Abstract
The plant pathogenic bacterium Pseudomonas syringae is divided into pathovars differing in host specificity, with P. syringae pv. syringae (Psy) and P. syringae pv. tomato (Pto) representing particularly divergent pathovars. P. syringae hrp/hrc genes encode a type III protein secretion system that appears to translocate Avr and Hop effector proteins into plant cells. DNA sequence analysis of the hrp/hrc regions in Psy 61, Psy B728a, and Pto DC3000 has revealed a Hrp pathogenicity island (Pai) with a tripartite mosaic structure. The hrp/hrc gene cluster is conserved in all three strains and is flanked by a unique exchangeable effector locus (EEL) and a conserved effector locus (CEL). The EELs begin 3 nt downstream of the stop codon of hrpK and end, after 2.5–7.3 kb of dissimilar intervening DNA with tRNALeu–queA–tgt sequences that are also found in Pseudomonas aeruginosa but without linkage to any Hrp Pai sequences. The EELs encode diverse putative effectors, including HopPsyA (HrmA) in Psy 61 and proteins similar to AvrPphE and the AvrB/AvrC/AvrPphC and AvrBsT/AvrRxv/YopJ protein families in Psy B728a. The EELs also contain mobile genetic element sequences and have a G + C content significantly lower than the rest of the Hrp Pai or the P. syringae genome. The CEL carries at least seven ORFs that are conserved between Psy B728a and Pto DC3000. Deletion of the Pto DC3000 EEL slightly reduces bacterial growth in tomato, whereas deletion of a large portion of the CEL strongly reduces growth and abolishes pathogenicity in tomato.
The plant pathogenic bacterium Pseudomonas syringae is noted for its diverse and host-specific interactions with plants (1). A specific strain may be assigned to one of at least 40 pathovars on the basis of its host range among different plant species and then further assigned to a race on the basis of differential interactions among cultivars of the host. In host plants the bacteria typically grow to high population levels in leaf intercellular spaces and then produce necrotic lesions. In nonhost plants or in host plants with race-specific resistance, the bacteria elicit the hypersensitive response (HR), a rapid, defense-associated programmed death of plant cells in contact with the pathogen (2). The ability to produce either of these reactions in plants appears to be directed by hrp (HR and pathogenicity) and hrc (HR and conserved) genes that encode a type III protein secretion pathway and by avr (avirulence) and hop (Hrp-dependent outer protein) genes that encode effector proteins injected into plant cells by the pathway (2). These effectors may also betray the parasite to the HR-triggering R-gene surveillance system of potential hosts (hence the avr designation), and plant breeding for resistance based on such gene-for-gene (avr–R) interactions may produce complex combinations of races and differential cultivars (3). hrp/hrc genes are probably universal among necrosis-causing Gram-negative plant pathogens, and they have been sequenced in P. syringae pv. syringae (Psy) 61, Erwinia amylovora Ea321, Xanthomonas campestris pv. vesicatoria (Xcv) 85–10, and Ralstonia solanacearum GMI1000 (2). On the basis of their distinct gene arrangements and regulatory components, the hrp/hrc gene clusters of these four bacteria can be divided into two groups: I (Pseudomonas and Erwinia) and II (Xanthomonas and Ralstonia). The discrepancy between the distribution of these groups and the phylogeny of the bacteria provides some evidence that hrp/hrc gene clusters have been horizontally acquired and therefore may represent pathogenicity islands (Pais) (2).
Hacker et al. (4) have defined Pais as gene clusters that (i) include many virulence genes, (ii) are selectively present in pathogenic strains, (iii) have different G + C content compared with host bacteria DNA, (iv) occupy large chromosomal regions, (v) are often flanked by direct repeats, (vi) are bordered by tRNA genes and/or cryptic mobile genetic elements, and (vii) are unstable. Some Pais have inserted into different genomic locations in the same species (5). Others reveal a mosaic structure indicative of multiple horizontal acquisitions (6). Genes encoding type III secretion systems are present in Pais in animal-pathogenic Salmonella spp. and Pseudomonas aeruginosa and on large plasmids in Yersinia and Shigella spp. Genes encoding effectors secreted by the pathway in these organisms are commonly linked to the pathway genes (7), although a noteworthy exception is sopE, which is carried by a temperate phage without apparent linkage to SPI1 in certain isolates of Salmonella typhimurium (8). Three avr/hop genes have already been shown to be linked to the hrp/hrc cluster in P. syringae: avrE and several other Hrp-regulated transcriptional units are linked to the hrpR border of the hrp cluster in P. syringae pv. tomato (Pto) DC3000 (9). avrPphE is adjacent to hrpY (hrpK) in P. syringae pv. phaseolicola (Pph) 1302A (10), and hopPsyA (hrmA) is adjacent to hrpK in Psy 61 (11). Other P. syringae avr genes are located elsewhere in the genome or on plasmids (12), including a plasmid-borne group of avr genes described as a Pai in Pph 1449B (13).
To better characterize the hrp/hrc genes and the genomic region associated with them in P. syringae, we have investigated three experimentally amenable strains that represent different levels of diversity in P. syringae: (i) Psy 61 is a weak pathogen of bean whose hrp gene cluster, cloned on cosmid pHIR11, contains all of the genes necessary for nonpathogenic bacteria such as Pseudomonas fluorescens and Escherichia coli to elicit the HR in tobacco and to secrete in culture the HrpZ harpin, a protein with unknown function that is secreted abundantly by the Hrp system (14). The pHIR11 hrp cluster has been completely sequenced (Fig. 1) (2), and the hopPsyA gene in the hypervariable region at the left edge of the cluster was shown to encode a protein that has an Avr phenotype, travels the Hrp pathway, and elicits cell death when expressed in tobacco cells (2, 15, 16). (ii) Psy B728a is in the same pathovar as strain 61 but is highly virulent and is a model for studying the role of the Hrp system in epiphytic fitness and pathogenicity (brown spot of bean) in the field (17). (iii) Pto DC3000 is a well-studied pathogen of Arabidopsis and tomato (causing bacterial speck) that is highly divergent from pathovar syringae strains. Analysis of rRNA operon restriction fragment length polymorphism (RFLP) patterns has indicated that Pto and Psy are distantly related and could be considered separate species (18). Thus, we were able to compare two strains in the same pathovar with a strain from a highly divergent pathovar.
