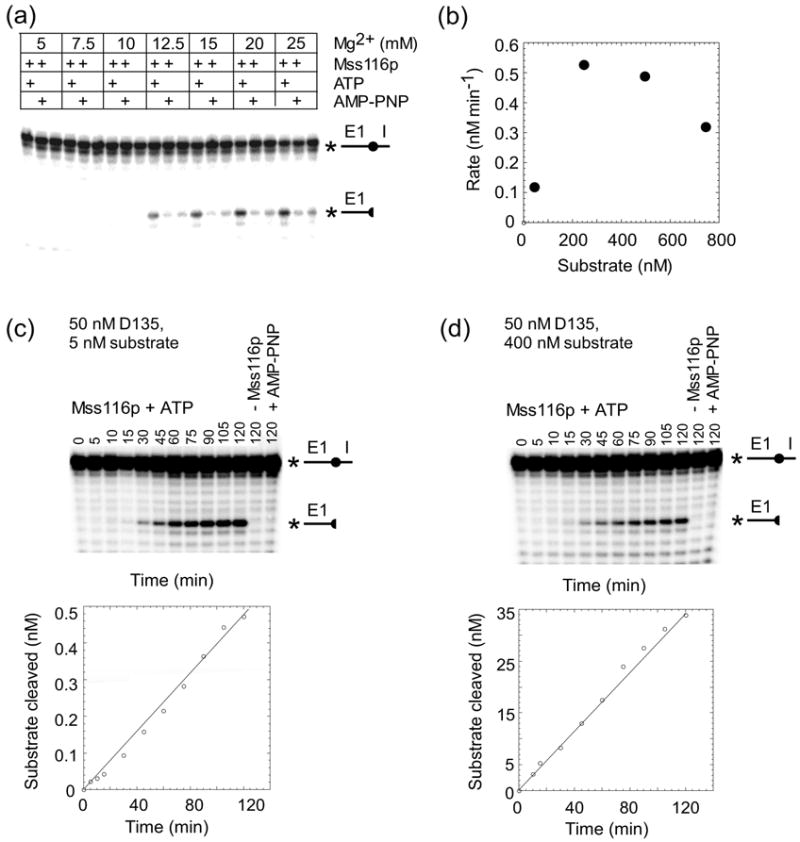
Mss116p promotes RNA-oligomer cleavage by the D135 ribozyme. (a) Mg2+-concentration dependence. D135 RNA (50 nM) was pre-incubated at 25°C with Mss116p (50 nM) for 5 min in reaction medium containing 100 mM KCl and the indicated Mg2+ concentrations in the presence or absence of 1 mM ATP or AMP-PNP. The reaction was initiated by adding 5′-32P-labeled RNA-oligomer substrate (5 mM), incubated for 90 min at 25°C, and terminated by adding 100 mM EDTA, followed by phenol-CIA extraction. The products were analyzed in a denaturing 20% polyacrylamide gel, which was dried and quantified with a phosphorimager. (b) Substrate-concentration dependence. For each substrate concentration (25, 250, 500, and 750 nM), a 2 h-time course were done as above with 50 nM D135 ribozyme and 50 nM Mss116p plus 1 mM ATP in reaction medium containing 100 mM KCl and 25 mM MgCl2 at 25°C. The plot shows the rate of cleavage as a function of RNA substrate concentration. (c) and (d) Time courses of D135-catalyzed RNA-cleavage with substoichiometric and saturating concentrations of RNA-oligomer substrate, respectively. 50 nM D135 RNA was pre-incubated with 50 nM Mss116p and 1 mM ATP in reaction medium containing 100 mM KCl and 25 mM Mg2+ for 5 min at 25°C, and the reaction was initiated by adding 5 or 400 nM RNA substrate in (c) or (d), respectively. The right-hand lanes show control reactions with the same amount of RNA substrate incubated for 2 h at 25°C in the absence of Mss116p or with 1 mM AMP-PNP instead of ATP. The plots beneath the gels show the appearance of product as a function of time. The 5′-32P-labeled RNA oligomer substrate and labeled cleavage product are depicted schematically to the right of the gels, with the circle indicating the 5′-exon-intron junction, and the star indicating the location of the label.
For subsequent experiments with D135 RNA, we used reaction medium containing 100 mM KCl, 25 mM Mg2+, and 25°C, both because of the high reaction rate compared to the RNA-only reaction under these conditions and because the relatively high Mg2+ concentration and low temperature were expected to maximize dependence upon an RNA chaperone to alleviate kinetic traps. Figure 10(b) shows that under these conditions with 50 nM each of D135 and Mss116p, the reaction was saturated at 250 nM substrate. At high substrate concentrations (750 nM), the reaction was inhibited, possibly reflecting competition between the substrate and D135 RNA for Mss116p binding.
Figures 10(c) and (d) show time courses for reactions with 50 nM D135 ribozyme at substoichiometric (5 nM) and saturating (400 nM) substrate concentrations. At both substrate concentrations, the reaction was stimulated by Mss116p plus ATP, but not by Mss116p in the absence of ATP or presence of AMP-PNP. The reaction rates with 5 and 400 nM substrate were 0.0042 and 0.3 nM min−1, respectively, with the reaction linear for at least 2 h in both cases.
The finding that Mss116p plus ATP stimulates the D135 ribozyme reaction at saturating substrate concentrations suggests that Mss116p acts by promoting a conformational change in the D135 ribozyme, likely acceleration of the rate-limiting disruption of a stable non-native or intermediate structure (“kinetic trap”). An alternative possibility that Mss116p plus ATP stimulates the reaction by promoting product release is unlikely because stimulation is also observed at substoichiometric substrate concentrations (i.e., single-turnover conditions; Figure 10(c)). Under these conditions, product release should not be rate-limiting, so long as the reaction equilibrium favors cleavage. The latter was expected for a hydrolysis reaction in aqueous solution, and we confirmed by separate experiments with RNA oligonucleotides corresponding to the cleavage products that the reverse reaction is undetectable under the conditions used in these experiments (data not shown).
Mss116p- or CYT-19-stimulated D135 ribozyme RNA cleavage requires an ATP-hydrolysis dependent step after substrate binding
Having established conditions under which Mss116p plus ATP stimulates RNA cleavage by the D135 ribozyme, we next wanted to test whether it could act productively on D135 RNA and then be removed by protease digestion, prior to adding RNA-oligomer substrate. Figures 11(a) shows such an experiment for Mss116p, and Figure 11(b) shows a parallel experiment for CYT-19. In both cases, protease digestion of the DEAD-box protein prior to the addition of substrate abolished stimulation by Mss116p. This result could reflect either that the putative RNA conformational change promoted by the DEAD-box protein can only occur after substrate binding or that the continued binding of the protein to the D135 RNA is needed to maintain the active structure of the ribozyme at the Mg2+ concentrations used in the experiments.
Figure 11.
