Abstract
Previous studies have shown that Delta-mediated Notch signaling regulates the number of early differentiating neurons. However, the role of Notch activation and Jagged-mediated signaling during late neurogenesis remains poorly defined. In the developing spinal cord of zebrafish, GABAergic Kolmer–Agduhr (KA′′) cells and motor neurons (MN) emerge sequentially from their progenitors in the p3 domain. Jagged2 is expressed uniformly in the pMN domain during late neurogenesis where Olig2 is required for its expression. We suggest that Jagged2 interacts ventrally with progenitors in the adjacent p3 domain, where it has a critical role in the maintenance of proliferating neural progenitors and in preventing differentiation of these progenitors as GABAergic KA′′ cells or secondary MN. This study identifies a critical role for Jagged–Notch signaling in the maintenance of proliferating neural precursors in a discrete compartment of the neural tube during the continuing growth and development of the vertebrate nervous system.
Keywords: neurogenesis, p3, progenitor, zebrafish, Jagged2
Molecular studies show that distinct subtypes of neurons are generated from different progenitors distributed along the dorsal–ventral axis of the spinal cord (1). The cellular diversity and complexity of neuronal organization in the CNS raises the question of how neuronal subtypes are generated at distinct locations and times by associating intrinsic factors with extrinsic signals during development (1–3). In the zebrafish embryo, distinct neurons, such as primary and secondary motor neurons (sMNs), GABAergic Kolmer–Agduhr (KA) cells, and Rohon–Beard (RB) sensory neurons emerge in a distinct spatiotemporal order from progenitors (4, 5). The MNs and GABAergic KA cells are generated from progenitors in discrete compartments of the ventral spinal cord that are likely to be analogous to the p2, pMN, and p3 progenitor zones described in other vertebrate model systems in refs. 1 and 6. To understand how specification of these neurons is regulated, we have examined how differentiation and proliferation of progenitors in discrete compartments of the ventral spinal cord is regulated by Notch signaling after a relatively early phase of “primary” neurogenesis.
Notch signaling inhibits neuronal differentiation in vertebrates and invertebrates during neurogenesis (3, 7–9) and suppresses oligodendrocyte development from oligodendrocyte precursors (OPC) during gliogenesis (10, 11). In the vertebrate CNS, Notch receptors and their ligands are expressed in proliferative zones of undifferentiated cells (12) and multiple notch genes are expressed throughout the developing zebrafish neural tube in all or most proliferative precursors (13). In zebrafish deltaA mutant embryos, neural precursors differentiate as early-born neurons, and the number of late-born neurons and glia is reduced (13). Jagged homologues have also been identified as ligands of Notch receptors (14). The concurrent knockdown of multiple Jagged homologues in zebrafish results in a phenotype that serves as a model for Alagille's syndrome in children (15). This study emphasized the role of Jagged–Notch signaling in liver development, and its function in the CNS development was not examined in detail. Although other studies have suggested Jagged functions in gliogenesis by inhibiting differentiation of OPC (10), its function in vertebrate neurogenesis remains poorly characterized.
We have investigated the role of Notch signaling in regulating the generation of late-born neurons, such as sMNs, and GABAergic KA′′ cells in the ventral spinal cord, where Jagged2 is expressed after an early wave of primary neurogenesis. We show that, as in early neurogenesis, Notch signaling continues to play a critical role in limiting the number of neurons that differentiate with different fates during late neurogenesis. Our study suggests that relatively uniform expression of Jagged2 in the dorsal–ventral compartment of the spinal cord, where MN progenitors are located, helps to activate Notch and maintain a population of proliferating progenitors in the most ventral part of the spinal cord. Furthermore, we suggest that, by preventing differentiation of proliferating progenitors for different lengths of time, Jagged2–Notch signaling may help in the diversification of cell fate and in continued growth of the spinal cord.
Results
The Number of Late-Born Neurons Is Limited by Jagged2-Mediated Signaling.
To understand the role that Jagged-mediated Notch signaling plays in neurogenesis, we compared expression of jagged1a, jagged1b, and jagged2 genes to the distribution of differentiating neurons. In the spinal cord, jagged2 transcripts are hard to detect until 14 h postfertilization (hpf) but they are easily detected at 28 hpf in the MN domain (Fig. 1 A and B). This domain is just dorsal to the most ventral part of the spinal cord where KA′′ cells, a subset of KA cells, differentiate. KA′ and KA′′ cells are distinguished by their location in the ventral spinal cord. The relatively dorsal KA′ cells are associated with olig2-expressing cells, identified as part of a fluorescent ventral population of spinal cord cells in Tg[olig2:egfp] embryos. The KA′′ cells are ventral to the fluorescent population identified in Tg[olig2:egfp] embryos (16). KA′ cells are located in the same dorsal–ventral compartment of the spinal cord as MNs and jagged2-expressing cells, whereas KA′′ cells are in a more ventral compartment, where jagged2 is not expressed (Fig. 1C). In contrast to relatively uniform jagged2 expression, jagged1b is expressed at high levels in a subset of ventral spinal cord cells at 28 hpf (Fig. 1D). It may also be expressed broadly at very low levels in more dorsal parts of the cord (data not shown). Expression of jagged1a is restricted to cells in a relatively dorsal compartment of spinal cord (data not shown).
Fig. 1.
