Abstract
Purpose
The aim of present study was to investigate the methylation and expression status of spleen tyrosine kinase (SYK) in human hepatocellular carcinoma (HCC) and to evaluate this information for its ability to predict disease prognosis. E-cadherin and TIMP-3 methylation was also analyzed here as control since both were associated with poor prognosis in some types of tumors.
Experimental Design
We analyzed the methylation status of SYK, E-cadherin and TIMP-3 in 124 cases of HCC and assessed the correlation of such methylations with clinicopathological parameters and prognosis after tumor resection.
Results
We found that SYK, E-cadherin, and TIMP-3 genes were methylated in 27%, 27%, and 42% of HCC neoplastic tissues, respectively. The loss of SYK mRNA or Syk protein expression was highly correlated with SYK gene methylation. The patients with methylated SYK in neoplastic tissues had a significantly lower overall survival rate after hepatectomy than those with unmethylated SYK. No significant difference in overall survival rates, however, was found between groups of patients with methylated and unmethylated E-cadherin or TIMP-3. Patients with negative Syk protein expression had a significantly lower overall survival rate than those with positive Syk protein expression. Multivariate analyses indicated that factors affecting overall survival were TNM stage, Child-Pugh classification, SYK methylation or Syk protein status.
Conclusions
Our results indicate that SYK methylation and loss of Syk expression in HCC neoplastic tissues are independent biomarkers of poor patient outcome and that determination of SYK methylation or Syk expression status may offer guidance for selecting appropriate treatments.
Keywords: DNA methylation, spleen tyrosine kinase, E-cadherin, TIMP-3, hepatocellular carcinoma, prognosis
Abbreviations: 5-aza-dc, 5-aza-2’-deoxycytidine; DNMT, DNA methyltransferase; HCC, hepatocellular carcinoma; PCR, polymerase chain reaction; MSP, methylation-specific PCR; RT-PCR, reverse transcriptase-PCR; SYK, spleen tyrosine kinase; TIMP, tissue inhibitor of metalloproteinase; MMP, matrix metalloproteinase; GGT, gamma-glutamyltransferase; AFP, α-fetoprotein
Hepatocellular carcinoma (HCC) is the third most common cause of cancer death in the world (1). China has one of the highest prevalence of HCC, largely because carriers of chronic hepatitis B account for more than 10% of its population (2). Although the incidence of HCC in the United States is relatively lower, the reported new cases have been increasing steadily (3). The prognosis for patients with HCC is generally poor, even after surgery or chemotherapy. The 5-year overall survival rate is between 35% and 41% after resection of primary tumors (4, 5) and between 47% and 61% after liver transplantation (6). Systemic chemotherapy gives a low response rate of only 10% to 20% and has shown no significant benefit with regard to overall survival (7). Given this poor therapeutic efficacy, the development of biomarkers for early detection and accurate prognosis of HCC is crucial for prescribing the most timely and effective treatment.
Although the etiology of HCC remains unclear, chronic infection with hepatitis B or C virus, chemical carcinogens (aflatoxins), and other environmental and host factors have been linked to hepatocarcinogenesis (8, 9). In China, most cases of HCC develop from liver cirrhosis with chronic infection of hepatitis B virus and/or chronic exposure to aflatoxin B1. In western countries, however, chronic alcoholism and chronic infection with hepatitis C virus are the major etiological factors. These various factors are believed to induce a spectrum of molecular alterations that contribute to the initiation and progression of HCC, including the genetic and epigenetic inactivation of tumor suppressor genes (8, 9). Similar to what has been shown in other tumor types, DNA methylation frequently occurs in HCC, represented by p16, p15, GSTP, E-cadherin, TIMP-3, APC, SOCS-1, RASSF1A, and 14-3-3δ (10–14). The prognostic value of methylation of these genes in HCC was either not systematically studied or was found not important in HCC.
The spleen tyrosine kinase (SYK) is a tumor/metastasis suppressor gene recently found to be silenced through DNA methylation in breast cancer (15) and T-lineage acute lymphoblastic leukemia (16). Loss of SYK expression has been implicated in increased invasiveness and proliferation of breast tumors (17). Concordantly, over-expression of SYK was shown to inhibit the invasiveness, proliferation, and motility of breast cancer cells (17–20). SYK was regarded as a novel regulator of metastatic behavior of melanoma cells (21). Decreased SYK expression in primary breast tumors was shown to predict shorter survival among cancer patients (22). Given that SYK methylation is primarily responsible for the loss of SYK expression, aberrant SYK promoter hypermethylation may serve as a valuable prognostic marker.
In this study, we correlated epigenetic alterations of SYK with clinical and pathological parameters to determine its prognostic value in HCC. Because methylation of E-cadherin and TIMP-3 have been shown to be associated with poor prognosis in gastric and esophageal cancer (23, 24), respectively, we also analyzed the E-cadherin and TIMP-3 methylation status in parallel to compare their prognostic value with that of SYK methylation.
PATIENTS AND METHODS
Cell Lines
Liver cancer cell lines HepG2 and Hep3B were purchased from the American Type Culture Collection (Manassas, VA) and maintained in recommended culture conditions. Cells were maintained at 37°C in a humidified environment containing 5% CO2.
Study Population and Tissue Samples
One hundred and twenty-four patients who were consecutively diagnosed with HCC and had undergone hepatectomy from 1998 to 2001 in a single group at the Department of Hepatobiliary Oncology, Sun Yat-sen University Cancer Center were enrolled in the study. Tissue samples, including 124 samples from primary tumors and 34 samples from matched adjacent non-neoplastic liver tissues, were archived in the institution’s liver tumor bank and stored at −80°C until use. All non-neoplastic and neoplastic samples were histologically confirmed. Neither chemotherapy nor radiation therapy was given before tumor excision. The tumor stages of HCC were classified according to the tumor-node-metastasis (TNM) criteria (25). The degree of underlying cirrhosis was graded based on the size of gross cirrhotic nodules and histological examination: (1) no cirrhosis: the liver was soft and smooth with no cirrhotic nodules. No pseudolobule formation was found microscopically; (2) mild cirrhosis: the largest nodule on liver surface was <0.4 cm, or cirrhosis was identified by microscopic examination; (3) moderate cirrhosis: the degree of cirrhosis was between mild and severe cirrhosis; (4) severe cirrhosis: the largest cirrhotic nodule on liver surface was >0.8 cm, or the liver was notably deformed and complicated by portal hypertension. The study protocol was approved by the Clinical Research Ethics Committee of Sun Yat-sen University Cancer Center.
