Abstract
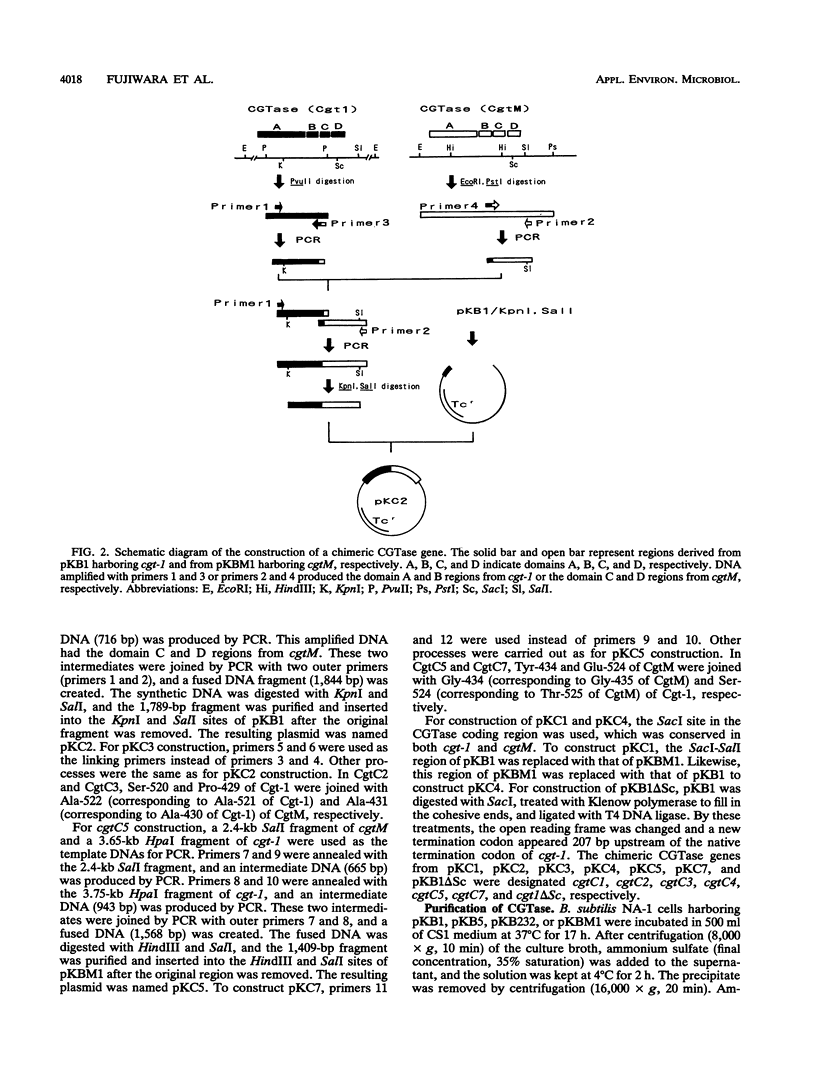
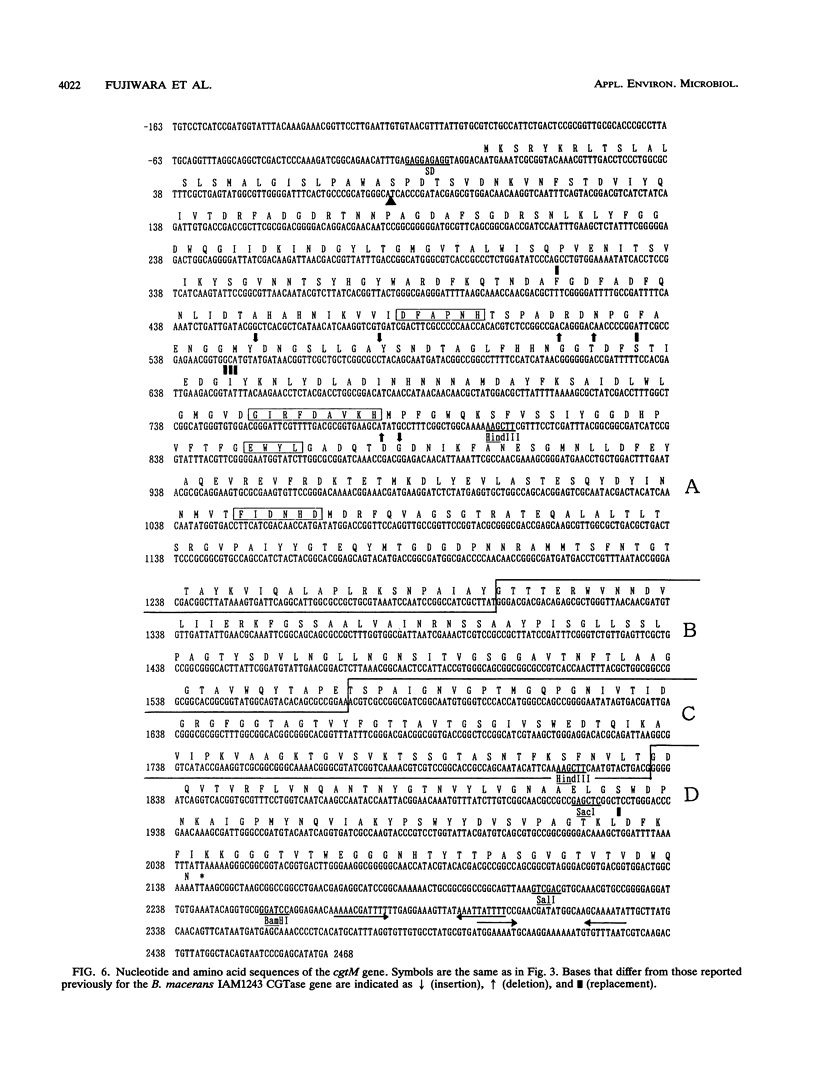
Cyclodextrin glucanotransferase (CGTase; EC 2.4.1.19) is produced mainly by Bacillus strains. CGTase from Bacillus macerans IFO3490 produces alpha-cyclodextrin as the major hydrolysis product from starch, whereas thermostable CGTase from Bacillus stearothermophilus NO2 produces alpha- and beta-cyclodextrins. To analyze the cyclization characteristics of CGTase, we cloned different types of CGTase genes and constructed chimeric genes. CGTase genes from these two strains were cloned in Bacillus subtilis NA-1 by using pTB523 as a vector plasmid, and their nucleotide sequences were determined. Three CGTase genes (cgt-1, cgt-5, and cgt-232) were isolated from B. stearothermophilus NO2. Nucleotide sequence analysis revealed that the three CGTase genes have different nucleotide sequences encoding the same amino acid sequence. Base substitutions were found at the third letter of five codons among the three genes. Each open reading frame was composed of 2,133 bases, encoding 711 amino acids containing 31 amino acids as a signal sequence. The molecular weight of the mature enzyme was estimated to be 75,374. The CGTase gene (cgtM) of B. macerans IFO3490 was composed of 2,142 bases, encoding 714 amino acids containing 27 residues as a signal sequence. The molecular weight of the mature enzyme was estimated to be 74,008. The sequence determined in this work was quite different from that reported previously by other workers. From data on the three-dimensional structure of a CGTase, seven kinds of chimeric CGTase genes were constructed by using cgt-1 from B. stearothermophilus NO2 and cgtM from B. macerans IFO3490. We examined the characteristics of these chimeric enzymes on cyclodextrin production and thermostability. It was found that the cyclization reaction was conferred by the NH2-terminal region of CGTase and that the thermostability of some chimeric enzymes was lower than that of the parental CGTases.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Argos P., Rossman M. G., Grau U. M., Zuber H., Frank G., Tratschin J. D. Thermal stability and protein structure. Biochemistry. 1979 Dec 11;18(25):5698–5703. doi: 10.1021/bi00592a028. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Crick F. H. The genetic code. 3. Sci Am. 1966 Oct;215(4):55–passim. doi: 10.1038/scientificamerican1066-55. [DOI] [PubMed] [Google Scholar]
