Biological processes occur in a temporal context, and the coordination of these processes with one another and with the external environmental cycle is critical for organismal fitness (1). Circadian rhythms, endogenous rhythms with periods of ≈24 h, have been described in almost all organisms, from bacteria to humans. An internal biological circadian clock controls many processes, notably the temporal expression of many genes. A substantial portion of the transcriptome is clock-regulated in fungi, plants, and animals (for examples, see refs. 2–4). In the most extreme example, Synechococcus elongatus PCC 7942, transcription rates of the entire genome are circadian-regulated (5). How might signals from a central oscillator be transduced into circadian transcription? In this issue of PNAS, Takai et al. (6) describe a histidine kinase/response regulator two-component module that transduces a signal emanating from the circadian clock to generate genome-wide oscillations in transcription.
In most systems studied to date, the circadian clock consists of a coupled transcription/translation feedback loop or loops (7). However, recent work has shown that S. elongatus provides a stunning exception to this generalization. Forward genetic analysis identified many mutations that affect clock function, such as period length, and these mutations were concentrated in a cluster of three kai (Japanese for cycle) genes (8). kaiB and kaiC orthologs are broadly conserved among cyanobacteria and are also found among nonphotosynthetic Eubacteria and Archaea (9). In S. elongatus, loss of function of any of the three kai genes results in arrhythmicity, whereas less severe mutations shorten or lengthen the period. kaiB and kaiC are cotranscribed in clock-regulated fashion, and kaiA transcription also cycles. Expression of these three genes is organized into a feedback loop, much like those that characterize clocks in multicellular eukaryotes (10). KaiC negatively regulates its own transcription, providing negative feedback, and KaiA enhances kaiBC expression, providing positive regulation. KaiB and KaiC protein levels also oscillate, although KaiA accumulates constitutively, and the three proteins assemble into a large complex termed the periodosome (Fig. 1). KaiC autophosphorylates, and this phosphorylation is enhanced by KaiA and negatively regulated by KaiB. The net phosphorylation state of KaiC oscillates with circadian period and is necessary for sustained rhythmicity; nonphosphorylatable KaiC assembles into a homohexamer but fails to assemble into the periodosome (11). Last year, Kondo and coworkers (12, 13) established that a temperature-compensated circadian rhythm was reconstituted in vitro in a simple mixture of three Kai proteins and ATP. Evidently, posttranslational modifications of purified Kai proteins that are catalyzed by those proteins themselves are sufficient to generate a sustainable circadian rhythm. Thus, the S. elongatus clock is fundamentally distinct from coupled transcription/translation based clocks described in multicellular organisms (7).
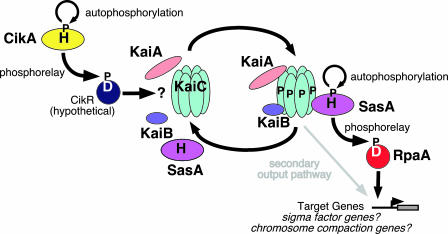
Fig. 1.
Environmental input is transmitted to the periodosome via the His kinase CikA and possibly an as-yet-unidentified cognate response regulator, CikR, or perhaps directly through a response regulator-like domain at the N terminus of KaiA. This stimulates the autophosphorylation of KaiC. SasA autophosphorylation is stimulated by phosphorylated KaiC, initiating the phosphorelay to its cognate response regulator, RpaA. Phosphorylated RpaA activates target genes that remain undefined.
If the Kai proteins themselves are sufficient to constitute an oscillator, what are the input mechanisms that synchronize the clock with the environment and what are the output pathways that confer temporal control on clock-controlled behaviors, such as organism-wide gene expression? Two-component signaling pathways play roles in both input to and output from the S. elongatus clock. Two-component signaling modules are commonly encountered in bacteria (and plants and fungi) and typically consist of a membrane-associated sensor histidine kinase that monitors an environmental signal (14). Upon stimulation, the kinase autophosphorylates on a His residue. The phosphate group is then transferred to an acceptor Asp residue in the receiver domain of a response regulator, thereby activating its transcriptional activator function, for example.