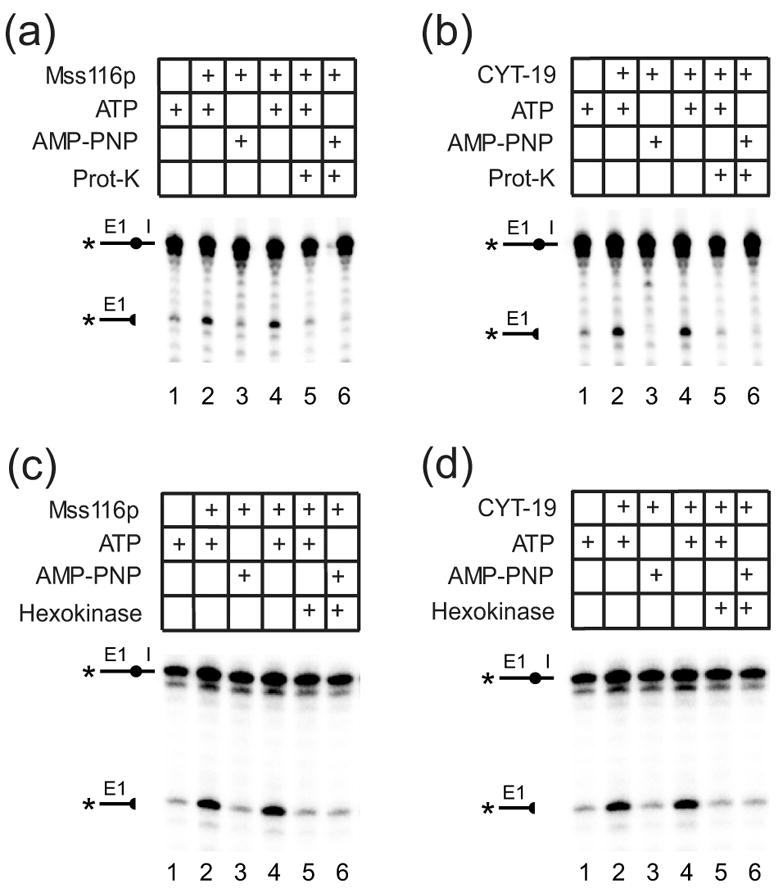
Protease sensitivity of DEAD-box protein-promoted D135-ribozyme cleavage reactions and requirement for ATP-hydrolysis after substrate binding. (a) and (b), protease sensitivity of D135 ribozyme cleavage promoted by (a) Mss116p or (b) CYT-19. Mss116p (100 nM) or CYT-19 (500 nM) were incubated with D135 RNA (100 nM) and RNA-oligomer substrate (5 nM) without or with 1 mM ATP or AMP-PNP in reaction medium containing 100 mM KCl and 25 mM MgCl2 at 25°C. Lanes 1-3, D135 RNA was pre-incubated in reaction medium for 90 min with ATP (lane 1), DEAD-box protein plus ATP (lane 2), or DEAD-box protein plus AMP-PNP (lane 3). Lanes 4 and 5, D135 RNA was pre-incubated for 60 min with DEAD-box protein plus ATP or AMP-PNP, respectively, then split with halves incubated for 30 min without (lane 4) or with 20 mg/ml proteinase K (prot-K) and 0.5% SDS (lane 5). Lane 6, D135 RNA was pre-incubated with DEAD-box protein for 60 min, then for 30 min with proteinase K plus SDS. After the pre-incubations, reactions were initiated by adding 5′-32P-labeled RNA oligomer, incubated for 30 min, then terminated, and products analyzed in a denaturing 20% polyacrylamide gel. (c) and (d), effect of ATP-depletion prior to substrate addition. Reactions were done as above, with D135 RNA pre-incubated with the indicated additions for 30 min, and then for an additional 15 min without or with addition of hexokinase plus glucose. Reactions were initiated by adding RNA-oligomer substrate, incubated for 120 min, and products analyzed as above.
To distinguish these possibilities, we carried out the experiment shown in Figure 11(c) and (d) in which the D135 ribozyme was again preincubated with Mss116p or CYT-19 plus ATP, and hexokinase plus glucose was then added to deplete ATP prior to initiating the reaction by adding the RNA-oligomer substrate. For both proteins, no stimulation was observed if ATP was depleted prior to substrate addition, indicating that the DEAD-box protein-promoted RNA conformational change must occur during or after substrate binding. A control experiment showed that the same amount of hexokinase added in the absence of glucose did not inhibit the Mss116p- or CYT-19-promoted reactions (data not shown). Together, these findings indicate that the requirement for the continued presence of Mss116p or CYT-19 is not due simply to RNA binding. It may reflect instead that substrate binding induces a required intermediate structure that is then acted upon by the DEAD-box protein plus ATP and/or that the active structure of D135 RNA is unstable and rapidly reverts to an inactive structure in the absence of DEAD-box protein plus ATP.
The yeast DEAD-box protein Ded1p can also promote group II intron splicing
Finally, the broad range of action and interchangeability of CYT-19 and Mss116p in many reactions prompted us to ask whether other DExH/D-box proteins might have an inherent RNA chaperone activity that enables them to promote the splicing of autocatalytic mt introns. As an initial test, we used the splicing reactions of group II introns aI5γ and bI1, which are stimulated by CYT-19 or Mss116p alone in the absence of partner proteins. The yeast DEAD-box protein Ded1p ordinarily functions in mRNA translation in the cytosol37,38 and may have other functions in RNA splicing and ribosome assembly in the nucleus,39 cellular compartments that do not contain group I or group II introns. As shown in Figure 12(a), Ded1p added at saturating concentrations (300 nM) stimulated the splicing of aI5γ, with rates and amplitudes similar to those for CYT-19 and Mss116p (kobs = 14–17 min−1, 17 nM precursor reacted after 120 min). Control lanes to the right of the gel show that the stimulation was not observed with AMP-PNP or ADP, indicating that the effect of Ded1p on aI5γ splicing requires ATP hydrolysis.
Figure 12.