- Hill D. E., Aldape R., Rozzell J. D. Nucleotide sequence of a cyclodextrin glucosyltransferase gene, cgtA, from Bacillus licheniformis. Nucleic Acids Res. 1990 Jan 11;18(1):199–199. doi: 10.1093/nar/18.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikai A. Thermostability and aliphatic index of globular proteins. J Biochem. 1980 Dec;88(6):1895–1898. [PubMed] [Google Scholar]
- Ikeda H., Shiozaki M. Nonhomologous recombination mediated by Escherichia coli DNA gyrase: possible involvement of DNA replication. Cold Spring Harb Symp Quant Biol. 1984;49:401–409. doi: 10.1101/sqb.1984.049.01.046. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Fujii M., Aramori I., Aiba S. Transformation of Bacillus stearothermophilus with plasmid DNA and characterization of shuttle vector plasmids between Bacillus stearothermophilus and Bacillus subtilis. J Bacteriol. 1982 Mar;149(3):824–830. doi: 10.1128/jb.149.3.824-830.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T., Himeno T., Aiba S. Effect of in vitro DNA rearrangement in the NH2-terminal region of the penicillinase gene from Bacillus licheniformis on the mode of expression in Bacillus subtilis. J Gen Microbiol. 1985 Jul;131(7):1753–1763. doi: 10.1099/00221287-131-7-1753. [DOI] [PubMed] [Google Scholar]
- Imanaka T., Tanaka T., Tsunekawa H., Aiba S. Cloning of the genes for penicillinase, penP and penI, of Bacillus licheniformis in some vector plasmids and their expression in Escherichia coli, Bacillus subtilis, and Bacillus licheniformis. J Bacteriol. 1981 Sep;147(3):776–786. doi: 10.1128/jb.147.3.776-786.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko T., Hamamoto T., Horikoshi K. Molecular cloning and nucleotide sequence of the cyclomaltodextrin glucanotransferase gene from the alkalophilic Bacillus sp. strain no. 38-2. J Gen Microbiol. 1988 Jan;134(1):97–105. doi: 10.1099/00221287-134-1-97. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Kudo T., Horikoshi K. Comparison of CD composition produced by chimeric CGTases. Agric Biol Chem. 1990 Jan;54(1):197–201. [PubMed] [Google Scholar]