Methylation-specific Polymerase Chain Reaction
A blinded methylation-specific polymerase chain reaction (MSP) analysis was carried out; no clinicopathological or follow-up data were revealed to the bench researchers until the MSP results were finalized. Genomic DNA was isolated from frozen tissue by digestion with proteinase K, followed by standard phenol/chloroform extraction and ethanol precipitation. Sodium bisulfite (Sigma, St. Louis, MO)–induced conversion of genomic DNA was performed as described previously (15). The modified DNA was subjected to a two-step MSP protocol to determine the methylation status of SYK, E-cadherin, and TIMP-3 promoter regions (15, 26, 27). Primers were designed to distinguish between bisulfite-sensitive and -resistant modifications of unmethylated and methylated cytosines, respectively. For the first-round MSP, a 30-μl reaction that contained 30 ng of bisulfite-treated DNA was processed in 40 thermal cycles. An aliquot (2 μl) of diluted (1:40) PCR product was subjected to the second-round PCR in another 30-μl reaction. For SYK gene, both methylation and unmethylation primers were included in the same reaction. For E-cadherin and TIMP-3, separate reactions for methylation and unmethylation detection were carried out. The primer sequences, PCR condition, and product sizes for each gene are listed in Table 1. To prepare the positive methylation control, 1 μg of genomic DNA from normal human liver was treated in vitro with SssI methyltransferase (NEB, Beverly, MA), yielding completely methylated DNA at all CpG-rich regions. Untreated genomic DNA was used as negative control. For positive (SssI-treated) or negative (SssI-non-treated) controls, 1 μg DNA each was modified by sodium bisulfite. Thirty ng of bisulfite-treated control DNA template underwent nested PCR amplication side-by-side with testing specimens. H2O was also used as negative control in nested MSP. The PCR products were visualized by agarose gel electrophoresis and ethidium bromide staining.
Table 1.
Summary of primer sequence and PCR amplification for methylation analysis
| Genes | Sense primer 5’ → 3’ | Antisense Primer 5’ → 3’ | Annealing Temp. (°C) | No. cycles | Size (bp) |
|---|---|---|---|---|---|
| External (stage 1) | External (stage 1) | ||||
| SYK | TTTAGGGAATATGTTATGTAGTG | CACATAATTTCAACACTTTTACC | 57 | 40 | 645 |
| E-cadherin | GTTTAGTTTTGGGGAGGGGTT | ACTACTACTCCAAAAACCCATAACTAA | 49 | 40 | 270 |
| TIMP-3 | GYGGTATTATTTTTTATAAGGATTTGA | ACCRAATAATATAACRCTAAACCCC | 56 | 40 | 185 |
| Internal methylated (stage 2) | Internal methylated (stage 2) | ||||
| SYK | CGATTTCGCGGGTTTCGTTC | AAAACGAACGCAACGCGAAAC | 57 | 24 | 243 |
| E-cadherin | TGTAGTTACGTATTTATTTTTAGTGGCGTC | CGAATACGATCGAATCGAACCG | 54 | 24 | 112 |
| TIMP-3 | CGTTTCGTTATTTTTTGTTTTCGGTTTC | CCGAAAACCCCGCCTCG | 60 | 27 | 116 |
| Internal un-methylated (stage 2) | Internal un-methylated (stage 2) | ||||
| SYK | ATTTTGTGGGTTTTGTTTGGTG | ACTTCCTTAACACACCCAAAC | 57 | 24 | 140 |
| E-Cadherin | TGGTTGTAGTTATGTATTTATTTTTAGTGGTGTT | ACACCAAATACAATCAAATCAAACCAAA | 54 | 24 | 120 |
| TIMP-3 | TTTTGTTTTGTTATTTTTTGTTTTTGGTTTT | CCCCCAAAAACCCCACCTCA | 60 | 27 | 122 |
Reverse Transcriptase-PCR
Reverse transcriptase-PCR (RT-PCR) was performed as described previously (15). Briefly, total RNA was prepared using an RNeasy kit (Qiagen, Valencia, CA) and reverse-transcribed using 18-mer oligo(dT) and Superscript II (Invitrogen, Carlsbad, CA). Reactions lacking RT were used to verify the absence of amplification from genomic DNA contamination. The cDNA templates were subjected to PCR amplification. To verify cDNA integrity, β2-microglobulin expression was analyzed as well. The primer sets were 5’-TGTCAAGGATAAGAACATCATAG-3’ (forward) and 5’-CACCACGTCATAGTAGTAATTG-3’ (reverse) for SYK, and 5’-ACCCCCACTGAAAAAGATGA-3’ (forward) and 5’-GCATCTTCAAACCTCCATGAT-3’ (reverse) for β2-microglobulin.
In some experiments, cells were treated for 5 days with a DNA methyltransferase (DNMT) inhibitor, 5-aza-2’-deoxycytidine (5-aza-dC; Sigma) at a final concentration of 2.0 μM. Cells were then collected for RNA extraction.
Immunohistochemical Assay
Formalin-fixed paraffin-embedded sections of HCC tumors and adjacent non-neoplastic liver tissues were subjected to immunostaining with an antibody against Syk using the rabbit EnVision Plus kit (DakoCytomation, Carpinteria, CA). Briefly, 5-pm thick tissue sections were deparaffinized, rehydrated, and subjected to antigen retrieval by boiling in sodium citrate buffer (10 mM, pH 6.0). The sections were incubated at 4°C overnight with Syk primary antibody (1:200 dilution; Cell Signaling, Beverly, MA) and then stained with 3,3’-diaminobenzidine. After visualization of immunoreactivity, the sections were counterstained with hematoxylin, and mounted. The immunostained sections were evaluated without any knowledge of the patients’ clinical information and status of MSP and RT-PCR of SYK. Normal liver tissues were taken as internal positive controls. The stains were graded as: (a) positive when immunoreactivity equivalent to that seen in normal liver cells or immunoreactivity moderately decreased; and (b) negative when weak immunoreactivity or no immunoreactivity.
Statistical Analysis
All clinicopathological and follow-up data were collected in a database. Overall survival times were measured from the date of resection of primary tumors to the date of death or of the last follow-up. Survival curves were constructed using the Kaplan-Meier method and compared using the log-rank test. The prognostic factors for survival after hepatectomy were elucidated by univariate and then multivariate analyses. The following variables were analyzed: patient sex, age, Child-Pugh classification, GGT (gamma-glutamyltransferase) level, AFP(α-fetoprotein) level, tumor size, tumor encapsulation status, presence of macro-tumor thrombus in the portal vein, presence of satellite nodules, degree of underlying cirrhosis, TNM stage, expression status of Syk protein and methylation status of SYK, E-cadherin, and TIMP-3 genes in tumor tissues. Significant prognostic factors found by univariate analysis were entered into a multivariate analysis using the Cox proportional-hazards model. The SPSS software package (Version 10.0; SPSS Inc., Chicago, IL) was used for the statistical analyses. P < 0.05 was considered to be statistically significant.
RESULTS
Promoter Hypermethylation Leads to SYK Silencing in HCC
SYK is expressed in many epithelial cell types. We began our study by analyzing SYK expression status in two liver cancer cell lines, HepG2 and Hep3B. RT-PCR showed that Hep3B but not HepG2 cells expressed SYK mRNA (Figure 1A). Since DNA methylation is primarily responsible for SYK gene silencing (15), we surmised that the SYK gene promoter might be methylated in HepG2 cells. To explore this possibility, we used MSP to measure both methylated and unmethylated SYK promoter (15). MSP analyses indicated that SYK was methylated in HepG2 but not in Hep3B (Figure 2A), consistent with the SYK expression status in these cells. To further substantiate that SYK methylation is primarily responsible for the loss of SYK expression, we treated HepG2 cells with a DNMT inhibitor, 5-aza-dc, to determine whether demethylation restored SYK expression. As shown in Figure 1B, 5-aza-dc reactivated SYK expression in the HepG2 line as detected by RT-PCR while not affecting that in Hep3B, suggesting that DNA methylation plays a causal role in the SYK loss of expression in HCC.