Forward genetic analysis identified the circadian input kinase (cikA) gene, which encodes an atypical sensor histidine kinase necessary for wild-type phase-resetting in response to dark or temperature pulses (10). CikA encodes a pseudoreceiver domain that negatively regulates histidine kinase activity; a cognate response regulator is anticipated but remains to be identified (10). Alternatively, CikA may directly target a response regulator-like domain at the N terminus of KaiA.
Yeast two-hybrid screening identified SasA, a sensory histidine kinase, as interacting physically with KaiC via a SasA domain that resembles a domain found in KaiB and responsible for its association with KaiC (15). SasA protein accumulates constitutively and assembles into the periodosome in vivo (16). Loss of SasA function greatly reduces the amplitude of oscillations of clock-controlled genes, often to arrhythmia (15), implicating it in circadian output. However, SasA’s cognate response regulator was not known.
Takai et al. (6) systematically disrupt 22 of 24 response regulator genes in the S. elongatus genome and show that disruption of one, rpaA, yields an arrhythmic transcription phenotype similar to that resulting from loss of SasA function. rpaA disruption dramatically reduces kaiBC promoter activity and mRNA and protein accumulation and results in arrhythmia in continuous light, much like the sasA-null phenotype. In wild type, there is a circadian rhythm in the state of KaiC phosphorylation. This rhythm is lost in either sasA or rpaA null mutants, and accumulated KaiC protein is phosphorylated constitutively.
The similarity in loss of function phenotypes associated with SasA and RpaA strongly suggests that they may form a two-component phosphorelay signaling module. Initial attempts to demonstrate phosphotransfer were unsuccessful. SasA autophosphorylates in vitro, consistent with earlier observations, and the addition of RpaA attenuates SasA autophosphorylation, although no phosphorylation of RpaA could be detected. However, in typical two-component modules, the kinase both phosphorylates and dephosphorylates the response regulator (14). It is possible that RpaA was phosphorylated and rapidly dephosphorylated by SasA in a futile cycle. Addition of KaiC stimulates SasA autophosphorylation and allows accumulation of phosphorylated RpaA. Does the phosphorylation status of KaiC, which oscillates in circadian fashion, modulate SasA and RpaA phosphorylation? This possibility was tested by collecting reaction mixtures of the three Kai proteins and ATP, which recapitulate a circadian rhythm in KaiC phosphorylation, at 4-h intervals and performing in vitro SasA–RpaA phosphorylation/phosphotransfer assays in the presence of each Kai protein mixture. The degree of SasA autophosphorylation and of RpaA phosphorylation is modulated in circadian fashion by the ratio of phosphorylated to non-phosphorylated KaiC and is maximal at the circadian phase when KaiC phosphorylation is increasing (although phase-leading relative to the peak in KaiC phosphorylation).
Circadian transcription of many genes is severely attenuated in mutants compromised in either SasA or RpaA function, although the effect is more severe in rpaA than in sasA null mutants. The latter retain transcription activity rhythms at lower light levels, although the phase angle difference between dawn- (e.g., purF) and dusk- (e.g., kaiBC) peaking genes is lost. Thus, some output pathway activity remains in sasA null mutants. It is possible that RpaA retains some activity in the absence of phosphorylation, or perhaps another histidine kinase can partially substitute for SasA signaling to RpaA. Signal cross-talk among two-component signaling modules is not well described. The almost complete arrhythmia of rpaA null mutants suggests that RpaA is necessary for almost all rhythmic promoter activity. However, residual rhythmicity can be detected in dim light in both sasA and rpaA null mutants. Thus, there are likely to be other minor output pathways.
Earlier work has demonstrated that clock function is adaptive for growth in diurnal cycles (1). Takai et al. (6) confirm and extend this observation by showing that rpaA null mutants, like sasA null mutants (15), grow as well as wild type under continuous light conditions but much more slowly than wild type under light–dark cycles.
These results demonstrate that the SasA–RpaA two-component module is the primary output pathway linking the Kai protein oscillator to circadian transcription with proper period and phase-relationship among genes and is essential for the maintenance of a robust oscillation in the state of KaiC phosphorylation in vivo. KaiC autophosphorylation itself is a critical modulator transmitting clock phase to the output module via its stimulation of SasA autophosphorylation and phosphotransfer or possibly through repression of the dephosphorylation activity.