Inhibition of Jagged2-mediated signaling causes an increase in the number of GABAergic KA′′ cells in the ventral spinal cord. (A–D) Expression of jagged2 and jagged1b in the ventral spinal cord at 28 hpf. Lateral views anterior to the left are shown. (A) Transcripts of jagged2 were detected in the ventral spinal cord. (B) Expression of jagged2 overlaps with ventral Islet1/2-positive (brown) MNs. (C) GABAergic KA′′ cells (green) are ventral to the jagged2 expression domain (red). (D) jagged1b is expressed in a subset of ventral cells. (E–H) Expression of islet2, islet1, and GAD67 in Con-MO- (E–H), J2-MO- (E′–H′) and J1b-MO- (E′′–H′′) injected embryos. Arrows indicate primary MNs, and arrowheads indicate KA′′ cells. (I–L) KA′′ cells are increased in J2-MO-injected embryos. Arrowheads identify GABAergic (red) KA′′ cells ventral to EGFP-expressing (green) cells in Con-MO- (I), J2-MO- (J), J1b-MO- (K), and J-MOs- (J2- and J1b-MO) (L) injected Tg[olig2:egfp] embryos. (M) KA′′ cells were counted between the 8th and the 10th somite. ∗∗∗, P < 0.001, n = 12. (N and O) Islet1/2-positive MNs and KA′′ cells are increased in the p3 domain of J2-MO-injected embryos at 28 hpf. White arrowheads identify GABAergic (red) KA′′ cells, and open arrowheads identify Islet1/2-positive (blue) MNs ventral to EGFP-expressing (green) cells in Con-MO- (N and N′) and J2-MO- (O and O′) injected Tg[olig2:egfp] embryos. (P and Q) Expression of zn5/DM-GRASP in Con-MO- (P and P′) and J2-MO- (Q and Q′) injected Tg[olig2:egfp] embryos at 30 hpf. Filled arrows identify sMNs (red) in the pMN domain, and open arrows identify ectopic sMNs in the p3 domain of J2-MO-injected Tg[olig2:egfp] embryos. Cross-sectional views of P and Q, at the position of the dashed lines, are displayed in p′ and q′, respectively. (Scale bars: in D for A–D, 25 μm; in H′′ for E–H′′, 25 μm; in L for I–L, 50 μm; in O′ for N and O, 25 μm; in Q′ for P–Q′, 25 μm.)
The generation of MNs and interneurons at distinct dorsal–ventral positions in the zebrafish spinal cord is reminiscent of the manner in which distinct types of neurons and glia are generated in a precise spatiotemporal manner from precursors arrayed along the dorsal–ventral axis of the neural tube in other vertebrates (6). On the basis of nomenclature used in other vertebrates and cell fate-specification studies in zebrafish, the olig2-expression domain where MN progenitors are located is referred to as the pMN domain. Dorsal to the pMN progenitors are the p2 progenitors, where V2 interneurons are generated. Ventral to the pMN progenitors are p3 progenitors, where interneurons in the most ventral domain of the spinal cord (V3 domain), including the KA′′ cells described above, are generated (6, 16, 17).
In the ventral spinal cord of the zebrafish embryo, primary MNs and sMNs, GABAergic KA′ and KA′′ cells, and GABAergic ventral longitudinal descending neurons (VeLDs) cells emerge in a distinct spatial and temporal order from progenitor cells (4, 5). To test whether Jagged-mediated signaling affects the specification of these ventral neurons, we attempted a loss-of-function study with morpholinos (MO) directed against jagged1b and jagged2 that are expressed in the ventral spinal cord.
jagged1b-MO (J1b-MO) and jagged2-MO (J2-MO), designed to interfere with splicing, led to the generation of alternative transcripts that were expected to be nonfunctional. Four base pair-mismatched MOs were used as controls, and these did not generate alternate transcripts [supporting information (SI) Fig. 6]. Increasing amounts of J1bATG-MO and J2ATG-MO, respectively (15), led to a specific reduction in the synthesis of Jagged1b and Jagged2 protein, (SI Fig. 6).
Morphologically, the neural tube and somites of embryos injected with MOs directed against jagged1b and jagged2 developed normally (15). Whole-mount in situ hybridization showed that, whereas primary MNs, marked by islet2 expression, developed normally at 21 hpf (Fig. 1 E and E′), an increase in the number of MNs was observed at 26 hpf in the J2-MO-injected embryo (Fig. 1 F and F′); the J1b-MO-injected embryo did not produce any obvious change (Fig. 1 E′′ and F′′). This result indicates that Jagged2 knockdown causes an increase in the number of MNs. A remarkable feature of some of the ectopic islet-positive cells is that they were located in the most ventral part of the spinal cord, suggesting that these ectopic MNs were located in the V3 rather than in the MN domain. Some cells in the V3 domains were also recognized by the zn5 antibody at 30 hpf (Fig. 1 P and Q), which labels a cell-surface protein, DM-GRASP, specifically present on sMNs and absent on primary MNs (18). This observation provided additional evidence that some of the ectopic neurons are MNs and furthermore it showed that they have specific characteristics of sMNs. The number of GABAergic neurons, identified by their expression of glutamic acid decarboxylase 67 (GAD67), also increased in the V3 domain in J2-MO-injected embryos (Fig. 1 H and H′). Immunohistochemistry with anti-GABA (Fig. 1 I–O) and anti-Islet1/2 antibody (Fig. 1 N and O) in Tg[olig2:egfp] embryos confirmed that the number of GABAergic cells and that of MNs increased in the p3 domain of J2-MO injected embryos. These GABAergic cells were likely to be KA cells. Furthermore, EGFP expression in the pMN domain of this transgenic line provided a strategy for determining specific changes in the number of GABAergic KA′′ interneurons located ventral to the EGFP-expression domain (Fig. 1 I–L). Although the number of KA′′ cells is variable, a significant increase (≈1.9 ± 0.2-fold, mean ± SD, P < 0.001) in their number between the 8th and 10th somite could be detected in the J2-MO-injected embryos compared with their number in Con-MO- or J1b-MO-injected embryos (Fig. 1M). The increase in the number of KA′′ cells, together with an increase in that of sMNs, indicated that Jagged2-mediated signaling limits the number of GABAergic neurons and sMNs.