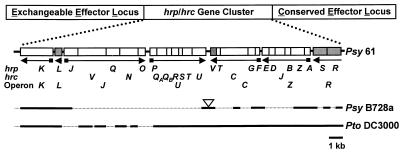
Figure 1.
The conserved arrangement of hrp/hrc genes within the Hrp Pais of Psy 61, Psy B728a, and Pto DC3000. Regions sequenced in B728a and DC3000 are indicated by lines beneath the strain 61 sequence. Known regulatory genes are shaded. Arrows indicate the direction of transcription, with small boxes denoting the presence of a Hrp box. The triangle denotes the 3.6-kb insert with phage genes in the B728a hrp/hrc region.
We report here the P. syringae Hrp Pai and its key characteristics, including (i) its tripartite mosaic architecture, (ii) a previously undescribed exchangeable effector locus, (iii) the tRNA insertion point in the ancestral Pseudomonas genome, and (iv) the differing contributions of the three regions of the Pai to the virulence of P. syringae.
Materials and Methods
Bacterial Strains, Culture Conditions, Plasmids, and DNA Manipulation Techniques.
Conditions for culturing E. coli and P. syringae strains have been described (16), as have the sources for Psy 61 (19), Psy B728a (17), and Pto DC3000 (19). Cloning and DNA manipulations were done in E. coli DH5α by using pBluescript II (Stratagene), pRK415 (20), and cosmid pCPP47 (21), according to standard procedures (22). Cosmid libraries of Pto DC3000 and Psy B728a genomic DNA were previously constructed (23). Oligonucleotide synthesis and DNA sequencing were performed at the Cornell Biotechnology Center. The nucleotide sequence of the Pto DC3000 hrp/hrc cluster was determined with subclones of pCPP2473, a cosmid selected from a genomic cosmid library on the basis of hybridization with the hrpK gene of Psy 61. The nucleotide sequence of the Psy B728a hrp/hrc cluster was determined in subclones of pCPP2346 and pCPP3017. These cosmids were selected from a genomic library on the basis of hybridization with the hrpC operon of 61. The left side of the Psy 61 exchangeable effector locus (EEL) region was cloned by PCR into pBSKSII+ XhoI and EcoRI sites by using the following primers: 5′-ATGACTCGAGGCGTGGATTCAGGCAAAT-3′ (which primes within queA and contains an XhoI site) and 5′-ATGAGAATTCTGCCGCCGCTTTCTCGTT-3′ (which primes within hopPsyA and contains an EcoRI site). Pfu DNA polymerase was used for all PCR experiments. DNA sequence data were managed and analyzed with the DNAStar program (Madison, WI), and databases were searched with the blastx, blastp, and blastn programs (24).
Mutant Construction and Analysis.
Large deletions in the Pto DC3000 Hrp Pai were constructed by subcloning border fragments into restriction sites on either side of an ΩSpr cassette in pRK415, electroporating the recombinant plasmids into DC3000, and then selecting and screening for marker exchange mutants as described (14). The following left and right side (Fig. 2 and 3) deletion border fragments were used (with residual gene fragments indicated): for CUCPB5110 left tgt-gueA-tRNALeu-ORF4′ (27 bp of ORF4) and right ORF1′-hrpK (396 bp of ORF1); and for CUCPB5115 left hrpS′-avrE′ (2,569 bp of avrE) and right ORF6 (156 bp upstream of ORF6 start codon). The later fragment was PCR-amplified by using primers 5′-CGCTCTAGACCAAGGACTGC-3′ (which primes in the ORF5–ORF6 intergenic region and contains an XbaI site) and 5′-CCAGAAGCTTCTGTTTTTGAGTC-3′ (which primes in ORF6 and contains a HindIII site). Mutant constructions were confirmed by Southern hybridizations using previously described conditions (23). The ability of mutants to secrete AvrPto was determined with anti-AvrPto antibodies and immunoblot analysis of cell fractions as previously described (16). Mutant CUCPB5115 was complemented with pCPP3016, which carries ORF2 through ORF10 in cosmid pCPP47, and was introduced from E. coli DH5α by triparental mating using helper strain E. coli DH5α(pRK600), as described (23).
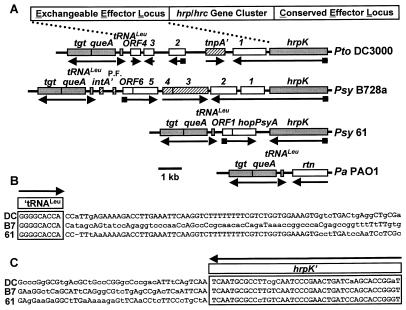
Figure 2.
The exchangeable effector loci (EELs) of Pto DC3000, Psy B728a, and Psy 61, the tgt-queA-tRNALeu locus in P. aeruginosa, and EEL border sequences. (A) EELs of the three P. syringae strains are shown aligned by their hrpK sequences and are compared with the tgt-queA-tRNALeu locus in P. aeruginosa (Pa PA01). Arrows indicate the direction of transcription, with small boxes denoting the presence of a Hrp box. Shaded regions are conserved, hatched regions denote mobile genetic elements, and open boxes denote genes that are completely dissimilar from each other. (B) The sequences of the DC3000 (DC), B728a (B7), and 61 EELs at the border with tRNALeu are shown with conserved nucleotides in uppercase. (C) The sequences of the EELs at the border with hrpK are shown as above.