Figure 1.
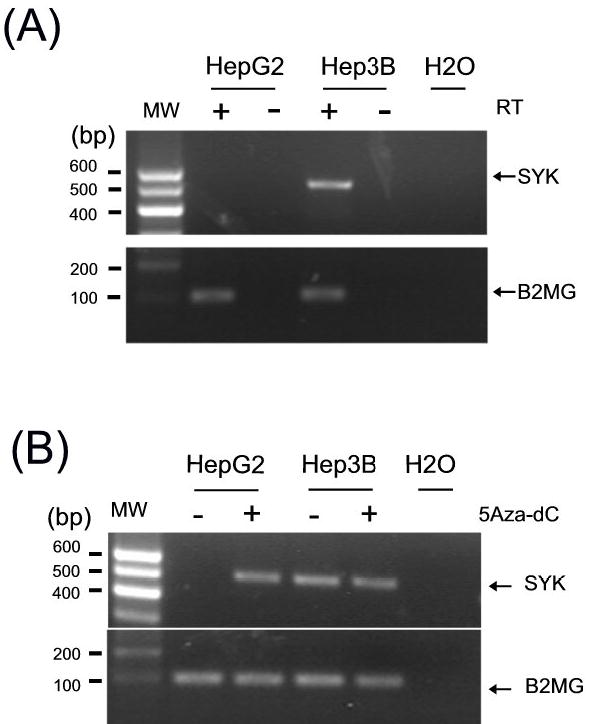
(A) Expression of SYK mRNA in HCC cell lines HepG2 and Hep3B. SYK mRNA expression was determined by RT-PCR. An RT-negative control (−) was used to rule out false positives resulting from contaminated genomic DNA. mRNA for β2-microglobulin (B2MG) was also analyzed to verify the RNA integrity. A blank control (H2O) was included in each PCR experiment. PCR products and a molecular weight (MW) marker were run on an agarose gel following by ethidium bromide staining. Bands of 507 bp and 115 bp are expected for SYK and β2-microglobulin transcripts, respectively. At least two independent experiments were carried out. (B) Restoration of SYK mRNA by treatment with DNMT1 inhibitor. HepG2 was treated for 5 days with (+) or without (−) 2.0 μM 5-aza-dC. As a control, Hep3B cells were processed in parallel. Total RNA was harvested and RT-PCR amplified as detailed in (A).
Figure 2.
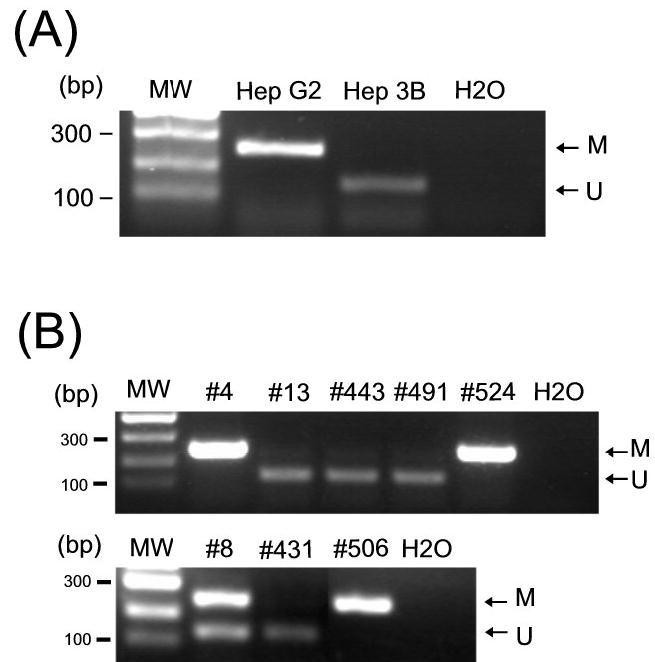
The SYK gene was hypermethylated in HCC cell lines (A) and primary tumors (B). DNA was treated with sodium bisulfite followed by the first round of PCR amplification. In the second round of PCR (nested), both methylation-specific and unmethylation-specific primers were used in the same reaction. DNA template-negative control (H2O) was also included. Products of 243 bp and 140 bp were expected for methylated (M) and unmethylated (U) DNA, respectively. The methylation status of three representative methylation-positive (#4, #524 and #506), one intermediate (#8), and four methylation-negative (#13, #443, #491, and #431) cases is shown in (B).
SYK Is Hypermethylated in Primary HCC
We next examined whether the epigenetic alteration of SYK observed in the HCC cell lines could be extrapolated to primary HCCs. All 124 patients included in the present study underwent surgical resection of primary tumors. The pathological diagnosis of all HCC cases was confirmed by histological reviews. We used MSP to evaluate the SYK methylation status in the 124 primary HCC tumors, in which 27 (21.8%) and 90 (72.6%) specimens were found to be SYK methylated and unmethylated, respectively. The remaining 7 cases (5.6%) showed amplification of both SYK methylation and unmethylation (Figure 2B). Coexistence of both methyaltion status in a given tumor could reflect the heterogeneity of HCC, although contamination from normal tissue DNA cannot be ruled out.
To ascertain whether SYK methylation leads to gene silencing in primary HCC, we used immunohistochemistry to assess the Syk protein expression in all 124 tumors (Figure 3). Immunohistochemical analyses showed that Syk protein was not expressed in 32 (25.8%) HCC cases; in this group, SYK was methylated, methylated/unmethylated, and unmethylated in 24, 5, and 3 cases, respectively. Among the remaining 92 (74.2%) Syk protein-positive cases, SYK was methylated, methylated/unmethylated, and unmethylated in 3, 2, and 87 cases, respectively. The correlation between SYK methylation and loss of Syk protein expression was highly significant (P < 0.001, Spearman test). The 3 cases in which Syk was expressed but methylated may reflect the heterogeneity of HCC; that is, methylation may occur in a subpopulation of neoplastic tissues that is readily detectable by MSP (31). The 3 cases in which Syk was not expressed but unmethylated suggest that there are other mechanisms to suppress SYK expression.
Figure 3.
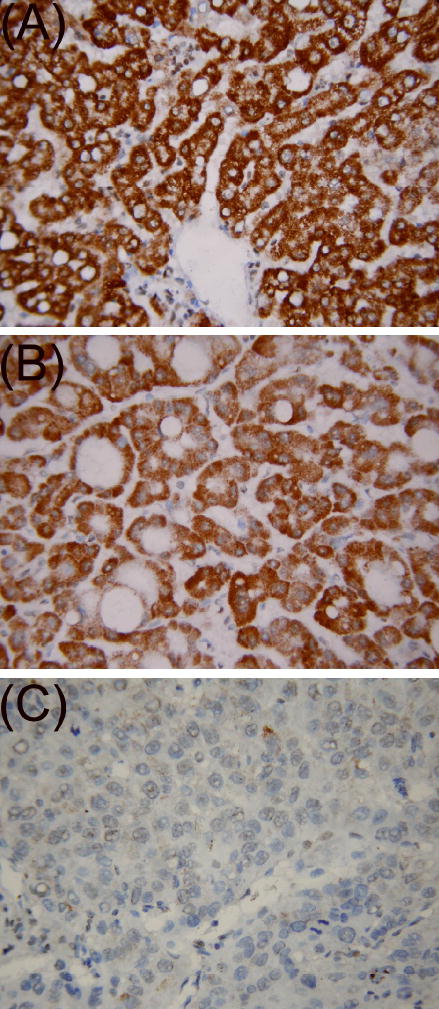
Immunohistochemical analyses of SYK gene product in primary HCC specimens. Staining of representative specimens of normal liver tissue (A), primary HCC with unmethylated SYK (B), and primary HCC with methylated SYK (C) is shown.