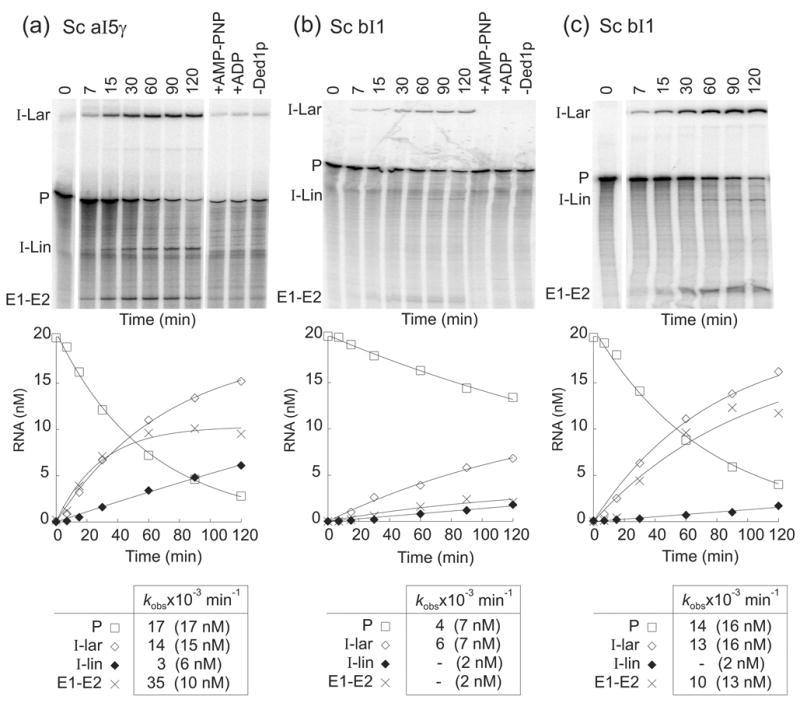
Ded1p-promoted splicing of group II introns aI5γ and bI1. Splicing time courses for (a) aI5γ with 300 nM Ded1p, and (b) and (c), bI1 with 300 and 600 nM Ded1p, respectively, were done using 20 nM 32P-labeled precursor RNA and 1 mM ATP in reaction medium containing 100 mM KCl and 8 mM MgCl2 at 30°C. Splicing products were analyzed in a denaturing 4% polyacrylamide gel, which was dried and quantified with a phosphorimager. Control lanes show precursor RNA incubated for 120 min without Ded1p (-Ded1p) or with Ded1p plus 1 mM AMP-PNP (+AMP-PNP) or 1 mM ADP (+ADP). The plots beneath the gels show disappearance of precursor RNA and appearance of products as a function of time, with the data fit to a single exponential. The table at the bottom summarizes kobs values, with the numbers in parentheses indicating the concentration of RNA (nM) reacted or produced after 120 min. kobss could not be determined for the low amounts of I-Lin and E1–E2 produced in panel (b) and I-Lin in panel (c) due to high errors in the curve fitting. The I-Lin band may contain a mixture of linear intron and broken lariat RNA. Abbreviations are as in Figure 7.
Figures 12(b) and (c) show that Ded1p also stimulated the splicing of the yeast bI1 intron in an ATP-hydrolysis-dependent manner. In this case, however, twice the amount of Ded1p (600 nM) was required to saturate the reaction and achieve a level of stimulation similar to aI5γ (kobs = 13–14 min−1, 16 nM precursor RNA reacted after 120 min). In preliminary experiments, the E. coli DEAD-box proteins DbpA and the vaccinia virus DExH-box protein NPH-II had no effect on the splicing of either intron, while the E. coli DEAD-box protein SrmB gave only minimal stimulation up to the highest protein concentrations tested (data not shown). Together, the above findings indicate that Ded1p has an inherent RNA chaperone activity that can be assayed by its effect on splicing group II intron splicing, but this activity appears more efficient for aI5γ than for bI1.
Discussion
Here, we expressed the yeast mt DEAD-box protein Mss116p in E. coli and characterized it biochemically. We find that the purified recombinant Mss116p is similar to other DEAD-box proteins in having RNA-dependent ATPase, ATP-dependent duplex-unwinding, and ATP-independent strand-annealing activities. Further, we show that Mss116p binds RNAs non-specifically, enabling it to use its biochemical activities to promote the in vitro splicing of structurally diverse group I and group II introns, as well as RNA cleavage by the D135 ribozyme. Our results suggest that, like the related N. crassa mt DEAD-box protein CYT-19, Mss116p promotes group I and group II intron splicing reactions primarily by using the energy of ATP hydrolysis to disrupt stable intermediate structures that are kinetic traps in RNA folding. However, we also find that Mss116p differs from CYT-19 in having additional ATP-hydrolysis-independent effects, particularly on group I intron splicing reactions, and is unable to stimulate splicing of at least one group I intron whose splicing is stimulated strongly by CYT-19. Similarly, a non-mitochondrial DEAD-box protein, yeast Ded1p, functions almost as efficiently as CYT-19 or Mss116p in promoting splicing of the yeast aI5γ group II intron, and less efficiently in splicing the bI1 group II intron. Our findings show that DEAD-box protein RNA chaperones are not completely interchangeable and likely have biochemical activities that were tuned by co-evolution to work optimally on specific RNA substrates.
Mss116p is a non-polar RNA helicase that also facilities strand annealing
The duplex-unwinding and strand-annealing activities of Mss116p are comparable to those of other DEAD-box proteins. Like many other DEAD-box proteins, Mss116p did not display a significant preference for single-stranded regions in a particular orientation with respect to the duplex, and the unwinding rate constants were within the same order-of-magnitude albeit slightly slower than those for Ded1p with identical substrates under identical conditions (data not shown).23 Notably, the functional binding of Mss116p to RNA substrates and the influence of single-stranded regions were significantly affected by Mg2+ concentration. At low Mg2+ concentrations (0.5 mM), where the substrates could be saturated with the enzyme, Mss116p unwound blunt-end duplexes as fast as those with single-stranded regions (Table 3), indicating that single-stranded regions do not affect the rate-limiting step for strand separation. Further, the lack of a preferred unwinding polarity by Mss116p and other DEAD-box proteins suggests that duplex unwinding by these enzymes does not involve directional translocation along one of the RNA strands.
At higher Mg2+ concentrations, however, a 5′-tailed substrate was unwound faster than a blunt-end substrate, with the difference in unwinding rate constants being ~10-fold at 5 mM Mg2+ and ~100-fold at 10 mM Mg2+, spanning the range of Mg2+ concentrations used in the group I and group II intron splicing reactions (Figure 2(a)). Further experiments indicated that these differences reflect that, with increasing Mg2+ concentration, the affinity of Mss116p for the blunt-end substrate decreases more strongly than that for the tailed substrate (data not shown). Collectively, these findings suggest that although single-stranded regions adjoining duplexes are not critical for the strand separation, they could significantly aid the binding of Mss116p to group I and group II intron RNAs under the conditions of the splicing reactions.
In addition to its duplex unwinding activity, Mss116p also promoted strand annealing with and without ATP. The enzyme accelerated the bimolecular rate constant for duplex formation by several orders of magnitude, close to the physical limit imposed by diffusion,22 and similar to the profound strand-annealing activity of the DEAD-box protein Ded1p.23 As for duplex unwinding, the rate constants for strand annealing by Mss116p decreased with increasing Mg2+ concentrations, but with less difference between a tailed and blunt-end substrate (Figure 2(b)). Unlike some DEAD-box proteins with strand-annealing activity, Mss116p does not have multiple RG clusters in its N- or C-terminal regions.23 It does, however, have a significant number of arginines and other charged amino acids in these regions, which could contribute to strand annealing.40
While the physiological ramifications of the strand-annealing activity of DEAD-box proteins remain to be investigated, we note that enzymes capable of both disrupting and facilitating formation of RNA structure may effectively chaperone a complex RNA into its active structure, without the need for numerous other factors. Moreover, the ratio of unwinding and annealing activities are likely to depend upon ATP and ADP concentrations and on features of the RNA structure, as shown previously for Ded1p.23 Thus, an enzyme like Mss116p that concurrently unwinds and anneals may be able to act on large complex RNAs, like group I and group II introns, at many different positions, yet elicit distinct effects at the various sites.