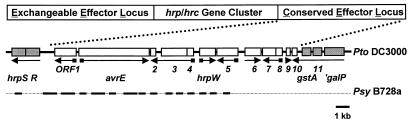
Figure 3.
The Hrp Pai conserved effector locus (CEL) of P. syringae. The Pto DC3000 CEL is shown with the corresponding fragments of Psy B728a that were sequenced aligned below. The nucleotide identity of the sequenced fragments in coding regions ranged from 72% to 83%. Arrows indicate the direction of transcription, with small boxes denoting the presence of a Hrp box.
T7 Expression Analysis.
Protein products of the Pto DC3000 EEL were analyzed by T7 polymerase-dependent expression using vector pET21 and E. coli BL21(DE3) as previously described (25). The following primer sets were used to PCR amplify each ORF from pCPP3091, which carries in pBSKSII+ a BamHI fragment containing tgt to hrcV: ORF1 5′-AGTAGGATCCTGAAATGTAGGGGCCCGG-3′ and 5′-AGTAAAGCTTATGATGCTGTTTCCAGTA-3′; ORF2 5′-AGTAGGATCCTCTCGAAGGAATGGAGCA-3′ and 5′-AGTAAAGCTTCGTGAAGATGCATTTCGC-3′; ORF3 5′-AGTAGGATCC-TAGTCACTGATCGAACGT-3′ and 5′-AGTACTCGA-GCCACGAAATAACACGGTA-3′; ORF4 5′-AGTAGGAT-CCCAGGACTGCCTTCCAGCG-3′ and 5′-AGTACTCGAG-CAGAGCGGCGTCCGTGGC-3′; tnpA 5′-AGTAGGATCC-AGAATTGTTGAAGAAATC-3′ and 5′-AGTAAAGCTTT-GCGCTGTTAACTCATCG-3′.
Plant Bioassays.
Tobacco (Nicotiana tabacum L. cv. Xanthi) and tomato (Lycopersicon esculentum Mill. cvs. Moneymaker and Rio Grande) were grown under greenhouse conditions and then maintained at 25°C with daylight and supplemental halide illumination for HR and virulence assays. Bacteria were grown overnight on King's medium B agar supplemented with appropriate antibiotics, suspended in 5 mM Mes at pH 5.6, and then infiltrated with a needleless syringe into the leaves of test plants at 108 colony-forming units (cfu)/ml for HR assays and 104 cfu/ml for pathogenicity assays (23). All assays were repeated at least four times on leaves from different plants. Bacterial growth in tomato leaves was assayed by excising disks from infiltrated areas with a cork borer, comminuting the tissue in 0.5 ml of 5 mM Mes, pH 5.6, with a Kontes Pellet Pestle (Fisher Scientific), and then dilution plating the homogenate on King's medium B agar with 50 μg/ml rifampicin and 2 μg/ml cycloheximide to determine bacterial populations. The mean and SE from three leaf samples were determined for each time point. The relative growth in planta of DC3000 and CUCPB5110 was similarly assayed in four independent experiments and the relative growth of DC3000, CUCPB5115, and CUCPB5115(pCPP3016), in three independent experiments. Although the final population levels achieved by DC3000 varied between experiments, the populations levels of the mutants relative to the wild type were the same as in the representative experiments presented below.
Results
The hrp/hrc Gene Clusters of Psy 61, Psy B728a, and Pto DC3000 Are Similarly Delimited by hrpK and hrpR.
To determine whether the hrp/hrc clusters from Psy B728a and Pto DC3000 were organized similarly to the previously characterized hrp/hrc cluster of Psy 61, we partially characterized two cosmids carrying hrp/hrc inserts. pCPP2346 carries the entire hrp/hrc cluster of B728a, and pCPP2473 carries the left half of the hrp/hrc cluster of DC3000. The right half of the DC3000 hrp/hrc cluster had been characterized previously (19). Sequencing the ends of several subclones derived from these cosmids provided fingerprints of the B728a and DC3000 hrp/hrc clusters, which indicated that both are arranged like that of strain 61 (Fig. 1). However, B728a contains between hrcU and hrpV a 3.6-kb insert with homologs of bacteriophage λ genes Ea59 (23% amino acid identity; E = 2e-7) and Ea31 (30% amino acid identity; E = 6e-8) (26), and the B728a hrcU ORF has 36 additional codons. A possible insertion of this size in several Psy strains that are highly virulent on bean was suggested by a previous restriction fragment length polymorphism analysis (27). Cosmid pCPP2346, which contains the B728a hrp/hrc region and flanking sequences (4 kb on the left and 13 kb on the right), enabled P. fluorescens to secrete the B728a HrpZ harpin in culture and to elicit the HR in tobacco leaves, however, confluent necrosis developed more slowly than with P. fluorescens(pHIR11) (data not shown). To further test the relatedness of the Psy 61 and B728a hrp/hrc gene clusters with an internal reference, we sequenced the B728a hrpA gene. Of the hrp/hrc genes that have been sequenced in Psy and Pto, hrpA, which encodes the major subunit of the Hrp pilus (28), is the least conserved (28% amino acid identity) (19). However, the hrpA genes of strains 61 and B728a were 100% identical, which further supports the close relationship of these strains and their Hrp systems.
An EEL Resides in the Hrp Pai between hrpK and tRNALeu.