We also measured the SYK methylation and expression status in matched normal liver tissues. Among the 124 cases, 34 had samples of matched adjacent pathologically non-neoplastic liver tissues that were used for MSP, RT-PCR and immunohistochemical analyses. SYK gene was found methylated, methylated/unmethylated, and unmethylated in 0, 3 and 31 in non-neoplastic specimens, respectively, in comparison with 6, 2 and 26 cases of neoplastic tissues, respectively. If the “methylation/unmethylation” was grouped into the “methylation-positive” category, the percentage of patients with positive methylation were 8.8% (3/34) and 23.5% (8/34), respectively, for neoplastic and non-neoplastic tissues. The difference in percentage of methylation-positive patients was not statistically significant (P = 0.186, Fisher test). The statistical significance could be reached if more samples were available. SYK methylation in non-neoplastic tissues was also observed in earlier studies that may represent DNA methylation in pre-malignant lesions (28, 29). Aging-related gene methylation could also be a contributor (30). The corresponding primary tumors of these 3 cases were found to have unmethylated SYK.
The expression of SYK mRNA as measured by RT-PCR and Syk protein by immunohistochemistry in the 34 cases was entirely consistent, indicating the SYK expressional control occurs at the transcriptional level. Both SYK mRNA and Syk protein were positive in all 34 matching non-neoplastic liver tissues. By contrast, 5 of the 34 primary HCCs expressed neither SYK mRNA nor Syk protein. Among the 29 SYK-positive HCCs, SYK was found methylated, methylated/unmethylated, and unmethylated in 1, 2, and 26 specimens, respectively. These numbers were in comparison with 5, 0, and 0 SYK-negative cases, respectively. Using Spearman correlation test, SYK methylation and SYK expression was strongly correlated (P < 0.001). Collectively, these results indicated that hypermethylation of SYK promoter was largely tumor-specific and responsible for the loss of SYK expression in HCC.
Like SYK, E-cadherin and TIMP-3 are thought to be tumor/metastasis suppressor genes. DNA methylation could lead to silencing of E-cadherin (27, 32) and TIMP-3 (13, 29, 33) in certain types of tumors, including HCC. Thus we also assessed the methylation status of E-cadherin and TIMP-3 genes in the 124 HCC cases. We found that E-cadherin was methylated, methylated/unmethylated, and unmethylated in 21, 9, and 82 cases, respectively (non-informative in 12 cases). TIMP-3 was methylated, methylated/unmethylated, and unmethylated in 32, 14, and 64 cases, respectively (non-informative in 14 cases) (Figure 4). Gene methylation is believed to be an aberrant alteration that is associated with neoplastic progression; amplification from unmethylation allele is likely contributed by common contaminant of normal tissues. Thus we classified cases with both methylation and unmethylation amplication into methylation-positive group in this study for clinical correlation analyses and prognostic evaluation. When this criteria is adopted, the percentage of patients with positive SYK methylation, E-cadherin and TIMP-3 gene became 27.4% (34/124), 26.8% (30/112) and 41.8% (46/110), respectively.
Figure 4.
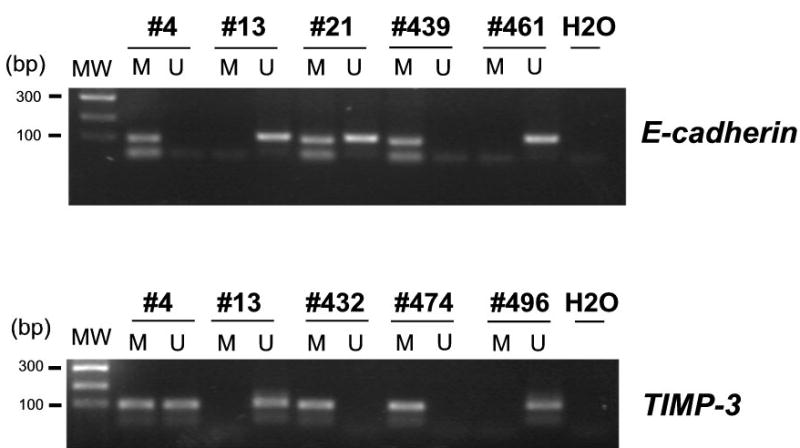
The E-cadherin and TIMP-3 genes were hypermethylated in primary HCC tumors. A two-step MSP protocol was used to analyze the gene methylation status. DNA was extracted from tissues, treated with sodium bisulfite, and then subjected to first-round PCR amplification. Then, in a nested PCR, methylation-specific or unmethylation-specific primers were used in separate reactions. For the E-cadherin gene, products of 112 bp and 120 bp were expected for methylated (M) and unmethylated (U) DNA, respectively. For the TIMP-3 gene, products of 116 bp and 122 bp were expected for methylated (M) and unmethylated (U) DNA, respectively.
Correlation of Gene Methylation with Clinicopathological Parameters
We next correlated the methylation status of SYK, E-cadherin, and TIMP-3 with 12 clinicopathological parameters, including patient gender, age, Hepatitis B infection status, Child-Pugh classification, GGT and AFP values, tumor size, status of macro tumor thrombus in the portal vein, satellite nodule, tumor capsule, degree of underlying cirrhosis, and TNM stage (Table 2). The patient age ranged from 23 to 76 years, with a median age of 48 years. Fourteen (11.3%) of the patients were women and 110 (88.7%) were men. Hepatitis B surface antigen (HbsAg) was detected in 115 patients (92.7%). Hepatitis C antibody was positive in only 2 cases (1.6%), whose HbsAg was negative. One hundred and five patients (84.7%) had histologically confirmed liver cirrhosis, and the remaining 19 (15.3%) did not. Tumor size ranged from 2 to 21 cm, with a median size of 7.5 cm. After a median follow-up of 2.6 years among 124 patients, 40 patients died of HCC and 8 patients died of other disease. Seventy-six patients were still alive at the time of last follow-up report. The 3- and 5-year overall survival rates were 58.3% and 40.9%, respectively. No significant correlation was observed among of SYK methylation and the above clinicopathological parameters. The percentage of patients with positive methylation of E-cadherin and TIMP-3 were significantly higher among those with Child-Pugh class B than those with Child-Pugh class A. In addition, E-cadherin methylation was significantly more frequent in patients with moderate or severe underlying cirrhosis, although the pathophysiological mechanism is not clear.
Table 2.