- Kimura K., Kataoka S., Ishii Y., Takano T., Yamane K. Nucleotide sequence of the beta-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. strain 1011 and similarity of its amino acid sequence to those of alpha-amylases. J Bacteriol. 1987 Sep;169(9):4399–4402. doi: 10.1128/jb.169.9.4399-4402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K., Kataoka S., Nakamura A., Takano T., Kobayashi S., Yamane K. Functions of the COOH-terminal region of cyclodextrin glucanotransferase of alkalophilic Bacillus sp. #1011: relation to catalyzing activity and pH stability. Biochem Biophys Res Commun. 1989 Jun 30;161(3):1273–1279. doi: 10.1016/0006-291x(89)91380-6. [DOI] [PubMed] [Google Scholar]
- Klein C., Schulz G. E. Structure of cyclodextrin glycosyltransferase refined at 2.0 A resolution. J Mol Biol. 1991 Feb 20;217(4):737–750. doi: 10.1016/0022-2836(91)90530-j. [DOI] [PubMed] [Google Scholar]
- Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol Rev. 1983 Mar;47(1):1–45. doi: 10.1128/mr.47.1.1-45.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriki T., Imanaka T. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J Gen Microbiol. 1989 Jun;135(6):1521–1528. doi: 10.1099/00221287-135-6-1521. [DOI] [PubMed] [Google Scholar]
- Kuriki T., Okada S., Imanaka T. New type of pullulanase from Bacillus stearothermophilus and molecular cloning and expression of the gene in Bacillus subtilis. J Bacteriol. 1988 Apr;170(4):1554–1559. doi: 10.1128/jb.170.4.1554-1559.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lejeune A., Sakaguchi K., Imanaka T. A spectrophotometric assay for the cyclization activity of cyclomaltohexaose (alpha-cyclodextrin) glucanotransferase. Anal Biochem. 1989 Aug 15;181(1):6–11. doi: 10.1016/0003-2697(89)90385-0. [DOI] [PubMed] [Google Scholar]
- MacGregor E. A., Svensson B. A super-secondary structure predicted to be common to several alpha-1,4-D-glucan-cleaving enzymes. Biochem J. 1989 Apr 1;259(1):145–152. doi: 10.1042/bj2590145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura M., Becktel W. J., Levitt M., Matthews B. W. Stabilization of phage T4 lysozyme by engineered disulfide bonds. Proc Natl Acad Sci U S A. 1989 Sep;86(17):6562–6566. doi: 10.1073/pnas.86.17.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura Y., Kusunoki M., Harada W., Kakudo M. Structure and possible catalytic residues of Taka-amylase A. J Biochem. 1984 Mar;95(3):697–702. doi: 10.1093/oxfordjournals.jbchem.a134659. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Nitschke L., Heeger K., Bender H., Schulz G. E. Molecular cloning, nucleotide sequence and expression in Escherichia coli of the beta-cyclodextrin glycosyltransferase gene from Bacillus circulans strain no. 8. Appl Microbiol Biotechnol. 1990 Aug;33(5):542–546. doi: 10.1007/BF00172548. [DOI] [PubMed] [Google Scholar]