Sequence analysis of the left side of the Psy 61, Psy B728a, and Pto DC3000 Hrp Pais revealed that the high percentage identity in hrpK sequences in these strains abruptly terminates 3 nt after the hrpK stop codon and then is restored near tRNALeu, queA, and tgt sequences after 2.5 kb (Psy 61), 7.3 kb (Psy B728a), or 5.9 kb (Pto DC3000) of dissimilar, intervening DNA (Fig. 2). The difference between Psy strains 61 and B728a in this region was particularly surprising. This region of the P. syringae Hrp Pai was given the EEL designation because it contained completely different effector protein genes (Table 1), which appear to be exchanged at this locus at a high frequency. In this regard, it is noteworthy that (i) ORF2 in the B728a EEL is a homolog of avrPphE, which is in a different location, immediately downstream of hrpK (hrpY), in Pph 1302A (10); (ii) hopPsyA (hrmA) is present in only a few Psy strains (11, 15); and (iii) ORF5 in the B728a EEL predicts a protein that is similar to Xanthomonas AvrBsT and possesses multiple motifs characteristic of the AvrRxv family (29) (data not shown). G + C content different from the genomic average is a hallmark of horizontally transferred genes, and the G + C contents of the ORFs in the three EELs are considerably lower than the average of 59–61% for P. syringae (30) (Table 1). They are also lower than hrpK (60%) and queA (63–64%). The ORFs in the Pto DC3000 EEL predict no products with similarity to known effector proteins; however, T7 polymerase-dependent expression revealed products in the size range predicted for ORF1, ORF3, and ORF4. Furthermore, the ORF1 protein is secreted in a hrp-dependent manner by E. coli(pCPP2156), which expresses an Erwinia chrysanthemi Hrp system that secretes P. syringae Avr proteins (31) (J.R.A., unpublished data). Several ORFs in these EELs are preceded by Hrp boxes indicative of HrpL-activated promoters (Fig. 1) (32), and the lack of intervening Rho-independent terminator sequences or promoters suggests that ORF1 in DC3000 and ORF1 and ORF2 in B728a are expressed from HrpL-activated promoters upstream of the respective hrpK genes.
Table 1.
ORFs and fragments of genetic elements in the EELs of Pto DC3000, Psy B728a, and Psy 61 and similarities with known avr genes and mobile genetic elements
| ORF or sequence |
% G+C | Size |
blastE value with representative similar sequence in database, or relevant feature |
|---|---|---|---|
| Pto* DC3000 | |||
| ORF1 | 55 | 466 aa | Hrp-secreted (J.R.A., unpublished data) |
| TnpA′ | 55 | 279 aa | 1e-125 P. stutzeri TnpA1 (52) |
| ORF2 | 51 | 241 aa | None |
| ORF3 | 53 | 138 aa | None |
| ORF4 | 47 | 136 aa | None |
| Psy* B728a | |||
| ORF1 | 51 | 323 aa | 9e-40 Pph* AvrPphC (53) |
| ORF2 | 58 | 382 aa | 1e-154 Pph AvrPphE (10) |
| ORF3 | 55 | 507 aa | 2e-63 E. coli L0015 (34) |
| ORF4 | 55 | 118 aa | 9e-9 E. coli L0014 (34) |
| ORF5 | 49 | 411 aa | 1e-4 Xcv AvrBsT (29) |
| ORF6 | 52 | 120 aa | None |
| B plasmid | 46 | 96 nt | 1e-25 Pph pAV511 (13) |
| IntA′ | 59 | 49 aa | 3e-5 E. coli CP4-like integrase (34) |
| Psy* 61 | |||
| HopPsyA | 53 | 375 aa | Hrp-secreted Avr (15, 16) |
| ORF1 | 57 | 112 aa | 6e-4 Yersinia pestis pCD1 Y0008 (54) |
Pathovar abbreviations correspond to the recommendations of Vivian and Mansfield (55) for uniform avr nomenclature.
The EELs of these three strains also contain sequences homologous to insertion sequences (ISs), transposases, phage integrase genes, and plasmids (Fig. 2 and Table 1). The Psy B728a ORF5 and ORF6 operon is bordered on the left side by sequences similar to those in a Pph plasmid that carries several avr genes (13) and by a sequence homologous to insertion elements that are typically found on plasmids, suggesting plasmid integration via an IS element in this region (33). Psy B728a ORF3 and ORF4 show similarity to sequences implicated in the horizontal acquisition of the LEE Pai by pathogenic E. coli strains (34). These Psy B728a ORFs are not preceded by Hrp boxes and are unlikely to encode effector proteins.
The left border of the EELs contains sequences similar to many tRNALeu genes and to E. coli queA and tgt queuosine biosynthesis genes (ca. 70% amino acid identity in predicted products). The EEL sequences terminate at the 3′ end of the P. syringae tRNA sequences, as is typical for Pais (35). Virtually identical tgt-queA-tRNALeu sequences are found in the genome of P. aeruginosa PAO1 (www.pseudomonas.com), which is also in the fluorescent pseudomonad group. But PAO1 is not a plant pathogen, and this tRNALeu in P. aeruginosa is not linked to any type III secretion system genes or other genes in the Hrp Pai (Fig. 2). Thus, this is the apparent point of insertion of the Hrp Pai in the ancestral Pseudomonas genome.
A CEL Is Located on the Right Side of the Hrp Pai in Psy B728a and Pto DC3000.
Previous studies of the region to the right of hrpR in DC3000 had revealed the existence of the avrE locus, which is composed of two transcriptional units (9), the 5′ sequences for the first 4 transcriptional units beyond hrpR (9), and the fourth transcriptional unit, which has been identified as the hrpW gene encoding a second harpin (23). Here, we completed the DNA sequence of the first 14 ORFs to the right of hrpR in Pto DC3000 and partially sequenced the corresponding region in Psy B728a (Fig. 3). Like the EEL, this region contains putative effector genes, e.g., avrE (9). Unlike the EEL, the ORFs in this region have an average G + C content of 58.0%, which is close to that of the hrp/hrc genes; the region contains no sequences similar to known mobile genetic elements, and it appears conserved between Psy and Pto (Fig. 3). Comparison of the regions sequenced in B728a and DC3000 revealed that the first 7 ORFs are arranged identically and have an average DNA sequence identity of 78%. Hence, this region was given the CEL designation.