Correlation of methylation of SYK, E-cadherin, and TIMP-3 genes with clinicopathological features in patients with HCC. Values of statistical significance are shown in bold face type.
|
% of patients showing a methylated gene |
||||
|---|---|---|---|---|
| Characteristics | No. patients | SYK | E-cadherin* | TIMP-3# |
| Gender | ||||
| Female | 14 | 42.9% (6/14) | 38.5% (5/13) | 53.8% (7/13) |
| Male | 110 | 25.5% (28/110) | 25.3% (25/99) | 40.2% (39/97) |
| P value | 0.205 | 0.329 | 0.382 | |
| Age (years) | ||||
| <40 | 35 | 25.7% (9/35) | 16.1% (5/31) | 38.7% (12/31) |
| 40–60 | 66 | 25.8% (17/66) | 29.0% (18/62) | 42.4% (25/59) |
| >60 | 23 | 34.8% (8/23) | 36.8% (7/19) | 45.0% (9/20) |
| P value | 0.681 | 0.231 | 0.899 | |
| HbsAg | ||||
| Negative | 11 | 18.2% (2/11) | 22.2%(2/9) | 30.0% (3/10) |
| Positive | 113 | 28.3% (32/113) | 27.2% (28/103) | 43% (43/100) |
| P value | 0.726 | 1.000 | 0.516 | |
| Child-Pugh classification$ | ||||
| A | 118 | 26.3% (31/118) | 24.5% (26/106) | 39.0% (41/105) |
| B | 6 | 50.0 (3/6) | 66.7% (4/6) | 100% (5/5) |
| P value | 0.344 | 0.043 | 0.011 | |
| GGT (U/L) | ||||
| <50 | 60 | 30.0% (18/60) | 20.0% (11/55) | 38.2% (21/55) |
| 50 – 100 | 38 | 28.9% (11/38) | 36.1% (13/36) | 44.1% (15/34) |
| >100 | 26 | 19.2% (5/26) | 28.6% (6/21) | 47.6% (10/21) |
| P value | 0.571 | 0.232 | 0.718 | |
| AFP (μg/L) | ||||
| <20 | 35 | 22.9% (8/35) | 38.7% (12/31) | 48.4% (15/31) |
| 20–400 | 43 | 27.9% (12/43) | 23.1% (9/39) | 42.5% (17/40) |
| >400 | 46 | 30.4% (14/46) | 21.4% (9/42) | 35.9% (14/39) |
| P value | 0.748 | 0.208 | 0.571 | |
| Tumor size (cm) | ||||
| <5 | 29 | 17.2% (5/29) | 38.5% (10/26) | 42.3% (11/26) |
| >=5 | 95 | 30.5% (29/95) | 23.3% (20/86) | 41.7% (35/84) |
| P value | 0.234 | 0.137 | 0.954 | |
| Tumor thrombus | ||||
| No | 103 | 28.2% (29/103) | 25.0% (23/92) | 39.3% (35/89) |
| Yes | 21 | 23.8% (5/21) | 35.0% (7/20) | 52.4% (11/21) |
| P value | 0.793 | 0.407 | 0.329 | |
| Satellite nodule | ||||
| No | 86 | 31.4% (27/86) | 27.8% (22/79) | 43.2% (32/74) |
| Yes | 38 | 18.4% (7/38) | 24.2% (8/33) | 38.9% (14/36) |
| P value | 0.190 | 0.817 | 0.686 | |
| Tumor capsule | ||||
| Yes | 45 | 26.7% (12/45) | 28.6% (12/42) | 38.1% (16/42) |
| No or incomplete | 79 | 27.8% (22/79) | 25.7% (18/70) | 44.1% (30/68) |
| P value | 0.877 | 0.826 | 0.557 | |
| Cirrhosis | ||||
| No | 19 | 21.1% (4/19) | 12.5% (2/16) | 38.9% (7/18) |
| Mild | 61 | 23.0% (14/61) | 21.4% (12/56) | 36.4% (20/55) |
| Moderate | 40 | 37.5% (15/40) | 36.1% (13/36) | 45.5% (15/33) |
| Severe | 4 | 25.0% (1/4) | 75.0% (3/4) | 100% (4/4) |
| P value | 0.383 | 0.032 | 0.09 | |
| TNM stage | ||||
| I | 68 | 33.8% (23/68) | 28.6% (18/63) | 43.9% (25/57) |
| II | 11 | 27.3% (3/11) | 20% (2/10) | 27.3% (3/11) |
| III | 45 | 17.8% (8/45) | 25.6% (10/39) | 42.9% (18/42) |
| P value | 0.173 | 0.834 | 0.585 | |
Methylation of E-cadherin was non-informative in 12 cases.
Methylation of TIMP-3 was non-informative in 14 cases.
No patients with Child-Pugh class C were studied.
Prognostic Value of Gene Methylation in HCC
The prognostic value of 11 widely used clinicopathological parameters and the methylation status of SYK, E-cadherin, and TIMP-3 were analyzed in the 124 HCC cases. Univariate analyses showed that Child-Pugh B classification, GGT level >100 U/L, the presence of macro tumor thrombus in the portal vein, presence of satellite nodule, presence of severe or moderate cirrhosis, and TNM stage >II predicted relative poor patient survival (Table 3).
Table 3.
Univariate analyses of overall survival rates among 124 HCC patients
|
Overall survival rate (%) |
||||
|---|---|---|---|---|
| Variable | No. patients | 3-Year | 5-Year | |
| Gender | ||||
| Female | 14 | 62.5 | 62.5 | 0.9282 |
| Male | 110 | 58.4 | 48.4 | |
| Age (years) | ||||
| <40 | 35 | 53.9 | 40.5 | 0.1248 |
| 40–60 | 66 | 66.3 | 59.7 | |
| >60 | 23 | 43.5 | 34.8 | |
| HbsAg | ||||
| Negative | 11 | 88.9 | 59.3 | 0.2965 |
| Positive | 113 | 55.7 | 48.8 | |
| Child-Pugh classification | ||||
| A | 118 | 59.6 | 50.0 | 0.0466 |
| B | 6 | 0.0 | 0.0 | |
| C | 0 | |||
| GGT (U/L) | ||||
| <50 | 60 | 73.6 | 53.4 | 0.0002 |
| 50 – 100 | 38 | 56.2 | 56.2 | |
| >100 | 26 | 21.3 | 0.0 | |
| AFP (μg/L) | ||||
| <20 | 35 | 76.7 | 61.4 | 0.1355 |
| 20–400 | 43 | 50.9 | 45.8 | |
| >400 | 46 | 50.8 | 40.6 | |
| Tumor size (cm) | ||||
| <5 | 29 | 70.4 | 61.6 | 0.2934 |
| 5–10 | 72 | 50.8 | 40.7 | |
| >10 | 23 | 65.7 | 65.7 | |
| Tumor thrombus | ||||
| No | 103 | 63.2 | 60.2 | 0.0033 |
| Yes | 21 | 33.5 | 0 | |
| Satellite nodule | ||||
| No | 86 | 68.4 | 65.0 | 0.0006 |
| Yes | 38 | 33.7 | 11.2 | |
| Tumor capsule | ||||
| Yes | 45 | 59.0 | 53.8 | 0.5433 |
| No/incomplete | 79 | 58.1 | 46.4 | |
| Cirrhosis | ||||
| No | 19 | 58.7 | 58.7 | 0.0010 |
| Mild | 61 | 71.5 | 61.2 | |
| Moderate | 40 | 32.3 | 16.1 | |
| Severe | 4 | 66.7 | 0.0 | |
| TNM stage | ||||
| I | 68 | 73.4 | 69.3 | 0.0001 |
| II | 11 | 53.3 | 53.3 | |
| III | 45 | 36.6 | 0.0 | |
| SYK protein expression | ||||
| Negative | 32 | 40.5 | 30.4 | 0.0405 |
| Positive | 92 | 65.7 | 55.6 | |
| SYK gene | ||||
| Methylated | 34 | 40.6 | 30.5 | 0.0288 |
| Unmethylated | 90 | 66.3 | 56.1 | |
| E-cadherin gene* | ||||
| Methylated | 30 | 59.9 | 48.0 | 0.8578 |
| Unmethylated | 82 | 59.3 | 50.2 | |
| TIMP-3 gene# | ||||
| Methylated | 46 | 62.0 | 41.8 | 0.6725 |
| Unmethylated | 64 | 55.2 | 50.6 | |
Methylation of E-Cadherin was non-informative in 12 cases.