RNA-binding properties of Mss116p
Like CYT-19, Mss116p binds a variety of group I and group II intron RNAs with similar affinities (Figure 3). This apparently non-specific RNA binding presumably reflects interaction at multiple sites in the intron RNAs and is key to enabling both DEAD-box proteins to act broadly as RNA chaperones to resolve different types of kinetic traps in diverse RNA and RNP substrates. Mss116p differs from CYT-19 in having a somewhat higher nucleotide-independent RNA-binding affinity, which is not significantly affected by AMP-PNP, whereas for CYT-19, AMP-PNP decreased the Kds for the same group I and group II intron RNAs to the same range found for Mss116p in the absence of nucleotide.9,20 The latter behavior, which is similar to that of other DExH/D-box proteins, is thought to reflect that AMP-PNP locks the two RecA-like domains of the core helicase region in a closed configuration that binds RNA.26 The minimal effect of AMP-PNP on the overall RNA-binding affinity of Mss116p may reflect either that the two RecA-like domains are more disposed toward the closed configuration in the absence of ATP or that there is a greater contribution to RNA binding from another region of the protein, such as the C-terminal domain. We also note that the Kds measured by equilibrium binding are presumably an average for binding at multiple different sites within group I and group II intron RNAs, and it remains possible that ATP, ADP, or AMP-PNP differentially affect the affinity of Mss116p for different types of sites without affecting the overall Kd values.
Mss116p plus ATP stimulates the splicing of both group I and group II introns
In vivo, Mss116p is required for the efficient splicing of all twelve S. cerevisiae mtDNA introns (eight group I and four group II introns10). These introns are structurally diverse, interact with a variety of different splicing factors for structural stabilization, and are likely subject to different types of kinetic traps.10 Previous studies suggested that the N. crassa mt LSU-ΔORF intron forms multiple different inactive structures that can be resolved by CYT-19, but are not sufficiently populated to be detected by chemical modification,9 and this may also be the case for many of the other introns acted on by Mss116p.
As expected from its function in vivo, we show here that Mss116p can act in conjunction with the N. crassa CYT-18 protein to promote the splicing of the N. crassa mt LSU-ΔORF intron, and that Mss116p alone promotes the splicing of the yeast aI5γ and bI1 group II introns. The ability of Mss116p to maximally promote group I and group II introns splicing is dependent upon ATP hydrolysis and presumably reflects primarily the ability of Mss116p to disrupt stable on- or off-pathway intermediates, enabling the intron RNAs to fold more rapidly into the final active structure, as first demonstrated for CYT-19 in group I intron splicing.9,16 In addition to acting as an RNA chaperone, an RNA helicase could also potentially stimulate group I or II intron splicing reactions with unfavorable equilibria by accelerating the release of ligated-exon products, which for both types of introns are involved in base-pairing interactions with the catalytic core. However, our findings for D135 RNA and the minimal effect of some other active RNA helicases on the rate of group II intron splicing (see Results) suggest that this mechanism does not make a major contribution, at least in those cases. Additionally, while RNA binding or strand annealing by DEAD-box proteins may contribute to the stimulation of group I and II intron splicing, as appears to be the case for Mss116p-promoted splicing of the N. crassa mt LSU-ΔORF intron in the presence of ADP, our results show that such ATP-hydrolysis-independent mechanisms alone are insufficient to maximally stimulate group I intron splicing or promote group II intron splicing.
Mss116p promotes RNA cleavage by the D135 ribozyme
In addition to the full-length aI5γ intron, we find that both Mss116p or CYT-19 plus ATP can promote RNA-oligomer cleavage by the aI5γ-derived D135 ribozyme in 100 mM KCl, 12.5–25 mM Mg2+ and 25°C, much closer to physiological conditions than had been used previously to study this RNA.34 Our results suggest that under these milder conditions D135 RNA forms stable non-native or intermediate structures, whose resolution is rate-limiting for RNA folding, and that this step is accelerated by either Mss116p or CYT-19, enabling the RNA to fold into the final active structure. For both DEAD-box proteins, the stimulation of RNA cleavage by D135 RNA is completely dependent upon ATP hydrolysis. However, protease digestion experiments indicate that both proteins are unable to stably pre-fold D135 RNA prior to the addition of substrate, and experiments in which ATP was depleted by using hexokinase and glucose show that the ATP-hydrolysis-dependent step must occur after substrate addition. These findings could reflect that that the DEAD-box proteins act on an intermediate structure that can form only after substrate binding, and/or that the active structure of D135 RNA, whose formation is promoted by the DEAD-box protein plus ATP, rapidly reverts to an inactive structure when the DEAD-box protein is removed or ATP hydrolysis is stopped.
We note that even under our milder conditions, the Mg2+-requirement for Mss116p-promoted D135 ribozyme cleavage remains higher than that for splicing of the full-length aI5γ intron (12.5 and 5 mM Mg2+, respectively), presumably reflecting a requirement for structural stabilization by the RNA segments that are missing from D135 RNA. These missing segments could also significantly change the RNA-folding pathway compared to that used by the full-length intron.
Mss116p has ATP-hydrolysis-independent effects on group I intron splicing reactions
Mss116p differs from CYT-19 in having ATP-hydrolysis-independent effects on group I intron splicing reactions, including a substantial contribution to stimulating CYT-18-dependent splicing of the N. crassa mt LSU-ΔORF intron in the presence of ADP, and inhibition of splicing of the T. thermophila LSU-ΔP5abc intron in the presence or absence of ATP. These effects likely reflect at least in part differences in the RNA-binding properties of Mss116p and CYT-19, such as Mss116p’s higher, non-specific RNA-binding affinity in the absence of nucleotides, or as yet unidentified differences in its affinity for single- and double-stranded RNA regions, specific sequences, or RNA structures, which may be modulated by different nucleotides.41,42 These differences may enable Mss116p to differentially stabilize active or inactive RNA structures, impede CYT-18-binding in the case of T. thermophila LSU-ΔP5abc intron, or function as an ATP-hydrolysis-independent RNA chaperone. In addition, while CYT-19 and Mss116p facilitate strand annealing to similar degrees (unpublished data), the two proteins may differ in the balance between duplex-unwinding and strand-annealing activities or how this balance is affected by different nucleotides. Although all the defects in an mss116 disruptant could be ameliorated by the expression of CYT-19, in most cases, the splicing defects were only partially suppressed, even when CYT-19 was expressed at higher levels than Mss116p.10 Our results raise the possibility that this partial suppression reflects differences in the biochemical activities of the two proteins, including the observed differences in RNA-binding properties, which may enhance Mss116p’s ability to promote reactions by mechanisms unavailable to or less efficient for CYT-19.
Notably, we did not observe similar ATP-hydrolysis-independent effects for the Mss116p-promoted splicing of the yeast aI5γ or bI1 group II introns, or for Mss116p-promoted RNA-cleavage by the aI5γ-derived D135 ribozyme. Further experiments will be required to assess whether this difference in the behavior of Mss116p toward group I and group II introns reflect some more specific interaction of Mss116p with group I intron RNAs, cumulative differences in the number or type of Mss116p-binding sites in the intron RNAs, differences in the nature of the rate-limiting step in group I and II intron splicing reactions, or are merely coincidental for the introns studied thus far.