Maintenance of jagged2 by Olig2 Is Necessary for Preventing Neural Progenitors from Premature Differentiation in the p3 Domain.
Examination of jagged2 expression with fluorescent whole-mount in situ hybridization in Tg[olig2:egfp] embryos shows that it is expressed in the EGFP-positive domain, where olig2 expressing precursors and their progeny are located (Fig. 2 A–C). Olig2 expressed in the pMN domain functions in the specification of MNs and in the promotion of pan-neuronal properties in vertebrates (17, 19). We hypothesized that Olig2 may also be required for jagged2 expression in the pMN domain, and through it, influence differentiation in the pMN domain and p3 domain. To test this hypothesis, we performed in situ hybridization and immunohistochemistry to examine the expression of jagged2, and to count the number of islet1-positive MNs and GABAergic interneurons in the ventral spinal cord of olig2-MO-injected embryos, respectively. MOs directed against olig2, shown in ref. 19 to be effective in knocking down Olig2 function, dramatically reduced the expression of jagged2 in this ventral domain of the spinal cord (Fig. 2 D and E). As expected, MNs decreased in the pMN domain, however, consistent with prior observations in J2-MO injected embryos, MNs could now be observed in the V3 domain (Fig. 3 F and G). Additionally, olig2-MO injected Tg[olig2:egfp] embryos had neither detectable EGFP nor GABA-expressing VeLD and KA′ cells in the ventral spinal cord at 26 hpf (Fig. 2 H and I). The number of KA′′ cells increased in the V3 domain (≈1.5 ± 0.1-fold, P < 0.001), whereas that of KA′ and VeLD cells decreased in the MN domain (≈0.1 ± 0.0-fold, P < 0.001) (Fig. 2J). These results suggest that Olig2 regulates jagged2 expression and/or the specification of cells that express this Notch ligand. They also indicate that differentiation of MNs and GABAergic neurons in the p3 domain is affected by the absence of a Notch ligand expressed in the relatively dorsal pMN domain. Furthermore, because KA′′ cells do not express EGFP in Tg[olig2:egfp] embryos, they are presumably not the progeny of Olig2-dependent precursors located at the pMN domain; rather, they are likely the progeny of neural precursors in the ventral-most p3 domain. The generation of these MNs and other cells, such as the KA′ and VeLD neurons, from both Olig2-expressing precursors in the pMN domain and precursors in the p3 domain, such as KA′′ neurons, is reminiscent of the manner in which distinct types of neurons are generated in a precise temporal and spatial manner from precursors arrayed along the dorsoventral axis of the neural tube during CNS development in other vertebrates as well (6).
Fig. 2.
Olig2 regulates jagged2 expression and its morphants display an increase in the number of MNs and KA′′ cells in the p3 domain. Lateral views anterior to the left are shown. (A–C) Expression of jagged2 in the pMN domain. Confocal images show EGFP-expressing cells (A) in Tg[olig2:egfp] embryos overlap with jagged2 transcripts (B) in a merged image (C). DAPI staining (blue) indicates nuclei in C. (D–G) Expression of jagged2 in the control- (D and E) and olig2-MO- (O2-MO) (F and G) injected embryo. E and F are higher magnifications of D and G, respectively. Upper brackets indicate the region expressing jagged2 (pMN domain), and lower brackets indicate the ventral region of spinal cord (p3 domain). (H and I) KA′′ cells in control- (H) and olig2-MO- (I) injected embryos at 26 hpf in Tg[olig2:egfp] embryos. Confocal images identify EGFP- (green) and GABA- (red) expressing cells. Arrowheads indicate KA′′ cells, and arrows identify GABAergic VeLD or KA′ cells. (J) Quantification of effects on GABAergic cells. KA′′, VeLD, and KA′ cells were counted between the 8th and the 10th somites. ∗∗∗, P < 0.001, n = 9. (Scale bar: in H for A–C, H, and I, 50 μm; in G for D and G, 100 μm.)
Fig. 3.
Inhibition of Notch signaling by XdnSu(H)myc at defined times reveals the period for selection of late-born MNs and GABAergic KA′′ cells. Lateral views anterior to the left are shown. GABA (A–E) and Islet1/2 (A′–E′) at 26 hpf after no heat shock (A and A′) or heat shock at 7 hpf (B and B′), 10 hpf (C and C′), 14 hpf (D and D′), and 17 hpf (E and E′) in Tg[hsp70:XdnSu(H)myc] embryos. (A′ and E′ Insets) High-magnification views of the merge image with A and E, respectively. Open arrowheads indicate MNs in the most ventral spinal cord. Filled arrowheads indicate KA′′ cells. (F and G), Quantification of effects on KA′′ cells. KA′′ cells (F), Islet1/2-expressing MNs, and RB sensory neurons (RB) (G) were counted between the 8th and the 10th somite (n = 9). The number of GABAergic neurons were counted in the ventral spinal cord of the 7-hpf heat shocked embryos, which was based on their location. (H–H′′) MNs at 48 hpf in wild-type embryo. Confocal images revealed Islet1/2-positive cells (green) and GABA-positive cells (red) in the ventral spinal cord. Open arrowheads indicate MNs in the most ventral spinal cord (H), and filled arrowheads indicate KA′′ cells (H′). (H′′) Merged image of H and H′. (Scale bars: in E for A–E and H–H′′, 25 μm; in E′ for A′–E′, 25 μm.)