Methylation of TIMP-3 was non-informative in 14 cases.
We also divided all cases into two groups according to the methylation status of SYK, E-cadherin, or TIMP-3 to determine whether these factors had prognostic value. Patients whose primary tumors exhibited SYK hypermethylation had lower rates of overall survival (P = 0.0288, log-rank test) after resection; the 3- and 5-year overall survival rates were 40.6% and 30.5%, respectively, for patients with tumors that showed SYK hypermethylation, compared with 66.3% and 56.1%, respectively, for those without SYK methylation. The status of E-cadherin (P = 0.8578) and TIMP-3 (P = 0.6725) methylation did not significantly influence patient survival (Table 3, Figure 5). In addition, prognostic value of the expression status of Syk protein was analyzed. Univariate analyses showed patients whose tumors exhibited negative Syk protein expression had lower rates of overall survival (P = 0.0405, log-rank test) after surgical resection; the 3- and 5-year overall survival rates were 40.5% and 30.4%, respectively, for patients with tumors that showed negative expression of Syk protein, compared with 65.7% and 55.6%, respectively, for those with positive expression of Syk protein.
Figure 5.
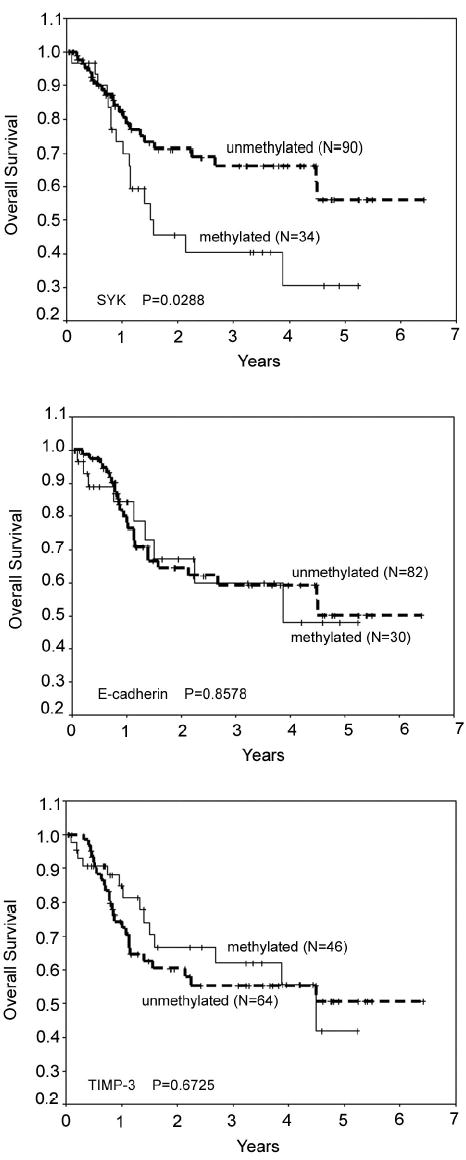
Overall survival of 124 HCC patients grouped according to methylation status of SYK (top panel), E-cadherin (middle panel), and TIMP-3 (bottom panel). Actuarial probabilities were calculated by the Kaplan-Meier method and compared with the log-rank test. After resection of primary tumors, patients with SYK hypermethylation in primary tumors had a worse overall survival rate than those without SYK methylation (P = 0.0288). No significant difference in survival was observed when analyzing the methylation status of E-cadherin (P = 0.8578) or TIMP-3 (P = 0.6725). Solid lines represent gene methylation and dashed line gene unmethylation.
The 6 clinicopathological factors and methylation status of SYK (or Syk protein status) found to be prognostic on univariate analysis were entered into a multivariate model to identify independent predictors of overall survival. Cox’s multivariate proportional-hazards model indicated that the factors significantly affecting overall survival were Child-Pugh classification (P = 0.038), TNM stage (P = 0.003), and SYK methylation status (P < 0.001) (Table 4). When we used the expression status of Syk protein to replace the methylation status of SYK in Cox’s multivariate model analysis, the factors significantly affecting overall survival were Child-Pugh classification (P = 0.040), TNM stage (P = 0.025), cirrhosis (P = 0.048) and Syk protein expression (P = 0.007). These data suggested that SYK gene methylation represented a surrogate for loss of SYK gene expression as an independent prognostic marker.
Table 4.
Cox’s multivariate analysis of factors contributing to long-term survival in HCC patients
| Variable | β | SE* | Hazard ratio (95% CI) | Pvalue |
|---|---|---|---|---|
| Child-Pugh classification | 1.612 | 0.776 | 5.013 (1.094–22.962) | 0.038 |
| GGT level | 0.422 | 0.217 | 1.524 (0.996–2.332) | 0.052 |
| Tumor thrombus | −0.451 | 0.492 | 0.637 (0.243–1.670) | 0.359 |
| Satellite nodule | 0.626 | 0.438 | 1.870 (0.793–4.413) | 0.153 |
| Cirrhosis | 0.455 | 0.265 | 1.575 (0.936–2.650) | 0.087 |
| TNM stage | 0.871 | 0.289 | 2.389 (1.357–4.207) | 0.003 |
| SYK methylation | −1.377 | 0.393 | 0.252 (0.117–0.545) | <0.001 |
SE, standard error; CI, confidence interval
DISCUSSION
In this study, we analyzed methylation of the SYK, E-cadherin, and TIMP-3 genes in 124 cases of HCC and correlated the methylation status with clinical and pathological features to determine whether these markers can predict disease outcomes. The E-cadherin and TIMP-3 tumor suppressor genes have been extensively studied and their suppressor activity has been characterized in several experimental settings (32–35). Our results support these two genes’ suppressor roles in HCC by showing methylation of E-cadherin and TIMP-3 in 26.8% and 41.8% of the cases, respectively. SYK, however, has been less well characterized. It was initially implicated as a tumor suppressor gene in breast cancer (17). SYK promoter methylation leading to gene silencing has been shown in breast cancer (15) and acute lymphoblastic leukemia (16). The loss of SYK expression is thought to contribute to tumor progression by promoting tumor invasion, proliferation, and motility. Here we showed that SYK hypermethylation was present in 27.4% of the HCCs and was associated with gene silencing. The tight correlation between SYK methylation and loss of SYK expression, together with the causal role of SYK methylation in gene silencing, indicates that epigenetic inactivation of SYK contributes to the progression of HCC. In this project, we explored the possibility of using SYK methylation as a prognostic marker in comparison to E-cadherin and TIMP-3 gene methylation.