DEAD-box proteins as general RNA chaperones
The pleiotropic defects in RNA metabolism in cyt-19 and mss116 mutants and the interchangeability of CYT-19 and Mss116p with each other in vitro and in vivo suggest that these DEAD-box proteins function as general RNA chaperones that bind non-specifically to diverse RNA and RNP substrates and have the ability to resolve different types of kinetic traps.9,10,16,17 At present, there is no indication that CYT-19 or Mss116p are targeted to RNA or RNP substrates by specific RNA-protein or protein-protein interactions, although given their large number of natural targets, this remains a possibility for cases that have not been investigated in detail.
Our finding that the yeast cytosolic/nuclear DEAD-box protein Ded1p can function similarly to CYT-19 and Mss116p to promote the splicing of the aI5γ and bI1 group II introns suggests that Ded1p also has an inherent RNA chaperone activity. This RNA chaperone activity could be related to the normal function of Ded1p and may help explain the complex phenotype of ded1 mutants.39 To our knowledge, it remains an open question whether Ded1p binds to its natural RNA substrates without the aid of co-factors or whether it is targeted to some or all of those substrates by specific RNA-protein or protein-protein interactions.
We and others have hypothesized that DExH/D-box proteins that function as RNA chaperones have distinct structural and functional characteristics that distinguish them from other DExH/D-box proteins, particularly those that are processive RNA helicases. These characteristics may include: (i) ability to bind RNA and RNP substrates non-specifically; (ii) high on and off rates for RNA binding by the catalytic domain, enabling multiple cycles of activity on the same RNA; (iii) duplex unwinding without preferred polarity and ability to unwind blunt-end substrates, enabling remodeling of diverse substrates and substrate regions; (iv) limited processivity of duplex unwinding and/or an optimal balance between duplex-unwinding and strand-annealing activities to avoid over disruption of RNA structure; and (v) ability to displace or remodel incorrectly bound proteins from RNP complexes.43,44
Finally, our findings show that while CYT-19, Mss116p, and Ded1p all have RNA chaperone activity, they differ in their ability to promote the splicing of different group I and group II intron RNAs. These differences likely reflect that the balance of RNA-binding, duplex-unwinding, and strand-annealing activities has been tuned by coevolution to function optimally only on specific RNA substrates. The group I and group II intron splicing assays that we developed may be useful both for identifying DExH/D-box proteins that have RNA chaperone activity and for relating differences in their biochemical activities to the ability to effect specific RNA structural transitions in natural RNA and RNP substrates.
Materials and Methods
Recombinant plasmids
pMAL-Mss116p contains the Mss116p coding sequence (codons 37-664) with an inframe N-terminal MalE fusion cloned downstream of a tac promoter in the expression vector pMAL-c2t. The latter is a derivative of pMAL-c2x (New England Biolabs; Ipswich, MA) in which the factor Xa protease cleavage site between MalE and the expressed protein was replaced by a tobacco etch virus (TEV) protease-cleavage site.45 To construct pMAL-Mss116p, the Mss116p ORF minus the mt targeting sequence (codons 1-36) was amplified by PCR of genomic DNA from yeast strain BY4704,46 using an N-terminal primer, which amplified from codon 37 and added a 5′-BamHI site, and a C-terminal primer, which amplified from the TAG stop codon and added a 3′-HindIII site. The resulting PCR product was then cloned between the BamHI and HindIII sites of the vector. The expressed MalE-Mss116p fusion protein is cleaved with TEV protease to yield the mature intramitochondrial form of Mss116p with two extra N-terminal amino acid residues (GlySer).
To construct pMAL-Mss116p-K158E, a fragment of the Mss116p ORF containing the K158E mutation was generated by PCR from pMAL-Mss116p DNA with primers mss116K-E (5′-GGGACCGGTACAGGTCTTACATTTGCCTTCTTGATTCC) and mss166r1 (5′-AGA AAGCTGGGTCATATATGTTGCTGTTTCTACTGGAG), digested with AgeI and MfeI, and swapped for the corresponding fragment of pMAL-Mss116p digested with the same enzymes.
RNA substrates for RNA-splicing and equilibrium-binding assays were synthesized by transcribing the following restriction enzyme-digested recombinant plasmids with phage T3 (N. crassa mt LSU-ΔORF, yeast aI1-ΔORF and aI2-ΔORF; Megascript kit; Ambion, Austin, TX) or T7 RNA polymerase (all other introns; Invitrogen, Carlsbad, CA; Megascript kit; Ambion): aI5γ, pJD20/HindIII;32 aI5γ-D135, pQL71/HindIII (gift of O. Federova and A. M. Pyle, Yale University, New Haven, CT); aI1-ΔORF, pSHΔAC/EcoRI;47 aI2-ΔORF, pSZD1/BstEII;48 bI1, pHrH154#4/EcoRI;10 Ll.LtrB-ΔORF, pGMΔORF/BamHI;49 N. crassa mt LSU-ΔORF, pBD5a/BanI;50 T. thermophila LSU-ΔP5abc, pBlueΔP5abc/XhoI (S. Mohr and A. M. Lambowitz, unpublished).
Protein expression and purification
Mss116p was expressed from pMAL-Mss116p in E. coli HMS174(DE3) or Rosetta 2 (Novagen/EMD Biosciences, Madison, WI). For HMS174(DE3), cells that had been freshly transformed with pMAL-Mss116p were inoculated into 10 ml of LB containing ampicillin (50 μg/ml) and grown with shaking at 200 rpm at 37°C until O.D.600 was ~1.0. This starter culture was then inoculated into 1 l of the same medium containing 0.2% (w/v) glucose and grown as above until O.D.600 = 0.6; then 0.3 mM IPTG was added to induce Mss116p expression and incubation continued overnight at 15°C. Cells were pelleted by centrifugation. For Rosetta 2, the concentration of ampicillin was 100 μg/ml and chloramphenicol was added at 50 μg/ml, and the cells were induced with 1 mM IPTG at 20°C. After centrifugation, the cell pellet was suspended in 30-ml buffer A (20 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM dithiothreitol (DTT)) with 0.5 M KCl and frozen at −80°C.
To isolate Mss116p, four frozen cell suspensions, each prepared from one liter of culture, were thawed at 25°C, incubated on ice with lysozyme (1 mg/ml; Sigma, St. Louis, MO) for 15 min, and sonicated (Branson Sonifier S-450A; 3 bursts of 15 sec at power setting 6 with double-stepped microtip on ice). After clarifying the lysates by centrifugation (18,000 x g, 30 min, 4°C), the supernatants were combined, and nucleic acids were precipitated by adding 10% (v/v) polyethylenenimine (PEI) in 100 μl increments to a final concentration of 0.4% (v/v), followed by centrifugation (15,000 x g, 15 min, 4°C). The supernatant was then either filtered (Puradisc 25AS; Whatman, Florham Park, New Jersey) or loaded directly onto a 2-ml amylose column (high-flow resin; New England BioLabs), which was washed sequentially with 50-ml of buffer A containing 0.5 M KCl, 1 or 1.5 M KCl, and 0.5 M KCl, then eluted with buffer A containing 0.5 M KCl and 20 mM maltose. Peak fractions containing the MalE-Mss116p fusion were pooled and digested overnight at 4°C with TEV protease (50–200 μg; purified as described by Kapust et al.51). The MalE segment was removed from the Mss116p preparation either by loading the cleaved sample onto a gravity-flow hydroxyapatite column (Bio-Gel HTP Gel; Bio-Rad Laboratories, Hercules, CA), followed by an amylose column, as described in the pMAL instruction manual (Method II; New England BioLabs), or by loading onto a 2-ml ceramic hydroxyapatite column (macro-prep ceramic hydroxyapatite type I, 40 μm particle size; Bio-Rad). For the latter column, the cleaved sample was diluted with an equal volume of buffer B (20 mM potassium phosphate, pH 7.3, 200 mM NaCl), loaded onto the column, washed with 80-ml buffer B, and eluted with 0.5 M potassium phosphate, pH 7.3. The N-terminal MalE segment does not bind to the column under these conditions, and the peak fractions contained only Mss116p. Purified Mss116p was dialyzed into buffer A containing 0.5 M KCl and 50% glycerol and stored on ice at 4°C. The purification yielded either ~0.5 mg (HMS174(DE3)) or ~1.5 mg (Rosetta 2) Mss116p per l of culture, and the protein was > 95% pure, as judged by Coomassie blue-stained SDS-polyacrylamide gels.