Notch Signaling Continues to Play a Role in the Selection of Late-Born Neurons.
Previous studies have shown that Notch signaling within neurogenic domains limits the number of cells that are permitted to differentiate as early neurons; this allows progenitors to differentiate later and to adopt alternate fates or to remain as undifferentiated progenitors (13, 20). To examine how Notch signaling affects differentiation of GABAergic KA′′ cells, we analyzed KA′′ cells in the mind bomb (mib) mutant embryos that are characterized by a wide range of phenotypes consistent with a broad reduction of Notch signaling from early development (20). All mutant embryos showed reduction of GABAergic neurons in the ventral spinal cord (SI Fig. 7). The reduction of GABAergic KA′′ cells in mib mutants is consistent with them being late differentiating neurons, because early failure of Notch signaling in these mutants typically results in an excess of early differentiating neurons and a deficit of late differentiating neurons.
To define when cells are selected to become KA′′ cells, and to compare the time of their selection by Notch signaling to selection of RB sensory neurons and MNs, we used a transgenic line that expresses a dominant-negative form of Xenopus Suppressor of Hairless [Su(H)] under the transcriptional control of the zebrafish heat shock 70 promoter {Tg[hsp70:XdnSu(H)myc]} (21). XdnSu(H)myc lacks its DNA binding domain, and it prevents transcriptional activation of target genes by the Notch intracellular domain (22). Notch signaling was inhibited at defined times by exposing Tg[hsp70:XdnSu(H)myc] embryos to heat shock at 7, 10, 14, and 17 hpf. KA′′ cells, RB sensory neurons and MNs were visualized by labeling with anti-GABA and anti-Islet1/2 antibodies (Fig. 3). As in the mib mutants, in which there is an early loss of Notch signaling, heat shock at early gastrulation (7 hpf) decreased the number of ventral GABAergic neurons, including KA′′ cells (Fig. 3B). However, the number of KA′′ cells increased when the embryos were heat shocked between the end of gastrulation (10 hpf, ≈1.8 ± 0.5-fold, P < 0.01) and the 10-somite stage (14 hpf, ≈1.5 ± 0.5-fold, P < 0.05) (Fig. 3 C, D, and F), and their number remained unchanged in the embryos heat shocked at the 17-somite stage (17 hpf) (Fig. 3 E and F). In contrast, the number of RB sensory neurons increased after heat shock at 7, 10, and 14 hpf but not after heat shock at 17 hpf (7 hpf, ≈2.7 ± 0.3-fold, P < 0.001; 10 hpf, ≈2.5 ± 0.4-fold, P < 0.005; 14 hpf, ≈2.9 ± 0.4-fold, P < 0.001; 17 hpf, ≈1.3 ± 0.3-fold, P < 0.05) (Fig. 3 A′–E′ and G). Finally, MNs increased after heat shock at all four time points (7 hpf, ≈1.3 ± 0.3-fold, P < 0.1; 10 hpf, ≈1.4 ± 0.2-fold, P < 0.01; 14 hpf, ≈1.5 ± 0.2-fold, P < 0.001; 17 hpf, ≈1.6 ± 0.2-fold, P < 0.001) (Fig. 3 A′–E′ and G).
The increase in the number of KA′′ interneurons, RB sensory neurons, and MNs after interference with Notch signaling at specific times shows that Notch, when signaling normally, limits how many cells adopt each of these fates at specific times in development. Consistent with KA′′ cells being late differentiating neurons, early inhibition of Notch signaling with heat shock in Tg[hsp70:XdnSu(H)myc] at 7 hpf reduces the number of KA′′ cells, whereas application of heat shock between 10 and 14 hpf increases the number of KA′′ cells. In contrast to KA′′ cells, heat shock at 7 hpf causes an increase in the number of RB neurons, consistent with these primary neurons being selected in one of the earliest waves of differentiation (13, 20). Furthermore, the increase in the number of RB cells observed after heat shock as late as 14 hpf indicates that Notch signaling does not just limit differentiation of RB neurons during an early wave of differentiation but that precursors are actively prevented from adopting this fate as late as 14 hpf. Whereas changes in the number of primary MNs were not distinguished from changes in the number of sMNs, the increase in the total number of MNs, observed at all stages when Notch signaling was inhibited, suggests that Notch activation may limit the number of precursors that are permitted to become MNs at all developmental stages that were examined. It should be noted, however, that ectopic MNs could be observed in the most ventral spinal cord (V3 domain) at 26 hpf when the Tg[hsp70:XdnSu(H)myc] embryos were heat shocked at 17 hpf (Compare Fig. 3E′ Inset with 3A′ Inset). A similar appearance of a population of V3 MNs was observed after Jagged2 and Olig2 knockdown (Fig. 1F′ and G′ and Fig. 2G). In the wild-type embryo, however, Islet1/2-expressing neurons could only be seen in the most ventral spinal cord at 48 hpf, not as early as 26 hpf (Fig. 3 H–H′′). This finding suggests that loss of Jagged2 function or relatively late interference with Notch signaling is accompanied by the premature differentiation of this specific population of MNs in the V3 domain.
Jagged2-Mediated Notch Signaling Is Required to Maintain a Population of Proliferating Neural Precursors Both in the pMN Domain and in the p3 Domain.