Puzzles remain: RpaA failed to interact with the kaiBC promoter in gel-shift experiments. Presumably, one class of RpaA target genes includes master transcription regulators, perhaps including sigma factors, and the effect on rhythmic kaiBC transcription is indirect. Other potential targets would be factors that regulate chromosome compaction, which is itself rhythmic (17). Nonetheless, this work fills a significant gap in our understanding of the cyanobacterial system and suggests obvious directions for further studies, with the identification of RpaA targets a critical next step.
Footnotes
Conflict of interest statement: No conflicts declared.
See companion article on page 12109.
References
- 1.Johnson C. H. Methods Enzymol. 2005;393:818–837. doi: 10.1016/S0076-6879(05)93043-7. [DOI] [PubMed] [Google Scholar]
- 2.Nowrousian M., Duffield G. E., Loros J. J., Dunlap J. C. Genetics. 2003;164:923–933. doi: 10.1093/genetics/164.3.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harmer S. L., Hogenesch J. B., Straume M., Chang H. S., Han B., Zhu T., Wang X., Kreps J. A., Kay S. A. Science. 2000;290:2110–2113. doi: 10.1126/science.290.5499.2110. [DOI] [PubMed] [Google Scholar]
- 4.Panda S., Antoch M. P., Miller B. H., Su A. I., Schook A. B., Straume M., Schultz P. G., Kay S. A., Takahashi J. S., Hogenesch J. G. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu Y., Tsinoremas N. F., Johnson C. H., Golden S. S., Ishiura M., Kondo T. Genes Dev. 1995;9:1469–1478. doi: 10.1101/gad.9.12.1469. [DOI] [PubMed] [Google Scholar]
- 6.N. Takai, M. Nakajima, T. Oyama, R. Kito, C. Sugita, M. Sugita, T. Kondo, H. Iwasaki. Proc. Natl. Acad. Sci. USA. 2006;103:12109–12114. [Google Scholar]
- 7.Bell-Pedersen D., Cassone V. M., Earnest D. J., Golden S. S., Hardin P. E., Thomas T. L., Zoran M. J. Nat. Rev. Genet. 2005;6:544–556. doi: 10.1038/nrg1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kondo T., Tsinoremas N. F., Golden S. S., Johnson C. H., Kutsuna S., Ishiura M. Science. 1994;266:1233–1236. doi: 10.1126/science.7973706. [DOI] [PubMed] [Google Scholar]
- 9.Dvornyk V., Vinogradova O., Nevo E. Proc. Natl. Acad. Sci. USA. 2003;100:2495–2500. doi: 10.1073/pnas.0130099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Golden S. S., Canales S. R. Nat. Rev. Microbiol. 2004;1:181–190. doi: 10.1038/nrmicro774. [DOI] [PubMed] [Google Scholar]
- 11.T. Nishiwaki, Y. Satomi, M. Nakajima, C. Lee, R. Kiyohara, H. Kageyama, Y. Kitayama, M. Temamoto, A. Yamaguchi, A. Hijikata, et al. Proc. Natl. Acad. Sci. USA. 2004;101:13927–13932. [Google Scholar]
- 12.Tomita J., Nakajima M., Kondo T., Iwasaki H. Science. 2005;307:251–253. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 13.Nakajima M., Imai K., Ito H., Nishiwaki T., Murayama Y., Iwasaki H., Oyama T., Kondo T. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 14.Stock A. M., Robinson V. L., Goudreau P. N. Annu. Rev. Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 15.Iwasaki H., Williams S. B., Kitayama Y., Ishiura M., Golden S. S., Kondo T. Cell. 2000;101:223–233. doi: 10.1016/S0092-8674(00)80832-6. [DOI] [PubMed] [Google Scholar]
- 16.Kageyama H., Kondo T., Iwasaki H. J. Biol. Chem. 2003;278:2388–2395. doi: 10.1074/jbc.M208899200. [DOI] [PubMed] [Google Scholar]
- 17.Mori T., Johnson C. H. Semin. Cell Dev. Biol. 2001;12:271–278. doi: 10.1006/scdb.2001.0254. [DOI] [PubMed] [Google Scholar]