CYT-18 was expressed and purified as described,27 but with an additional final gel-filtration through a Superdex 200 column (GE Healthcare Biosciences Corp., Piscataway, NJ) in 100 mM KCl, 25 mM Tris-HCl, pH 7.5 followed by addition of 10% glycerol for storage. The CYT-19 protein used in this study for comparison with Mss116p was expressed from a matching MalE-fusion construct and purified by essentially the same method developed for Mss116p (M.D. and A.M.L., unpublished data).
Ded1p and NPH-II were purified as described.52
Preparation of nucleotides and Mg2+ concentrations in assays
Just prior to each experiment, ATP, AMP-PNP, ADP and GTP were mixed with equimolar MgCl2 purchased as a 1 M stock solution (Molecular Biology Grade; Sigma). The Mg2+ concentration indicated for all assays is for Mg2+ that is not bound to nucleotide (e.g., 0.5 mM Mg2+ corresponds to an overall concentration of 1.5 mM Mg2+ with 1 mM ATP or other nucleotide; 10 mM Mg2+ corresponds to an overall concentration of 11 mM Mg2+ with 1 mM nucleotide).
ATPase assays
ATPase assays were done at 30°C in 20 μl of reaction medium containing 100 mM KCl, 8 mM Mg2+, 10 mM Tris-HCl, pH 7.5, 10% glycerol, 50 nM Mss116p, 10 μCi [γ-32P]ATP (10 Ci/mmol; Perkin Elmer, Wellesley, MA), and saturating unlabeled ATP (3 mM). Group I or group II intron RNAs or homopolymers (Sigma) were added as described for individual experiments. At each time point, a 1-μl portion was removed and applied to a PEI-cellulose thin-layer chromatography plate (20 x 20 cm; Scientific Adsorbents Inc., Atlanta, GA), which was developed with 0.75 M LiCl and 1 M formic acid, then dried, and quantified with a phosphorimager.
Duplex-unwinding and strand-annealing reactions
Duplex-unwinding and strand-annealing assays were done in 30 μl of reaction medium using substrates prepared as described.23,53 Reactions with 16-bp duplex substrates contained 50 mM KCl, 0.5 mM MgCl2, 40 mM Tris-HCl, pH 8.0, 2 mM DTT, 1 unit/μl RNasin, 0.01% NP-40, 0.5 nM 32P labeled RNA substrate and ATP, as indicated, and were done at 24°C. Reactions with 13-bp duplex substrates contained 100 mM KCl, 20 mM Tris-HCl, pH 7.5, MgCl2 and ATP as indicated, 2 mM DTT, 1 unit/μl RNasin, 0.01% NP-40, and 0.1 nM 32P-labeled RNA substrate and were done at 30°C. For unwinding reactions, Mss116p (400 nM) was pre-incubated with the RNA substrate under reaction conditions in the absence of ATP for 5 min. Longer pre-incubation did not alter the reaction kinetics (data not shown). Reactions were initiated by adding an equimolar mixture of MgCl2 and ATP to the indicated ATP concentrations. For annealing reactions, Mss116p (400 nM) was added to the reaction mixture (including ATP, where applicable), and the reactions were initiated by adding the denatured strands (0.5 nM). For both unwinding and annealing reactions, 3-μl portions were removed at the indicated times, and the reactions stopped by adding 3 μl of a solution containing 1% SDS, 50 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue, and 20% glycerol on ice. The samples were then analyzed by electrophoresis in a non-denaturing 15% polyacrylamide gel, which was run at ~100V/cm. After drying the gel, radiolabeled RNAs were visualized and quantified with a phosphorimager, using ImageQuant 5.2. software (Molecular Dynamics). The fractions of single-stranded and duplex RNAs were determined from the relative amount of radioactivity in the respective bands, and unwinding and annealing rate constants were calculated as described.23
RNA-binding assays
Equilibrium-binding assays were done by incubating 5 pM 32P-labeled RNA with increasing concentrations of Mss116p at 30°C in reaction medium containing 100 mM KCl, 5 or 8 mM MgCl2, 10 mM Tris-HCl, pH 7.5, and 0.1 mg/ml bovine serum albumin to stabilize Mss116p at low concentrations. The solutions were then filtered through nitrocellulose (Bio-Rad) backed by nylon (Hybond-N; GE Healthcare Biosciences Corp.), and the retained radioactivity was quantified with a phosphorimager.
RNA-splicing assays
Group I intron splicing reactions were done with 100 nM Mss116p, 100 nM CYT-18 dimer, and 20 nM 32P-labeled precursor RNA at 25°C (N. crassa mt LSU- ORF intron) or 30°C (T. thermophila LSU-ΔP5abc intron) in 100 μl of reaction medium containing 100 mM KCl, 5 mM MgCl2, 25 mM Tris-HCl, pH 7.5, 10% glycerol, 0.2 U/μl SUPERasin (Ambion) plus 1 mM GTP and 1 mM ATP, AMP-PNP or ADP. For time courses, reactions were initiated by adding CYT-18, 10-μl portions were removed at different times, and the reactions were terminated by adding 20 μl of 75 mM EDTA, followed by extraction with phenol/chloroform/isoamyl alcohol (25:24:1).
Standard conditions for group II intron splicing reactions were 100 nM Mss116p and 20 nM 32P-labeled precursor RNA at 30°C in 100 μl of reaction medium containing 100 mM KCl, 8 mM MgCl2, 10 mM Tris-HCl, pH 7.5, 10% glycerol, plus 1 mM ATP, AMP-PNP, or ADP and 0.2 U/μl SUPERasin. The reactions were initiated by adding MSS116p, 8-μl portions were removed at different times, and the reactions were terminated by adding 25 μl 0.1 M EDTA, 0.2 M Tris-HCl, pH 7.5, 0.1% SDS, followed by extraction with phenol/chloroform/isoamyl alcohol (25:24:1).
Group I and group II intron splicing reactions were analyzed in denaturing 4% polyacrylamide gels, which were dried and quantified with a phosphorimager. Phosphorimager counts were corrected for background and normalized based on the number of U-residues to calculate the molar fraction of total RNA in each band. In most cases, the data from splicing time courses were fit to equations with a single exponential to obtain kobs.