We demonstrated that jagged2 expressed in the pMN domain is necessary for limiting the number of the GABAergic neurons generated in the ventral-most p3 domain, and that Notch signaling is required to limit the number of KA′′ cells after 10 and 14 hpf but before heat shock at 17 hpf becomes effective at interfering with Notch signaling in the Tg[hsp70:XdnSu(H)myc] embryos. Next, we wanted to examine when KA′′ cells are born and to determine whether ectopic KA′′ cells that differentiate when Jagged-mediated Notch signaling is impaired are produced by neural precursors born at the same time as other KA′′ cells or whether ectopic KA′′ cells are produced by aberrant differentiation of precursors born at a later stage.
To address when GABAergic KA′′ cells are generated, we exposed embryos to 10 mM BrdU at several stages and labeled them with anti-GABA antibody at 26 hpf (Fig. 4). All KA′′ cells were labeled with BrdU when Con-MO-injected embryos were exposed to BrdU at mid-gastrulation (9 hpf) (Fig. 4 A–C and A′–C′), whereas none of them were labeled with BrdU when the embryos were exposed to BrdU at the 14-somite stage (16 hpf) (Fig. 4 D–F and D′–F′). This finding implies that KA′′ cells are born from precursors that are still at S phase at 9 hpf; they are not normally produced by precursors that are still dividing and at S phase at 16 hpf. According to previous cell lineage analysis, deep cell clones divide synchronously until the end of gastrulation (cell cycle 16, ≈9 hpf), and the earliest differentiating primary neurons are born at cell cycle 16; sMNs and interneurons are born after cell cycle 17 (≈12 hpf to ≈18 hpf) and later divisions (23). Because KA′′ cell precursors are still in S phase at 9 hpf and have completed S phase by 16 hpf, they are likely to be generated during cell cycle 17 like many other secondary neurons. In the J2-MO-injected embryo, however, some GABA-positive KA′′ cells were labeled with BrdU when the embryos were exposed to BrdU at 16 hpf (Fig. 4 G–I and G′–I′). Such BrdU-positive KA′′ cells, thought to be ectopic, were located between other BrdU-negative KA′′ cells (Fig. 4 I and I′). The aberrant appearance of BrdU-positive KA′′ cells, sometimes adjacent to other BrdU-positive non-KA′′ cells, suggested that, after division of a neural progenitor at ≈16 hpf, one daughter had aberrantly differentiated into a KA′′ cell, instead of remaining undifferentiated, when Jagged2-mediated Notch signaling was impaired. The additional KA′′ cells could represent premature differentiation of a postmitotic population or the differentiation of neural progenitors that would otherwise have continued dividing. In the later case, the increase in the number of KA′′ cells in the embryos with impaired Jagged2-mediated Notch signaling would be accompanied by a reduction in the number of dividing neural precursors after ≈17 hpf.
Fig. 4.
BrdU-labeling assay reveals the period for selection of KA′′ cells. Lateral views anterior to the left are shown. (A–I) Confocal images identify GABA- (red) and BrdU-labeled nuclei (green) in Con-MO- (A–F) and J2-MO- (G–I) injected embryo at 26 hpf. Embryos were exposed to BrdU at 9 hpf (A–C) or at the 14 somite stage (14ss) (D–I). A′–C′, D′–F′, and G′–I′ show higher magnification views of the area outlined with a dashed rectangle in C, F, and I, respectively. KA′′ cell precursors in control embryos incorporate BrdU when exposed at 9 hpf but not at the 14-somite stage (A–F and A′–F′). (G–I) Ectopic KA′′ cells from precursors that divide after the 14ss. Arrowheads indicate BrdU-positive KA′′ cells. Whereas no BrdU-positive KA′′ cells were identified in Con-MO-injected embryos exposed late to BrdU at the 14ss (D–F), BrdU-positive KA′′ cells can be seen in J2-MO-injected embryos exposed to BrdU at the 14ss (G–I). (Scale bar: in I for A–I, 25 μm.)
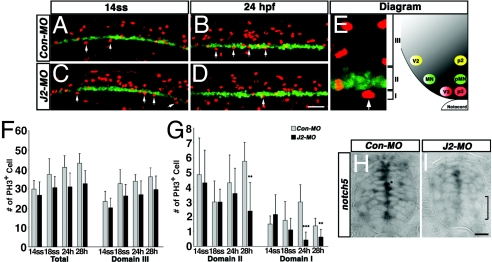
To determine whether dividing precursors are reduced after knockdown of jagged2 function, embryos were labeled at several stages, 14-so (16 hpf), 18-so (18 hpf), and 24 and 28 hpf (Fig. 5), with an antibody that recognizes phosphorylated histone H3 on Ser-10, an established marker for chromosome condensation during mitosis. Changes were scored in the whole width of the cord and specifically in domain I (p3 domain, ventral to the EGFP-positive cells in Tg[olig2:egfp] embryos), domain II (pMN domain, the EGFP-positive cells in Tg[olig2:egfp] embryos), and domain III (dorsal to the EGFP-positive cells in Tg[olig2:egfp] embryos) (Fig. 5E). The overall rate of cell division in all three domains (≈0.9 ± 0.2-fold, P < 0.001), and that, specifically in domain III (≈0.8 ± 0.2-fold, P < 0.01), decreased slightly but insignificantly after knockdown of Jagged2 (Fig. 5F). However, the rate of cell division significantly decreased both in domain II (pMN) at 28 hpf (≈0.4 ± 0.3-fold, P < 0.005), and in domain I (p3) at 24 hpf and 28 hpf (24 hpf, ≈0.1 ± 0.1 fold, P < 0.001; 28 hpf, ≈0.6 ± 0.1-fold, P < 0.005) (Fig. 5G). Moreover, transverse sections showed that the transcripts of notch5, which are normally expressed in proliferative precursors, were specifically diminished in the ventral spinal cord of J2-MO injected embryos (Fig. 5 H and I). These observations confirmed that Jagged2-mediated signaling is required to maintain a population of proliferating neural precursors both in the Olig2-positive pMN domain and in the most ventral p3 domain.