The precise border of the CEL remains undefined, and we found no sequences that were repeated in the EEL border of the Hrp Pai. ORF7 and ORF8 are likely to be part of the CEL, based on the presence of an upstream Hrp box (Fig. 3). However, the region beyond ORF10 probably is not in the CEL because the product of the next ORF shows homology to a family of bacterial GstA proteins (e.g., 28% identity with E. coli GstA over 204 amino acids; E = 1e-8) (36), and glutathione S-transferase activity is common in nonpathogenic fluorescent pseudomonads (37). The presence of a galP homolog (38% identity over 256 amino acids, based on incomplete sequence, to E. coli GalP; E = 2e-42) (36) in this region further suggests that it is beyond the CEL.
Several other features of this region in B728a and DC3000 are noteworthy. (i) Both strains have a 1-kb intergenic region between hrpR and ORF1 that is distinguished by low sequence identity (44%) but that contains three inverted repeats that could form stem-loop structures affecting expression of the hrpRS operon. (ii) ORF1 is most similar to E. coli murein lytic transglycosylase MltD (38% identity over 324 amino acids; E = 4e-56). (iii) ORF2 is 42% identical over 130 amino acids with Erwinia amylovora DspF (E = 9e-24), a candidate chaperone (38, 39). (iv) The ORF5 protein is secreted in a hrp-dependent manner by E. coli(pCPP2156), but mutation with an ΩSpr cassette has little effect on either HR elicitation in tobacco or pathogenicity in tomato (A.O.C., unpublished data). (v) Six operons in this region are preceded by Hrp boxes (9) (Fig. 3), which is characteristic of known avr genes in P. syringae (14). Thus, the CEL carries multiple candidate effectors.
Deletion of the Pto DC3000 EEL Partially Reduces Pathogen Fitness in Planta.
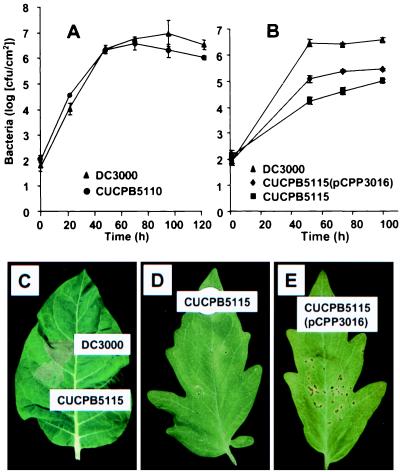
A mutation was constructed in DC3000 that replaced all of the ORFs between hrpK and tRNALeu with an ΩSpr cassette (Fig. 2). This Pto mutant, CUCPB5110, was tested for its ability to elicit the HR in tobacco and to cause disease in tomato. The mutant retained the ability to elicit the HR and to produce disease symptoms (data not shown), but it failed to reach population levels as high as the parental strain in tomato (Fig. 4A).
Figure 4.
Plant interaction phenotypes of Pto mutants carrying deletions of the EEL (CUCPB5110) and CEL (CUCPB5115). (A) Growth in tomato of DC3000 and CUCPB5110 (mean and SE). (B) Growth in tomato of DC3000, CUCPB5115, and CUCPB5115(pCPP3016) (mean and SE). (C) HR collapse in tobacco leaf tissue 24 hr after infiltration with 107 cfu/ml DC3000 and CUCPB5115. (D) Absence of disease symptoms in tomato leaf 4 days after inoculation with 104 cfu/ml CUCPB5115. (E) Disease symptoms typical of wild type in tomato leaf 4 days after inoculation with 104 cfu/ml CUCPB5115(pCPP3016).
A Large Deletion in the Pto DC3000 CEL Abolishes Pathogenicity.
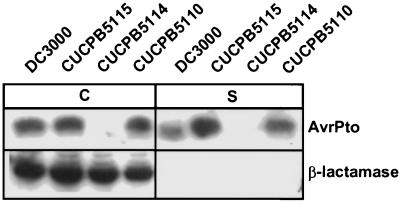
A mutation was constructed in DC3000 that replaced avrE through ORF5 with an ΩSpr cassette. This deleted all of the CEL ORFs that were both partially characterized and likely to encode effectors. This Pto mutant, CUCPB5115, still elicited the HR in tobacco, but tissue collapse was delayed ca. 5 hr (Fig. 4C). The mutant no longer elicited disease symptoms in tomato when infiltrated at a concentration of 104 cfu/ml, and growth in planta was strongly reduced (Fig. 4B). However, the mutant elicited an HR dependent on the tomato Pto R gene that was indistinguishable from the wild-type in tests involving PtoS (susceptible) and PtoR (resistant) Rio Grande tomato lines (data not shown). Plasmid pCPP3016, which carries ORF2 through ORF10, fully restored the ability of CUCPB5115 to cause disease symptoms and partially restored the ability of the mutant to multiply in tomato leaves (Fig. 4 B and E). Deletion of the hrp/hrc cluster abolishes HR and pathogenicity phenotypes in Pto DC3000 (40). To confirm that the large deletions in Pto mutants CUCPB5110 and CUCPB5115 did not disrupt Hrp secretion functions, we compared the ability of these mutants, the DC3000 hrp/hrc deletion mutant, and wild-type DC3000 to make and secrete AvrPto in culture while retaining a cytoplasmic marker composed of β-lactamase lacking its signal peptide. AvrPto provided an ideal subject for this test because it is a well-studied effector protein that is secreted in culture and injected into host cells in planta (2, 16). Only the hrp/hrc deletion cluster mutant was impaired in AvrPto production and secretion (Fig. 5).
Figure 5.