The main focus of this study was to identify accurate biomarkers of prognosis for HCC patients after hepatectomy. Several clinicopathological features and molecular markers, with varied predictive power, have been linked to HCC prognosis. They include clinical indices (tumor size, tumor number and vascular invasion, underlying liver cirrhosis, Child-Pugh classification, and tumor microvessel density) (36–39) and molecular markers (p27 expression and p53 mutation) (40, 41). In this study, the prognostic value of SYK, E-cadherin, and TIMP-3 methylation in tumor cells was investigated. Although methylation of E-cadherin and TIMP-3 have been shown to predict a worse prognosis in node-positive diffuse gastric cancer and in esophageal adenocarcinoma, respectively (23, 24), we did not find any correlation between either E-cadherin or TIMP-3 methylation and HCC patient survival. In contrast, methylation of SYK in HCC tissues predicted poor overall survival after hepatectomy on univariate analysis. Furthermore, Cox’s multivariate proportional-hazard model confirmed that methylation of SYK in HCC was an independent and strong predictor of overall survival of these patients. SYK methylation appears to be a more powerful biomarker for risk prediction in HCC than other classic clinicopathological features, such as TNM staging and Child-Pugh classification (Table 4). It remains to be seen whether the use of SYK methylation as a prognostic tool can be extended to other tumor types, such as breast carcinoma. An earlier study indicated that in breast cancer patients, low SYK mRNA expression in tumors predicted short survival time (22). Presuming that the loss of SYK expression results from DNA methylation, SYK methylation is conceivably suitable for use as a biomarker of breast cancer prognosis.
The association between SYK methylation and poor survival rates suggests that SYK plays an important role in HCC progression. Since this study included only Chinese patients, it is not known the prognostic value of SYK methylation can be extended to HCC cases resulting from other etiological factors. It has been reported that rates of p16 methylation in HCC vary significantly among different geographic locations (e.g., it is present in 34.4% of cases from China and Egypt but only 12.2% of those from the United States and Europe). Similar geographic variations have been observed for estrogen receptor-α methylation and CpG island methylator phenotype (CIMP) (42). Whether SYK methylation has such geographic and ethnic variation and whether SYK methylation is associated with certain etiological factors need to be further investigated.
Since CpG island methylation is a reversible epigenetic change, the use of demethylation agents presents a novel therapeutic opportunity (43). Early clinical trials with demethylation compounds, such as 5-azacytidine and 5-aza-dc, have shown disappointing results in solid tumors. Their use in hematological malignancies, however, has yielded promising responses (44, 45), despite their high toxicity and chemical instability. The therapeutic outcome could be compromised without knowledge on the methylation status of tumor-related genes; demethylation agents should be effective only for patients with epigenetic inactivation of key tumor suppressor genes. Therefore, sensitive detection and a better understanding of the frequency of gene methylation must be obtained before the use of such demethylation drugs can be optimized. The present study showed that one, two, and all three of the SYK, E-cadherin, and TIMP-3 genes were methylated in 38.7% (48/124), 17.7% (22/124), and 4.8% (6/124) of our HCC cases, respectively. Thus, 61.3% of the HCC patients had at least one of the three genes methylated. They may benefit from the demethylation-based therapy. Furthermore, a new generation of demethylation drugs that are more chemically stable, such as zebularine, could be more effective clinically and may be applicable in solid tumors (46).
In conclusion, the present data demonstrate that the SYK gene can be silenced through epigenetic pathway and that positive methylation of SYK is an adverse prognostic factor among HCC patients. This information can be used to identify high-risk HCC patients who may benefit from adjuvant or more aggressive therapy after resection of primary tumors. It also justifies further studies of novel demethylating agents in the treatment of HCC.
Acknowledgments
The authors thank Drs. Jiehua He, and Kaichuan Feng for their technical supports.
Footnotes
Supported by grants from China Medical Board of New York, Inc. (98-677), National Natural Science Foundation of China (30540047), Department of Science and Technology of Guangdong, China (2002B30107), and National Institutes of Health (CA100278).
References
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Wild CP, Hall AJ. Primary prevention of hepatocellular carcinoma in developing countries. Mutat Res. 2000;462:381–93. doi: 10.1016/s1383-5742(00)00027-2. [DOI] [PubMed] [Google Scholar]
- 3.El-Serag HB, Mason AC. Rising incidence of hepatocellular carcinoma in the United States. N Engl J Med. 1999;340:745–50. doi: 10.1056/NEJM199903113401001. [DOI] [PubMed] [Google Scholar]
- 4.Lai EC, Fan ST, Lo CM, et al. Hepatic resection for hepatocellular carcinoma. An audit of 343 patients. Ann Surg. 1995;221:291–8. doi: 10.1097/00000658-199503000-00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vauthey JN, Klimstra D, Franceschi D, et al. Factors affecting long-term outcome after hepatic resection for hepatocellular carcinoma. Am J Surg. 1995;169:28–34. doi: 10.1016/s0002-9610(99)80106-8. [DOI] [PubMed] [Google Scholar]
- 6.Yoo HY, Patt CH, Geschwind JF, Thuluvath PJ. The outcome of liver transplantation in patients with hepatocellular carcinoma in the United States between 1988 and 2001: 5-year survival has improved significantly with time. J Clin Oncol. 2003;21:4329–35. doi: 10.1200/JCO.2003.11.137. [DOI] [PubMed] [Google Scholar]
- 7.Yeo W, Mok TS, Zee B, et al. A randomized phase III study of doxorubicin versus cisplatin/interferon alpha-2b/doxorubicin/fluorouracil (PIAF) combination chemotherapy for unresectable hepatocellular carcinoma. J Natl Cancer Inst. 2005;97:1532–8. doi: 10.1093/jnci/dji315. [DOI] [PubMed] [Google Scholar]
- 8.Thorgeirsson SS, Grisham JW. Molecular pathogenesis of human hepatocellular carcinoma. Nat Genet. 2002;31:339–46. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 9.Feitelson MA, Sun B, Satiroglu Tufan NL, Liu J, Pan J, Lian Z. Genetic mechanisms of hepatocarcinogenesis. Oncogene. 2002;21:2593–604. doi: 10.1038/sj.onc.1205434. [DOI] [PubMed] [Google Scholar]
- 10.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–8. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang B, Guo M, Herman JG, Clark DP. Aberrant promoter methylation profiles of tumor suppressor genes in hepatocellular carcinoma. Am J Pathol. 2003;163:1101–7. doi: 10.1016/S0002-9440(10)63469-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schagdarsurengin U, Wilkens L, Steinemann D, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866–71. doi: 10.1038/sj.onc.1206338. [DOI] [PubMed] [Google Scholar]