Fig. 5.
Inhibition of Jagged2-mediated Notch signaling causes a reduction in the number of the proliferative neural precursors in the ventral spinal cord. (A–D) Reduction in the number of mitotic cells labeled with anti-phospho-histone H3 (PH3) antibody. Lateral views anterior to the left are shown. Confocal images identify PH3-labeled nuclei (red) and EGFP (green) in Con-MO- (A and B) and J2-MO- (C and D) injected Tg[olig2:egfp] embryos at the 14 somite stage (14ss) (A and C) and at 24 hpf (B and D). Arrows indicate the PH3-positive cells in the p3 domain. (E) The diagram shows comparable zebrafish (Left) and mouse (Right) progenitor and neuronal domains. (F–G) Quantification of effects on mitotic cells. PH3-labeled nuclei were counted between the 8th and the 12th somite from 18ss, 24-hpf, and 28-hpf embryos, and between the fifth and the ninth somite of 14ss embryos in three different domains: domain I, ventral to the EGFP-expressing domain (p3); domain II, within the EGFP-expressing domain (pMN); and domain III, above the EGFP-expressing domain. ∗∗, P < 0.05 vs. Con-MO-injected embryos at 28 hpf, n = 8; ∗∗∗, P < 0.001 vs. Con-MO-injected embryos at 24 hpf, n = 8. (H and I) Expression of notch5. Transverse sections show that expression of notch5 was decreased in the ventral spinal cord of J2-MO-injected embryo (bracket in I) compared with Con-MO-injected embryo (H). (Scale bar: in D for A–D, 50 μm; in I for H and I, 25 μm.)
Discussion
Late-born neurons, including sMNs and some GABAergic neurons, have special requirements for their specification. Generation of these late-born neurons requires regulatory mechanisms that maintain neural progenitor cells in an uncommitted or undifferentiated state until appropriate times, when release from such mechanisms permits the progenitors to differentiate and adopt appropriate fates (24). Our results indicate that Jagged2-mediated Notch signaling is part of such a mechanism and plays an active role in the maintenance of dividing precursors in the ventral spinal cord until it is time for them to adopt specific fates.
Jagged2-Mediated Extrinsic Signal from the pMN to the p3 Domain Is Necessary to Maintain the Proliferating Neural Progenitor.
This study examined the role of Jagged2 in regulating neurogenesis in the ventral spinal cord and showed that Jagged2 knockdown results in an increase in sMNs and KA′′ cells, a subset of GABAergic KA cells that differentiate from the most ventral p3 progenitors. Previous studies have emphasized a role for Delta–Notch signaling in mediating competitive interactions that select cells that will differentiate as neurons from a pool of progenitors (3, 13, 20, 25, 26). In zebrafish the selected cells form a heterogeneous population and are characterized by relatively high levels of Delta expression. One role suggested for Jagged2, in preventing generation of premature sMNs or supernumerary KA′′ cells, is slightly different. The relatively uniform expression of Jagged2 in the pMN domain, activates Notch and prevents differentiation of progenitors in the more ventral p3 domain, where it maintains a proliferating progenitor population that is essential for generation of late-born neurons during growth and diversification of the nervous system.
Neurogenesis in the Most Ventral Spinal Cord.
Heat shock-induced expression of XdnSu(H)myc at 7 hpf reduced the number of KA′′ cells, whereas heat shock between 10 and 14 h increased the number of KA′′ cells, and heat shock at 17 hpf had little effect on the number of KA′′ cells. These observations suggested early Notch signaling maintains a population of undifferentiated progenitors that later becomes competent to differentiate as KA′′ cells. Later, when the p3 progenitors are competent to differentiate as KA′′ cells, Notch signaling limits the number of cells that adopt this fate. BrdU incorporation studies showed that KA′′ cells incorporate BrdU at 9 hpf but not at 16 hpf. This suggested that KA′′ cell progenitors are still dividing when most primary neurons are born in cell cycle 16 but are born soon after, like many other secondary neurons, at cell cycle 17. Whereas KA′′ cells in control embryos did not incorporate BrdU at 16 hpf, after Jagged2 knockdown, many KA′′ cells incorporated BrdU at 16 hpf. This suggested that many supernumerary KA′′ cells were not being specified from the same population of postmitotic p3 progenitors from which KA′′ cells are normally specified. Rather, p3 progenitors that have undergone an additional round of division, and would otherwise not have differentiated as KA′′ cells, aberrantly adopt this fate in the absence of Jagged2 function.
sMNs generated after Jagged2 knockdown also had some unusual features: Many ectopic MNs were observed in the most ventral, V3, domain after Jagged2 knockdown at 26 hpf, where MNs are only observed in the wild-type embryos at 48 h. Their early appearance within this domain at 26 hpf suggested p3 progenitors that would normally have remained undifferentiated were prematurely adopting this fate in the absence of Jagged2 function. The ectopic KA′′ cells and MNs in the V3 domain, together with the observation that the number of dividing cells is significantly reduced in the ventral spinal cord, suggested that many of these cells are likely generated by p3 progenitors that would have continued dividing in the presence of Jagged2 expression.
How Is V3 Cell Fate Determined by Jagged2 Expressed in the pMN Domain?