Immunoblot analysis of AvrPto secretion by Pto DC3000 derivatives with deletions affecting the three major regions of the Hrp Pai. Bacteria were grown in Hrp-inducing minimal medium at pH 5.5 and 22°C to an OD600 of 0.35 and then separated into cell-bound (C) and supernatant (S) fractions by centrifugation. Proteins were then resolved by SDS/PAGE, blotted, and immunostained with antibodies against AvrPto and β-lactamase as described (18), except that supernatant fractions were concentrated 3-fold relative to cell-bound fractions before loading. Pto DC3000, CUCPB5115 (CEL deletion), CUCPB5114 (hrp/hrc deletion), and CUCPB5110 (EEL deletion) all carried pCPP2318, which expresses β-lactamase without a signal peptide as a cytoplasmic marker.
Discussion
We have determined that the P. syringae hrp/hrc genes are part of a Hrp Pai that has three distinct loci: an EEL, the hrp/hrc gene cluster, and a CEL. The EEL harbors exchangeable effector genes and makes only a quantitative contribution to parasitic fitness in host plants. The hrp/hrc locus encodes the Hrp secretion system and is required for effector protein delivery, parasitism, and pathogenicity. The CEL makes no discernible contribution to Hrp secretion functions but contributes strongly to parasitic fitness and is required for Pto pathogenicity in tomato. The Hrp Pai of P. syringae has several properties of Pais possessed by animal pathogens (4), including the presence of many virulence-associated genes (several with relatively low G + C content) in a large (ca. 50-kb) chromosomal region linked to a tRNA locus and absent from the corresponding locus in a closely related species. In addition, the EEL portion of the Hrp Pai is unstable and contains many sequences related to mobile genetic elements.
The EEL is a novel feature of known Pais, which is likely involved in fine-tuning the parasitic fitness of P. syringae strains with various plant hosts. By comparing closely and distantly related strains of P. syringae, we were able to establish the high instability of this locus and the contrasting high conservation of its border sequences. No single mechanism can explain the high instability, as we found fragments related to phages, insertion sequences, and plasmids in the Psy and Pto EELs, and insertion sequences were recently reported in the corresponding region of three other P. syringae strains (41). The mechanism or significance of the localization of the EELs between tRNALeu and hrpK sequences in the Hrp Pais also is unclear. Pto DC3000 carries at least one other effector gene, avrPto, that is located elsewhere in the genome (42), many P. syringae avr genes are located on plasmids (12), and the EEL ORFs represent a mix of widespread (e.g., avrRxv family) and seemingly rare (e.g., hopPsyA) effector genes. The G + C content of the EEL ORFs is significantly lower than that of the rest of the Hrp Pai and the P. syringae genome. Although certain genes in the non-EEL portions of the Hrp Pai, such as hrpA, are highly divergent, they have a high G + C content, and there is no evidence that they have been horizontally transferred separately from the rest of the Hrp Pai. The relatively low G + C content of the ORFs in the EELs (and of other P. syringae avr genes) suggests that these genes may be horizontally acquired from a wider pool of pathogenic bacteria than just P. syringae (43). Indeed, the avrRxv family of genes is found in a wide range of plant and animal pathogens (29). The weak effect on parasitic fitness of deleting the Pto DC3000 EEL, or of mutating hopPsyA (hrmA) in Psy 61 (44), is typical of mutations in individual avr genes and presumably results from redundancy in the effector protein system (12).
The functions of hrpK and of the CEL ORF1 are unclear but warrant discussion. These two ORFs reside just outside the hrpL and hrpR delimited cluster of operons containing both hrp and hrc genes and thereby spatially separate the three regions of the Hrp Pai (Figs. 1, 2, and 3). hrpK mutants have a variable Hrp phenotype (10, 45), and a Psy B728a hrpK mutant still secretes HrpZ (J.R.A., unpublished data), which suggests that HrpK may be an effector protein. Nevertheless, the HrpK proteins of Psy 61 and Pto DC3000 are 79% identical and therefore are more conserved than many Hrp secretion system components. It is also noteworthy that hrpK appears to be in an operon with other effector genes in Psy B728a and Pto DC3000. In contrast, the CEL ORF1 may contribute (weakly or redundantly) to Hrp secretion functions by promoting penetration of the system through the bacterial peptidoglycan layer. The ORF1 product has extensive homology with E. coli MltD and shares a lysozyme-like domain with the product of ipgF (46), a Shigella flexneri gene that is also located between loci encoding a type III secretion system and effector proteins (47). Mutations in these genes in Pto and S. flexneri have no obvious phenotype (9, 47), as is typical for genes encoding peptidoglycan hydrolases (48).
The loss of pathogenicity in Pto mutant CUCPB5115, with an avrE-ORF5 deletion in the CEL, was surprising because pathogenicity is retained in DC3000 mutants in which the corresponding operons are individually disrupted (9, 23) (A.O.C., unpublished data). In assessing the possible function of this region and the conservation of its constituent genes, it should be noted that avrE is unlike other avr genes found in Pto in that it confers avirulence to P. syringae pv. glycinea on all tested soybean cultivars and it has a homolog (dspE) in E. amylovora that is required for pathogenicity (9, 49). Although the CEL is required for pathogenicity, it is not essential for type III effector protein secretion because the mutant still secretes AvrPto. It also appears to play no essential role in type III translocation of effector proteins into plant cells because the mutant still elicits the HR in nonhost tobacco and in a PtoR-resistance tomato line, and pHIR11, which lacks this region, appears capable of translocating several Avr proteins (50, 51). The conservation of this region in the divergent pathovars Psy and Pto, and its importance in disease, suggests that the products of the CEL may be redundantly involved in a common, essential aspect of pathogenesis.