DEAD-box protein stimulated RNA-cleavage by the D135 ribozyme
D135 RNA (50 nM) was pre-incubated with Mss116p (50 nM) for 5 min in reaction medium containing 100 mM KCl, 10 mM Tris-HCl, pH 7.5, and different Mg2+ concentrations (for determination of the Mg2+ optimum) or 25 mM Mg2+ (all other reactions) in the presence or absence of 1 mM ATP, AMP-PNP, or ADP. Reactions were initiated by adding 5′-[32P]-labeled RNA-oligomer substrate (5′-CGUGGUGGGACAUUUUCGAGCGGU), incubated at 25°C, and terminated by adding 100 mM EDTA, followed by phenol-CIA extraction. The products were analyzed in a denaturing 20% polyacrylamide gel, which was dried and quantified with a phosphorimager. To test protease-sensitivity, 20 mg/ml proteinase K (prot-K; Ambion) plus 0.5% SDS were added to some reactions. ATP was depleted by adding hexokinase (0.002 U/μl; Roche Diagnostics, Indianapolis, IN) and glucose (200 mM) and incubating for 15 min at 25°C. ATP depletion was confirmed thin-layer-chromatograpy of trace amounts of [γ-32P]-ATP added just prior to the addition of hexokinase plus glucose.
Acknowledgments
We thank Olga Federova and Anna Marie Pyle (Yale) for the D135 ribozyme construct, Paul Paukstelis (UT Austin) for preparation of CYT-18, and Rick Russell (UT Austin) for very insightful suggestions and comments on the manuscript. Mark Del Campo is the recipient of NIH post-doctoral fellowship F01-GM76961. This work was supported by NIH grant GM37951 to A.M.L. and GM067700 to E.J.
Abbreviations used
- AMP-PNP
5′ adenylyl imidodiphosphate
- DTT
dithiothreitol
- mt
mitochondrial
- PEI
polyethylenenimine
- SER
spliced-exon reopening
- TEV
tobacco etch virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. Cold Spring Harbor Laboratory Press: Plainview, NY; 1999. pp. 451–485. [Google Scholar]
- 2.Lehmann K, Schmidt U. Group II introns: structure and catalytic versatility of large natural ribozymes. Crit Rev Biochem Mol Biol. 2003;38:249–303. doi: 10.1080/713609236. [DOI] [PubMed] [Google Scholar]
- 3.Pyle AM, Lambowitz AM. Group II introns: ribozymes that splice RNA and invade DNA. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 3. Cold Spring Harbor Laboratory Press; Plainview, NY: 2006. pp. 469–505. [Google Scholar]
- 4.Caprara MG, Nilsen TW. RNA: versatility in form and function. Nat Struct Biol. 2000;7:831–833. doi: 10.1038/82816. [DOI] [PubMed] [Google Scholar]
- 5.Weeks KM. Protein-facilitated RNA folding. Curr Opin Struct Biol. 1997;7:336–342. doi: 10.1016/s0959-440x(97)80048-6. [DOI] [PubMed] [Google Scholar]
- 6.Karpel RL, Miller NS, Fresco JR. Mechanistic studies of ribonucleic acid renaturation by a helix-destabilizing protein. Biochemistry. 1982;21:2102–2108. doi: 10.1021/bi00538a019. [DOI] [PubMed] [Google Scholar]
- 7.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 8.Schroeder R, Barta A, Semrad K. Strategies for RNA folding and assembly. Nat Rev Mol Cell Biol. 2004;5:908–919. doi: 10.1038/nrm1497. [DOI] [PubMed] [Google Scholar]
- 9.Mohr S, Stryker JM, Lambowitz AM. A DEAD-box protein functions as an ATP-dependent RNA chaperone in group I intron splicing. Cell. 2002;109:769–779. doi: 10.1016/s0092-8674(02)00771-7. [DOI] [PubMed] [Google Scholar]
- 10.Huang HR, Rowe CE, Mohr S, Jiang Y, Lambowitz AM, Perlman PS. The splicing of yeast mitochondrial group I and group II introns requires a DEAD-box protein with RNA chaperone function. Proc Natl Acad Sci USA. 2005;102:163–168. doi: 10.1073/pnas.0407896101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanner NK, Linder P. DExD/H box RNA helicases: from generic motors to specific dissociation functions. Mol Cell. 2001;8:251–262. doi: 10.1016/s1097-2765(01)00329-x. [DOI] [PubMed] [Google Scholar]
- 12.Silverman E, Edwalds-Gilbert G, Lin RJ. DExD/H-box proteins and their partners: helping RNA helicases unwind. Gene. 2003;312:1–16. doi: 10.1016/s0378-1119(03)00626-7. [DOI] [PubMed] [Google Scholar]
- 13.Rocak S, Linder P. DEAD-box proteins: the driving forces behind RNA metabolism. Nat Rev Mol Cell Biol. 2004;5:232–241. doi: 10.1038/nrm1335. [DOI] [PubMed] [Google Scholar]
- 14.Akins RA, Lambowitz AM. A protein required for splicing group I introns in Neurospora mitochondria is mitochondrial tyrosyl-tRNA synthetase or a derivative thereof. Cell. 1987;50:331–345. doi: 10.1016/0092-8674(87)90488-0. [DOI] [PubMed] [Google Scholar]
- 15.Caprara MG, Mohr G, Lambowitz AM. A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J Mol Biol. 1996;257:512–531. doi: 10.1006/jmbi.1996.0182. [DOI] [PubMed] [Google Scholar]
- 16.Tijerina P, Bhaskaran HP, Russell R. Non-specific binding to structured RNA and preferential unwinding of an exposed helix by the CYT-19 protein, a DEAD box RNA chaperone. Proc Natl Acad Sci USA. 2006 doi: 10.1073/pnas.0603127103. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bertrand H, Bridge P, Collins RA, Garriga G, Lambowitz AM. RNA splicing in Neurospora mitochondria. Characterization of new nuclear mutants with defects in splicing the mitochondrial large rRNA. Cell. 1982;29:517–526. doi: 10.1016/0092-8674(82)90168-4. [DOI] [PubMed] [Google Scholar]
- 18.Henderson MF. Master’s Thesis. St. Louis University; St. Louis, Missouri: 1981. The effect of cyt-4-1, cyt-18-1 and cyt-19-1 mutations on RNA processing in Neurospora mitochondria. [Google Scholar]
- 19.Séraphin B, Simon M, Boulet A, Faye G. Mitochondrial splicing requires a protein from a novel helicase family. Nature. 1989;337:84–87. doi: 10.1038/337084a0. [DOI] [PubMed] [Google Scholar]
- 20.Mohr S, Matsuura M, Perlman PS, Lambowitz AM. A DEAD-box protein alone promotes group II intron splicing and reverse splicing by acting as an RNA chaperone. Proc Natl Acad Sci USA. 2006;103:3569–3574. doi: 10.1073/pnas.0600332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 22.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu Rev Biophys Biophys Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 23.Yang Q, Jankowsky E. ATP- and ADP-dependent modulation of RNA unwinding and strand annealing activities by the DEAD-box protein DED1. Biochemistry. 2005;44:13591–13601. doi: 10.1021/bi0508946. [DOI] [PubMed] [Google Scholar]