This study suggests Jagged2 knockdown allows p3 progenitors that are still dividing at 16 hpf to aberrantly differentiate as KA′′ cells, and it allows some p3 progenitors to prematurely differentiate as MNs in the V3 domain. How does Jagged2 expressed in the pMN domain influence the fate of p3 progenitors in a more ventral domain? The simplest possibility is that Jagged2 expressed in the pMN domain directly activates Notch in adjacent p3 progenitors. Notch activation at this stage prevents differentiation of the p3 progenitors, keeps them in a proliferative state and allows them to differentiate and adopt alternate fates later in development. However, we have not yet identified a Notch target gene expressed in the p3 domain that effectively or selectively reports Notch activation by Jagged2. Expression of a Notch target gene her4, for example, whose expression we have used to monitor Notch activity in ref. 27, is not expressed at high levels in the most ventral spinal cord at the stages we examined and there are no significant changes in its expression after knockdown of Jagged2 (SI Fig. 8). In the absence of such a reporter or experiments demonstrating a direct link between Jagged2 expression in the pMN domain and Notch activation in the p3 domain, it remains formally possible that Notch activation by Jagged2 in the pMN domain indirectly influences fate in the more ventral p3 domain by first inducing expression of a factor in the pMN domain that then influences fate in the more ventral p3 domain. It should, however, be noted that although the pMN and p3 domains are described as distinct progenitor domains, they nevertheless represent adjacent cell populations between which direct interactions mediated by Jagged2 and Notch remain plausible, and additional secreted factors need not be invoked to account for effects of Jagged2 on cell fate in a more ventral domain.
Temporal Regulation of Notch Signaling May Regulate Cell Diversity Fate in the Ventral Cord.
Knockdown of Jagged2 generates an aberrant population of late differentiating KA′′ cells and a prematurely differentiating population of sMNs in the V3 domain. However, inhibition of Notch signaling by heat shock-induced XdnSu(H)myc expression specifically increases KA′′ cells when heat shock is applied between 10 and 14 hpf, and premature sMN are induced only when heat shock is applied at 17 hpf. These observations suggest that, whereas activation of Notch by Jagged2 over a protracted period may normally prevent aberrant differentiation of progenitors as both KA′′ cells and sMNs, failure of Notch signaling leaves cells vulnerable to differentiate with distinct fates at different times during development. In a similar manner, physiological mechanisms that temporally regulate Notch signaling in progenitors could facilitate their differentiation with distinct fates during development. Although knockdown of Jagged2 causes premature differentiation of sMNs in the V3 domain, it is not obvious that the normal differentiation of sMNs in the V3 domain at 48 hpf is accompanied by reduced Jagged2 expression in the pMN domain. Future experiments will determine, however, whether differentiation of such late differentiating neurons is normally associated with intrinsic or extrinsic factors that attenuate the function or ability to respond to Notch ligands, like Jagged2, during development.
Although the conservation of signaling molecules reflects conserved functions of the Notch-signaling pathway during evolution and development, individual subtypes of Notch receptors and ligands, like Delta or Jagged, are likely to contribute in unique ways to building the overall neural architecture. Our results imply that Jagged2 signaling not only plays a role in regulating the number of neurons by preventing uncommitted precursors from acquiring a neuronal fate, it also plays an active role in vivo in the maintenance of dividing precursors that contribute to the increase in neurons during growth of the nervous system. Furthermore, Jagged2-mediated Notch signaling may also help diversify vertebrate neural cell fates by determining when progenitors stop dividing and differentiate, because differentiation at distinct times may facilitate acquisition of distinct fates.
Materials and Methods
Fish Lines and Mutants.
Zebrafish were maintained as described by Yeo et al. (28). AB* wild-type, mibta52b mutant (20), Tg[hsp70:XdnSu(H)myc] (21), and Tg[olig2:egfp] (16) were used.
Morpholino Injections.
Spliced jagged1b-MO (5′-AAATCAAGACTCACCGTCGTCCGCA-3′; Gene Tools, Philomath, OR), spliced jagged2-MO (5′-TCAGAGCTCTCACCTTCGTCCACAT-3′; Gene Tools), jagged1b mismatched control-MO (5′-AAATCtAGACTCtgCGTCGTCCGgA-3′; Gene Tools), jagged2 mismatched control-MO (5′-TCtGAGCTCTCtcCTTCGTCCAgAT-3′; Gene Tools), and jagged1b-MO (15), jagged2-MO (15), olig2-MO (Open Biosystems, Huntsville, AL) (2.5–4 ng) were injected into two-cell-stage zebrafish embryos as described in Yeo et al. (28). Embryos were studied at 14–28 hpf.
Whole-Mount in Situ Hybridization.
Whole-mount in situ hybridization was performed as described by Yeo et al. (28). Antisense riboprobes were transcribed from cDNA for zebrafish jagged1b, jagged2, islet1, islet2, GAD67, and notch5. Fluorescent in situ was performed as according to the manufacturer's protocol (Roche, Indianapolis, IN). Photos were taken with a differential interference contrast microscope (Axioplan2; Carl Zeiss, Oberkochen, Germany).
Immunohistochemistry.
Immunohistochemistry was performed as described by Yeo et al. (28). Primary antibodies used were as follows: anti-GABA antibody (Sigma, St. Louis, MO), anti-BrdU (Sigma), anti-phospho-histone H3 (Upstate, Lake Placid, NY), zn5/DM-GRASP, and anit-Islet1/2 (Developmental Studies Hybridoma Bank, Iowa City, IA). Secondary antibodies used were as follows: Alexa Fluor 488-conjugated goat anti-mouse, Alexa Fluor 488-conjugated goat anti-rabbit IgG, Alexa Fluor 405-conjugated goat anti-mouse IgG, and Alexa Fluor 568-conjugated goat anti-rabbit IgG (Molecular Probes, Eugene, OR). Photos were taken with a confocal laser scanning microscope (LSM 510; Carl Zeiss).