The similar G + C content and codon usage of the hrp/hrc genes, the genes in the CEL, and total P. syringae genomic DNA suggest that the Hrp Pai was acquired early in the evolution of P. syringae. Although, the EEL region may have similarly developed early in the radiation of P. syringae into its many pathovars, races, and strains, the apparent instability that is discussed above suggests ongoing rapid evolution at this locus. Indeed, many P. syringae avr genes are associated with mobile genetic elements, regardless of their location (43). Thus, it appears that Hrp-mediated pathogenicity in P. syringae is collectively dependent on a set of genes that are universal among divergent pathovars and on another set that varies among strains even in the same pathovar. The later are presumably acquired and lost in response to opposing selection pressures to promote parasitism while evading host R-gene surveillance systems. What effector proteins do to make plants susceptible to P. syringae remains unknown, but exploring those effector proteins that are universal is likely to be particularly fruitful.
Acknowledgments
We thank Adela Ramos for partial sequencing of the Pto DC3000 hrpJ and hrpU operons, Angela R. Hill for cloning and sequencing the Psy 61 tRNALeu–EEL border region, Cathy Zumoff for screening the Pto DC3000 cosmid library, and Kent Loeffler for photography. This work was supported by National Science Foundation Grant MCB 97–35303-4488 (A.C.) and the National Research Initiative Competitive Grants Program of the U.S. Department of Agriculture Grant 98–35303-6464 (J.R.A.).
Abbreviations
- Psy
Pseudomonas syringae pv. syringae
- Pto
P. syringae pv. tomato
- Pph
P. syringae pv. phaseolicola
- HR
hypersensitive response
- hrp
HR and pathogenicity
- hrc
HR and conserved
- Pai
pathogenicity island
- EEL
exchangeable effector locus
- CEL
conserved effector locus
- Hop
Hrp-dependent outer protein
- Avr
avirulence
- cfu
colony-forming units
Footnotes
References
- 1.Hirano S S, Upper C D. Annu Rev Phytopathol. 1990;28:155–177. [Google Scholar]
- 2.Alfano J R, Collmer A. J Bacteriol. 1997;179:5655–5662. doi: 10.1128/jb.179.18.5655-5662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keen N T. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- 4.Hacker J, Blum-Oehler G, Muhldorfer I, Tschape H. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 5.Wieler L H, McDaniel T K, Whittam T S, Kaper J B. FEMS Microbiol Lett. 1997;156:49–53. doi: 10.1111/j.1574-6968.1997.tb12704.x. [DOI] [PubMed] [Google Scholar]
- 6.Hensel M, Nikolaus T, Egelseer C. Mol Microbiol. 1999;31:489–498. doi: 10.1046/j.1365-2958.1999.01190.x. [DOI] [PubMed] [Google Scholar]
- 7.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirold S, Rabsch W, Rohde M, Stender S, Tschape H, Russmann H, Igwe E, Hardt W D. Proc Natl Acad Sci USA. 1999;96:9845–9850. doi: 10.1073/pnas.96.17.9845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lorang J M, Keen N T. Mol Plant–Microbe Interact. 1995;8:49–57. doi: 10.1094/mpmi-8-0049. [DOI] [PubMed] [Google Scholar]
- 10.Mansfield J, Jenner C, Hockenhull R, Bennett M A, Stewart R. Mol Plant–Microbe Interact. 1994;7:726–739. doi: 10.1094/mpmi-7-0726. [DOI] [PubMed] [Google Scholar]
- 11.Heu S, Hutcheson S W. Mol Plant–Microbe Interact. 1993;6:553–564. doi: 10.1094/mpmi-6-553. [DOI] [PubMed] [Google Scholar]
- 12.Leach J E, White F F. Annu Rev Phytopathol. 1996;34:153–179. doi: 10.1146/annurev.phyto.34.1.153. [DOI] [PubMed] [Google Scholar]
- 13.Jackson R W, Athanassopoulos E, Tsiamis G, Mansfield J W, Sesma A, Arnold D L, Gibbon M J, Murillo J, Taylor J D, Vivian A. Proc Natl Acad Sci USA. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alfano J R, Bauer D W, Milos T M, Collmer A. Mol Microbiol. 1996;19:715–728. doi: 10.1046/j.1365-2958.1996.415946.x. [DOI] [PubMed] [Google Scholar]
- 15.Alfano J R, Kim H-S, Delaney T P, Collmer A. Mol Plant–Microbe Interact. 1997;10:580–588. doi: 10.1094/MPMI.1997.10.5.580. [DOI] [PubMed] [Google Scholar]
- 16.van Dijk K, Fouts D E, Rehm A H, Hill A R, Collmer A, Alfano J R. J Bacteriol. 1999;181:4790–4797. doi: 10.1128/jb.181.16.4790-4797.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirano S, Charkowski A O, Collmer A, Willis D K, Upper C D. Proc Natl Acad Sci USA. 1999;96:9851–9856. doi: 10.1073/pnas.96.17.9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manceau C, Horvais A. Appl Environ Microbiol. 1997;63:498–505. doi: 10.1128/aem.63.2.498-505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston G, Huang H-C, He S Y, Collmer A. Mol Plant–Microbe Interact. 1995;8:717–732. doi: 10.1094/mpmi-8-0717. [DOI] [PubMed] [Google Scholar]
- 20.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 21.Bauer D W, Collmer A. Mol Plant–Microbe Interact. 1997;10:369–379. doi: 10.1094/MPMI.1997.10.3.369. [DOI] [PubMed] [Google Scholar]
- 22.Ausubel F M, Brent R, Kingston R, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current Protocols in Molecular Biology. New York: Wiley; 1994. [Google Scholar]