- PULLEY A. O., FRENCH D. Studies on the Schardinger dextrins. XI. The isolation of new Schardinger dextrins. Biochem Biophys Res Commun. 1961 May 15;5:11–15. doi: 10.1016/0006-291x(61)90071-7. [DOI] [PubMed] [Google Scholar]
- Perry L. J., Wetzel R. Disulfide bond engineered into T4 lysozyme: stabilization of the protein toward thermal inactivation. Science. 1984 Nov 2;226(4674):555–557. doi: 10.1126/science.6387910. [DOI] [PubMed] [Google Scholar]
- Randall L. L., Hardy S. J. Correlation of competence for export with lack of tertiary structure of the mature species: a study in vivo of maltose-binding protein in E. coli. Cell. 1986 Sep 12;46(6):921–928. doi: 10.1016/0092-8674(86)90074-7. [DOI] [PubMed] [Google Scholar]
- Roberts J. M., Buck L. B., Axel R. A structure for amplified DNA. Cell. 1983 May;33(1):53–63. doi: 10.1016/0092-8674(83)90334-3. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985 Oct;150(1):76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- Svensson B., Jespersen H., Sierks M. R., MacGregor E. A. Sequence homology between putative raw-starch binding domains from different starch-degrading enzymes. Biochem J. 1989 Nov 15;264(1):309–311. doi: 10.1042/bj2640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Imanaka T., Aiba S. Nucleotide sequence and promoter region for the neutral protease gene from Bacillus stearothermophilus. J Bacteriol. 1985 Sep;163(3):824–831. doi: 10.1128/jb.163.3.824-831.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano T., Fukuda M., Monma M., Kobayashi S., Kainuma K., Yamane K. Molecular cloning, DNA nucleotide sequencing, and expression in Bacillus subtilis cells of the Bacillus macerans cyclodextrin glucanotransferase gene. J Bacteriol. 1986 Jun;166(3):1118–1122. doi: 10.1128/jb.166.3.1118-1122.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton J. M. Disulphide bridges in globular proteins. J Mol Biol. 1981 Sep 15;151(2):261–287. doi: 10.1016/0022-2836(81)90515-5. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Yon J., Fried M. Precise gene fusion by PCR. Nucleic Acids Res. 1989 Jun 26;17(12):4895–4895. doi: 10.1093/nar/17.12.4895. [DOI] [PMC free article] [PubMed] [Google Scholar]