Supplementary Material
Acknowledgments
We thank B. Appel and J. Shin (both at Vanderbilt University, Nashville, TN) for providing essential reagents; Igor B. Dawid, M. J. Kim, and D. Kwon for critical reading of the manuscript; S. Chandrasekharappa, T. Oda, and C. H. Kim for contributions to the early stages of this work; and the members of the A.B.C. Laboratory. This work was supported by the Intramural Research Program of the National Institutes of Health/National Institute of Child Health and Human Development.
Abbreviations
- KA
Kolmer–Agduhr
- hpf
h postfertilization
- MN
motor neurons
- MO
morpholino
- RB
Rohon–Beard sensory neurons
- sMN
secondary MN
- Su(H)
Suppressor of Hairless
- V3 domain
ventral spinal cord domain
- VeLD
ventral longitudinal descending interneurons.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0607062104/DC1.
References
- 1.Briscoe J, Ericson J. Curr Opin Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 2.Jessell TM. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 3.Artavanis-Tsakonas S, Rand MD, Lake RJ. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 4.Bernhardt RR, Patel CK, Wilson SW, Kuwada JY. J Comp Neurol. 1992;326:263–272. doi: 10.1002/cne.903260208. [DOI] [PubMed] [Google Scholar]
- 5.Westerfield M, McMurray JV, Eisen JS. J Neurosci. 1986;6:2267–2277. doi: 10.1523/JNEUROSCI.06-08-02267.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Briscoe J, Pierani A, Jessell TM, Ericson J. Cell. 2000;101:435–445. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- 7.Fortini ME, Rebay I, Caron LA, Artavanis-Tsakonas S. Nature. 1993;365:555–557. doi: 10.1038/365555a0. [DOI] [PubMed] [Google Scholar]
- 8.Struhl G, Fitzgerald K, Greenwald I. Cell. 1993;74:331–345. doi: 10.1016/0092-8674(93)90424-o. [DOI] [PubMed] [Google Scholar]
- 9.Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- 10.Wang S, Sdrulla AD, diSibio G, Bush G, Nofziger D, Hicks C, Weinmaster G, Barres BA. Neuron. 1998;21:63–75. doi: 10.1016/s0896-6273(00)80515-2. [DOI] [PubMed] [Google Scholar]
- 11.Park HC, Appel B. Development (Cambridge, UK) 2003;130:3747–3755. doi: 10.1242/dev.00576. [DOI] [PubMed] [Google Scholar]
- 12.Lindsell CE, Boulter J, diSibio G, Gossler A, Weinmaster G. Mol Cell Neurosci. 1996;8:14–27. doi: 10.1006/mcne.1996.0040. [DOI] [PubMed] [Google Scholar]
- 13.Appel B, Givan LA, Eisen JS. BMC Dev Biol. 2001;1:13. doi: 10.1186/1471-213X-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindsell CE, Shawber CJ, Boulter J, Weinmaster G. Cell. 1995;80:909–917. doi: 10.1016/0092-8674(95)90294-5. [DOI] [PubMed] [Google Scholar]
- 15.Lorent K, Yeo SY, Oda T, Chandrasekharappa S, Chitnis A, Matthews RP, Pack M. Development (Cambridge, UK) 2004;131:5753–5766. doi: 10.1242/dev.01411. [DOI] [PubMed] [Google Scholar]
- 16.Park HC, Shin J, Appel B. Development (Cambridge, UK) 2004;131:5959–5969. doi: 10.1242/dev.01456. [DOI] [PubMed] [Google Scholar]
- 17.Novitch BG, Chen AI, Jessell TM. Neuron. 2001;31:773–789. doi: 10.1016/s0896-6273(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 18.Fashena D, Westerfield M. J Comp Neurol. 1999;406:415–424. doi: 10.1002/(sici)1096-9861(19990412)406:3<415::aid-cne9>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 19.Park HC, Mehta A, Richardson JS, Appel B. Dev Biol. 2002;248:356–368. doi: 10.1006/dbio.2002.0738. [DOI] [PubMed] [Google Scholar]
- 20.Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- 21.Latimer AJ, Shin J, Appel B. Dev Dyn. 2005;232:1098–1104. doi: 10.1002/dvdy.20264. [DOI] [PubMed] [Google Scholar]
- 22.Wettstein DA, Turner DA, Kintner C. Development (Cambridge, UK) 1995;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- 23.Kimmel CB, Warga RM, Kane DA. Development (Cambridge, UK) 1994;120:265–276. doi: 10.1242/dev.120.2.265. [DOI] [PubMed] [Google Scholar]
- 24.Anderson DJ. Neuron. 2001;30:19–35. doi: 10.1016/s0896-6273(01)00260-4. [DOI] [PubMed] [Google Scholar]
- 25.Skeath JB, Thor S. Curr Opin Neurobiol. 2003;13:8–15. doi: 10.1016/s0959-4388(03)00007-2. [DOI] [PubMed] [Google Scholar]
- 26.Riley BB, Chiang MY, Farmer L, Heck R. Development (Cambridge, UK) 1999;126:5669–5678. doi: 10.1242/dev.126.24.5669. [DOI] [PubMed] [Google Scholar]
- 27.Yeo SY, Kim M, Kim HS, Huh TL, Chitnis AB. Dev Biol. 2007;301:555–567. doi: 10.1016/j.ydbio.2006.10.020. [DOI] [PubMed] [Google Scholar]
- 28.Yeo SY, Little MH, Yamada T, Miyashita T, Halloran MC, Kuwada JY, Huh TL, Okamoto H. Dev Biol. 2001;230:1–17. doi: 10.1006/dbio.2000.0105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.