- 24.Rogers GW, Jr, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J Biol Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 25.Cordin O, Tanner NK, Doere M, Linder P, Banroques J. The newly discovered Q motif of DEAD-box RNA helicases regulates RNA-binding and helicase activity. EMBO J. 2004;23:2478–2487. doi: 10.1038/sj.emboj.7600272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 27.Paukstelis PJ, Coon R, Madabusi L, Nowakowski J, Monzingo A, Robertus J, Lambowitz AM. A tyrosyl-tRNA synthetase adapted to function in group I intron splicing by acquiring a new RNA binding surface. Mol Cell. 2005;17:417–428. doi: 10.1016/j.molcel.2004.12.026. [DOI] [PubMed] [Google Scholar]
- 28.Motojima F, Yoshida M. Discrimination of ATP, ADP, and AMPPNP by chaperonin GroEL: hexokinase treatment revealed the exclusive role of ATP. J Biol Chem. 2003;278:26648–26654. doi: 10.1074/jbc.M300806200. [DOI] [PubMed] [Google Scholar]
- 29.Joyce GF, van der Horst G, Inoue T. Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res. 1989;17:7879–7889. doi: 10.1093/nar/17.19.7879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohr G, Caprara MG, Guo Q, Lambowitz AM. A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature. 1994;370:147–150. doi: 10.1038/370147a0. [DOI] [PubMed] [Google Scholar]
- 31.Daniels DL, Michels WJ, Jr, Pyle AM. Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J Mol Biol. 1996;256:31–49. doi: 10.1006/jmbi.1996.0066. [DOI] [PubMed] [Google Scholar]
- 32.Jarrell KA, Peebles CL, Dietrich RC, Romiti SL, Perlman PS. Group II intron self-splicing. Alternative reaction conditions yield novel products. J Biol Chem. 1988;263:3432–3439. [PubMed] [Google Scholar]
- 33.Hiller R, Hetzer M, Schweyen RJ, Mueller MW. Transposition and exon shuffling by group II intron RNA molecules in pieces. J Mol Biol. 2000;297:301–308. doi: 10.1006/jmbi.2000.3582. [DOI] [PubMed] [Google Scholar]
- 34.Qin PZ, Pyle AM. Stopped-flow fluorescence spectroscopy of a group II intron ribozyme reveals that domain 1 is an independent folding unit with a requirement for specific Mg2+ ions in the tertiary structure. Biochemistry. 1997;36:4718–4730. doi: 10.1021/bi962665c. [DOI] [PubMed] [Google Scholar]
- 35.Swisher JF, Su LJ, Brenowitz M, Anderson VE, Pyle AM. Productive folding to the native state by a group II intron ribozyme. J Mol Biol. 2002;315:297–310. doi: 10.1006/jmbi.2001.5233. [DOI] [PubMed] [Google Scholar]
- 36.Fedorova O, Su LJ, Pyle AM. Group II introns: highly specific endonucleases with modular structures and diverse catalytic functions. Methods. 2002;28:323–335. doi: 10.1016/s1046-2023(02)00239-6. [DOI] [PubMed] [Google Scholar]
- 37.Chuang RY, Weaver PL, Liu Z, Chang TH. Requirement of the DEAD-Box protein ded1p for messenger RNA translation. Science. 1997;275:1468–1471. doi: 10.1126/science.275.5305.1468. [DOI] [PubMed] [Google Scholar]
- 38.de la Cruz J, Iost I, Kressler D, Linder P. The p20 and Ded1 proteins have antagonistic roles in eIF4E-dependent translation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:5201–5206. doi: 10.1073/pnas.94.10.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Linder P. Yeast RNA helicases of the DEAD-box family involved in translation initiation. Biol Cell. 2003;95:157–167. doi: 10.1016/s0248-4900(03)00032-7. [DOI] [PubMed] [Google Scholar]
- 40.Pontius BW, Berg P. Renaturation of complementary DNA strands mediated by purified mammalian heterogeneous nuclear ribonucleoprotein A1 protein: implications for a mechanism for rapid molecular assembly. Proc Natl Acad Sci USA. 1990;87:8403–8407. doi: 10.1073/pnas.87.21.8403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 1 A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry. 1998;37:2180–2193. doi: 10.1021/bi972430g. [DOI] [PubMed] [Google Scholar]
- 42.Lorsch JR, Herschlag D. The DEAD box protein eIF4A. 2 A cycle of nucleotide and RNA-dependent conformational changes. Biochemistry. 1998;37:2194–2206. doi: 10.1021/bi9724319. [DOI] [PubMed] [Google Scholar]
- 43.Bowers HA, Maroney PA, Fairman ME, Kastner B, Lührmann R, Nilsen TW, Jankowsky E. Discriminatory RNP remodeling by the DEAD-box protein DED1. RNA. 2006;12:903–912. doi: 10.1261/rna.2323406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankowsky E, Bowers H. Remodeling of ribonucleoprotein complexes with DExH/D RNA helicases. Nucleic Acids Res. 2006 doi: 10.1093/nar/gkl410. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kristelly R, Earnest BT, Krishnamoorthy L, Tesmer JJ. Preliminary structure analysis of the DH/PH domains of leukemia-associated RhoGEF. Acta Crystallogr D Biol Crystallogr. 2003;59:1859–1862. doi: 10.1107/s0907444903018067. [DOI] [PubMed] [Google Scholar]
- 46.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 47.Hebbar SK, Belcher SM, Perlman PS. A maturase-encoding group IIA intron of yeast mitochondria self-splices in vitro. Nucleic Acids Res. 1992;20:1747–1754. doi: 10.1093/nar/20.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zimmerly S, Guo H, Eskes R, Yang J, Perlman PS, Lambowitz AM. A group II intron RNA is a catalytic component of a DNA endonuclease involved in intron mobility. Cell. 1995;83:529–538. doi: 10.1016/0092-8674(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 49.Matsuura M, Saldanha R, Ma H, Wank H, Yang J, Mohr G, Cavanagh S, Dunny GM, Belfort M, Lambowitz AM. A bacterial group II intron encoding reverse transcriptase, maturase, and DNA endonuclease activities: biochemical demonstration of maturase activity and insertion of new genetic information within the intron. Genes Dev. 1997;11:2910–2924. doi: 10.1101/gad.11.21.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guo Q, Akins RA, Garriga G, Lambowitz AM. Structural analysis of the Neurospora mitochondrial large rRNA intron and construction of a mini-intron that shows protein-dependent splicing. J Biol Chem. 1991;266:1809–1819. [PubMed] [Google Scholar]
- 51.Kapust RB, Tozser J, Fox JD, Anderson DE, Cherry S, Copeland TD, Waugh DS. Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 2001;14:993–1000. doi: 10.1093/protein/14.12.993. [DOI] [PubMed] [Google Scholar]
- 52.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D "RNA helicases" without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 53.Jankowsky E, Gross CH, Shuman S, Pyle AM. The DExH protein NPH-II is a processive and directional motor for unwinding RNA. Nature. 2000;403:447–451. doi: 10.1038/35000239. [DOI] [PubMed] [Google Scholar]