- 23.Charkowski A O, Alfano J R, Preston G, Yuan J, He S Y, Collmer A. J Bacteriol. 1998;180:5211–5217. doi: 10.1128/jb.180.19.5211-5217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang H-C, Lin R-W, Chang C-J, Collmer A, Deng W-L. Mol Plant–Microbe Interact. 1995;8:733–746. doi: 10.1094/mpmi-8-0733. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix R W, Roberts J W, Stahl F W, Weisberg R A, editors. Lambda II. Plainview, NY: Cold Spring Harbor Lab. Press; 1983. [Google Scholar]
- 27.Legard D E, Aquadro C F, Hunter J E. Appl Environ Microbiol. 1993;59:4180–4188. doi: 10.1128/aem.59.12.4180-4188.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ciesiolka L D, Hwin T, Gearlds J D, Minsavage G V, Saenz R, Bravo M, Handley V, Conover S M, Zhang H, Caporgno J, Phengrasamy N B, Toms A O, Stall R E, Whalen M C. Mol Plant–Microbe Interact. 1999;12:35–44. doi: 10.1094/MPMI.1999.12.1.35. [DOI] [PubMed] [Google Scholar]
- 30.Palleroni N J. In: Bergey's Manual of Systematic Bacteriology. Krieg N R, Holt J G, editors. Baltimore: Williams and Wilkins; 1984. pp. 141–199. [Google Scholar]
- 31.Ham J H, Bauer D W, Fouts D E, Collmer A. Proc Natl Acad Sci USA. 1998;95:10206–10211. doi: 10.1073/pnas.95.17.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Y, Hutcheson S. J Bacteriol. 1994;176:3089–3091. doi: 10.1128/jb.176.10.3089-3091.1994. , and correction (1994) 176, 6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szabo L J, Mills D. J Bacteriol. 1984;157:821–827. doi: 10.1128/jb.157.3.821-827.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perna N T, Mayhew G F, Posfai G, Elliott S, Donnenberg M S, Kaper J B, Blattner F R. Infect Immun. 1998;66:3810–3817. doi: 10.1128/iai.66.8.3810-3817.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hou Y-M. Trends Biochem Sci. 1999;24:295–298. doi: 10.1016/s0968-0004(99)01428-0. [DOI] [PubMed] [Google Scholar]
- 36.Blattner F R, Plunkett G, 3rd, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, et al. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 37.Zablotowicz R M, Hoagland R E, Locke M A, Hickey W J. Appl Environ Microbiol. 1995;61:1054–1060. doi: 10.1128/aem.61.3.1054-1060.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bogdanove A J, Bauer D W, Beer S V. J Bacteriol. 1998;180:2244–2247. doi: 10.1128/jb.180.8.2244-2247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gaudriault S, Malandrin L, Paulin J-P, Barny M-A. Mol Microbiol. 1997;26:1057–1069. doi: 10.1046/j.1365-2958.1997.6442015.x. [DOI] [PubMed] [Google Scholar]
- 40.Collmer A, Alfano J R, Anderson D M, Badel J L, Deng W-L, Fouts D E, Rehm A H, Rojas C M, Schneewind O, van Dijk K. In: Biology of Plant-Microbe Interactions. de Wit P J G M, Bisseling T, Stiekema W, editors. Vol. 2. St. Paul: Int. Soc. Molecular Plant-Microbe Interactions; 2000. pp. 65–70. [Google Scholar]
- 41.Inoue Y, Takikawa Y. Ann Phytopathol Soc Japan. 1999;65:100–109. [Google Scholar]
- 42.Ronald P C, Salmeron J M, Carland F M, Staskawicz B J. J Bacteriol. 1992;174:1604–1611. doi: 10.1128/jb.174.5.1604-1611.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J F, Charkowski A O, Alfano J R, Collmer A, Beer S V. Mol Plant–Microbe Interact. 1998;11:1247–1252. [Google Scholar]
- 44.Huang H-C, Hutcheson S W, Collmer A. Mol Plant–Microbe Interact. 1991;4:469–476. doi: 10.1094/mpmi-6-515. [DOI] [PubMed] [Google Scholar]
- 45.Bozso Z, Ott P G, Kecskes M L, Klement Z. Physiol Mol Plant Pathol. 1999;55:215–223. [Google Scholar]
- 46.Mushegian A R, Fullner K J, Koonin E V, Nester E W. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allaoui A, Menard R, Sansonetti P J, Parsot P. Infect Immun. 1993;61:1707–1714. doi: 10.1128/iai.61.5.1707-1714.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dijkstra A J, Keck W. J Bacteriol. 1996;178:5555–5562. doi: 10.1128/jb.178.19.5555-5562.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bogdanove A J, Kim J F, Wei Z, Kolchinsky P, Charkowski A O, Conlin A K, Collmer A, Beer S V. Proc Natl Acad Sci USA. 1998;95:1325–1330. doi: 10.1073/pnas.95.3.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gopalan S, Bauer D W, Alfano J R, Loniello A O, He S Y, Collmer A. Plant Cell. 1996;8:1095–1105. doi: 10.1105/tpc.8.7.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pirhonen M U, Lidell M C, Rowley D L, Lee S W, Jin S, Liang Y, Silverstone S, Keen N T, Hutcheson S W. Mol Plant–Microbe Interact. 1996;9:252–260. doi: 10.1094/mpmi-9-0252. [DOI] [PubMed] [Google Scholar]
- 52.Bosch R, Garcia-Valdes E, Moore E R B. Gene. 1999;236:149–157. doi: 10.1016/s0378-1119(99)00241-3. [DOI] [PubMed] [Google Scholar]
- 53.Yucel I, Slaymaker D, Boyd J, Murillo J, Buzzell R I, Keen N T. Mol Plant–Microbe Interact. 1994;7:677–679. doi: 10.1094/mpmi-7-0677. [DOI] [PubMed] [Google Scholar]
- 54.Perry R D, Straley S C, Fetherston J D, Rose D J, Gregor J, Blattner F R. Infect Immun. 1998;66:4611–4623. doi: 10.1128/iai.66.10.4611-4623.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vivian A, Mansfield J. Mol Plant–Microbe Interact. 1993;6:9–10. [Google Scholar]