- 14.Iwata N, Yamamoto H, Sasaki S, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 15.Yuan Y, Mendez R, Sahin A, Dai JL. Hypermethylation leads to silencing of the SYK gene in human breast cancer. Cancer Res. 2001;61:5558–61. [PubMed] [Google Scholar]
- 16.Goodman PA, Burkhardt N, Juran B, Tibbles HE, Uckun FM. Hypermethylation of the spleen tyrosine kinase promoter in T-lineage acute lymphoblastic leukemia. Oncogene. 2003;22:2504–14. doi: 10.1038/sj.onc.1206313. [DOI] [PubMed] [Google Scholar]
- 17.Coopman PJ, Do MT, Barth M, et al. The Syk tyrosine kinase suppresses malignant growth of human breast cancer cells. Nature. 2000;406:742–7. doi: 10.1038/35021086. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Duke L, Zhang PS, et al. Alternative splicing disrupts a nuclear localization signal in spleen tyrosine kinase that is required for invasion suppression in breast cancer. Cancer Res. 2003;63:4724–30. [PubMed] [Google Scholar]
- 19.Mahabeleshwar GH, Kundu GC. Syk, a protein-tyrosine kinase, suppresses the cell motility and nuclear factor kappa B-mediated secretion of urokinase type plasminogen activator by inhibiting the phosphatidylinositol 3'-kinase activity in breast cancer cells. J Biol Chem. 2003;278:6209–21. doi: 10.1074/jbc.M208905200. [DOI] [PubMed] [Google Scholar]
- 20.Moroni M, Soldatenkov V, Zhang L, et al. Progressive loss of Syk and abnormal proliferation in breast cancer cells. Cancer Res. 2004;64:7346–54. doi: 10.1158/0008-5472.CAN-03-3520. [DOI] [PubMed] [Google Scholar]
- 21.Hoeller C, Thallinger C, Pratscher B, et al. The non-receptor-associated tyrosine kinase Syk is a regulator of metastatic behavior in human melanoma cells. J Invest Dermatol. 2005;124:1293–9. doi: 10.1111/j.0022-202X.2005.23685.x. [DOI] [PubMed] [Google Scholar]
- 22.Toyama T, Iwase H, Yamashita H, et al. Reduced expression of the Syk gene is correlated with poor prognosis in human breast cancer. Cancer Lett. 2003;189:97–102. doi: 10.1016/s0304-3835(02)00463-9. [DOI] [PubMed] [Google Scholar]
- 23.Graziano F, Arduini F, Ruzzo A, et al. Prognostic analysis of E-cadherin gene promoter hypermethylation in patients with surgically resected, node-positive, diffuse gastric cancer. Clin Cancer Res. 2004;10:2784–9. doi: 10.1158/1078-0432.ccr-03-0320. [DOI] [PubMed] [Google Scholar]
- 24.Darnton SJ, Hardie LJ, Muc RS, Wild CP, Casson AG. Tissue inhibitor of metalloproteinase-3 (TIMP-3) gene is methylated in the development of esophageal adenocarcinoma: Loss of expression correlates with poor prognosis. Int J Cancer. 2005;115:351–8. doi: 10.1002/ijc.20830. [DOI] [PubMed] [Google Scholar]
- 25.Sobin LH, Wittekind C, editors. UICC TNM classification of malignant tumors. 6th ed. New York: John Wiley & Sons; 2002. pp. 81–6. [Google Scholar]
- 26.House MG, Guo M, Iacobuzio-Donahue C, Herman JG. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–8. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 27.Corn PG, Smith BD, Ruckdeschel ES, Douglas D, Baylin SB, Herman JG. E-cadherin expression is silenced by 5' CpG island methylation in acute leukemia. Clin Cancer Res. 2000;6:4243–8. [PubMed] [Google Scholar]
- 28.Yuan Y, Liu H, Sahin A, Dai JL. Reactivation of SYK expression by inhibition of DNA methylation suppresses breast cancer cell invasiveness. Int J Cancer. 2005;113:654–9. doi: 10.1002/ijc.20628. [DOI] [PubMed] [Google Scholar]
- 29.Zochbauer-Muller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61:249–55. [PubMed] [Google Scholar]
- 30.Ahuja N, Li Q, Mohan AL, Baylin SB, Issa JP. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res. 1998;58:5489–94. [PubMed] [Google Scholar]
- 31.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–6. doi: 10.1073/pnas.93.18.9821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Graff JR, Herman JG, Lapidus RG, et al. E-cadherin expression is silenced by DNA hypermethylation in human breast and prostate carcinomas. Cancer Res. 1995;55:5195–9. [PubMed] [Google Scholar]
- 33.Bachman KE, Herman JG, Corn PG, et al. Methylation-associated silencing of the tissue inhibitor of metalloproteinase-3 gene suggest a suppressor role in kidney, brain, and other human cancers. Cancer Res. 1999;59:798–802. [PubMed] [Google Scholar]
- 34.Wei Y, Van Nhieu JT, Prigent S, Srivatanakul P, Tiollais P, Buendia MA. Altered expression of E-cadherin in hepatocellular carcinoma: correlations with genetic alterations, beta-catenin expression, and clinical features. Hepatology. 2002;36:692–701. doi: 10.1053/jhep.2002.35342. [DOI] [PubMed] [Google Scholar]
- 35.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–9. [PubMed] [Google Scholar]
- 36.Yamamoto M, Takasaki K, Otsubo T, et al. Favorable surgical outcomes in patients with early hepatocellular carcinoma. Ann Surg. 2004;239:395–9. doi: 10.1097/01.sla.0000114215.03112.e0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bilimoria MM, Lauwers GY, Doherty DA, et al. Underlying liver disease, not tumor factors, predicts long-term survival after resection of hepatocellular carcinoma. Arch Surg. 2001;136:528–35. doi: 10.1001/archsurg.136.5.528. [DOI] [PubMed] [Google Scholar]
- 38.Vauthey JN, Lauwers GY, Esnaola NF, et al. Simplified staging for hepatocellular carcinoma. J Clin Oncol. 2002;20:1527–36. doi: 10.1200/JCO.2002.20.6.1527. [DOI] [PubMed] [Google Scholar]
- 39.Poon RT, Ng IO, Lau C, et al. Tumor microvessel density as a predictor of recurrence after resection of hepatocellular carcinoma: a prospective study. J Clin Oncol. 2002;20:1775–85. doi: 10.1200/JCO.2002.07.089. [DOI] [PubMed] [Google Scholar]
- 40.Fiorentino M, Altimari A, D'Errico A, et al. Acquired expression of p27 is a favorable prognostic indicator in patients with hepatocellular carcinoma. Clin Cancer Res. 2000;6:3966–72. [PubMed] [Google Scholar]
- 41.Honda K, Sbisa E, Tullo A, et al. p53 mutation is a poor prognostic indicator for survival in patients with hepatocellular carcinoma undergoing surgical tumour ablation. Br J Cancer. 1998;77:776–82. doi: 10.1038/bjc.1998.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shen L, Ahuja N, Shen Y, et al. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–61. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 43.Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496–503. doi: 10.1038/sj.onc.1205602. [DOI] [PubMed] [Google Scholar]
- 44.Santini V, Kantarjian HM, Issa JP. Changes in DNA methylation in neoplasia: pathophysiology and therapeutic implications. Ann Intern Med. 2001;134:573–86. doi: 10.7326/0003-4819-134-7-200104030-00011. [DOI] [PubMed] [Google Scholar]
- 45.Issa JP, Garcia-Manero G, Giles FJ, et al. Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2'-deoxycytidine (decitabine) in hematopoietic malignancies. Blood. 2004;103:1635–40. doi: 10.1182/blood-2003-03-0687. [DOI] [PubMed] [Google Scholar]
- 46.Cheng JC, Weisenberger DJ, Gonzales FA, et al. Continuous zebularine treatment effectively sustains demethylation in human bladder cancer cells. Mol Cell Biol. 2004;24:1270–8. doi: 10.1128/MCB.24.3.1270-1278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]


