Abstract
The nonpathogenic human virus adeno-associated virus type 2 (AAV) has evolved the potentially unique strategy to establish latency by site-specifically integrating its genome into human chromosome 19 (19q13.3-qter) at a locus designated AAVS1. This nonhomologous, site-specific recombination of viral DNA with the human genome provides a basis for developing targeted gene therapy vectors. To assess whether the region surrounding AAVS1 might have contributed to the selection of the specific integration site, we have investigated this locus. Here, we show that AAVS1 is closely linked to the slow skeletal troponin T gene, TNNT1, which has been mapped previously to 19q13.4. In support of this idea, we demonstrate that site-specific AAV DNA integration can result in the formation of TNNT1-AAV junctions. The question now arises whether muscle represents a natural target tissue for latent AAV infection. This possibility is of additional interest in view of recent observations that muscle tissue is particularly well suited for AAV-mediated gene transfer. The question also occurs whether latent infection by AAV can lead to phenotypic changes of the multinucleated muscle fiber cells.
The human parvovirus adeno-associated virus type 2 (AAV) has adopted the strategy of alternate life styles to ensure a successful relationship with its host (1). This nonpathogenic virus productively replicates only in the presence of helper virus co- or superinfection (2). In the absence of such helper conditions, AAV has evolved a strategy potentially unique among eukaryotic viruses. On infection of the non-coinfected host cell, AAV establishes latency by integrating its genome site-specifically into a region on the q arm of human chromosome 19 (19q13.3-qter) (3, 4), designated AAVS1.
The 4.7-kb linear single-stranded genome contains two ORFs and is flanked by inverted terminal repeats (ITRs) (5). The right ORF encodes three capsid proteins, and the left ORF encodes the four nonstructural proteins (Rep proteins) that are involved in regulating all aspects of the viral life style (2).
The 145-nt ITRs are the only viral sequences required in cis for DNA replication, packaging of the viral genome into the capsid, and site-specific DNA integration. Within the ITRs, a Rep binding site (RBS) allows for specific recruitment of the large Rep proteins (i.e., Rep68 and Rep78) to the origin of replication (6–8). A Rep-specific endonuclease site (terminal resolution site, TRS) is separated from the RBS by a 13-nt spacer (9–11). Together, RBS and TRS can act as a minimal origin for Rep-mediated DNA replication (12).
To dissect the requirements for site specificity in targeted AAV DNA integration, an 8.2-kb fragment of AAVS1 has been isolated from embryonic lung fibroblast (WI38) DNA, and a 4-kb sequence was determined (13). It was shown by using an episomal model system that site specificity is determined by cellular sequences (14) and that a 33-nt sequence is necessary and sufficient for this targeted nonhomologous recombination event to occur (15). This 33-nt chromosomal sequence is similar to the minimal viral origin of DNA replication, consisting of an RBS and a TRS (16), leading to the concept that Rep-mediated DNA replication is involved in the integration mechanism. Complementing this idea was the observation that the viral Rep proteins are required for site-specific integration (17, 18). Biochemical assays further showed that the Rep proteins can specifically interact with the viral and cellular RBS and TRS motifs (8, 19, 20) to mediate replication and potentially targeted integration of AAV into AAVS1 (16, 21). Through isolation and analysis of several viral–cellular junctions, it became apparent that, although all of the junctions contained AAVS1 sequences, the immediate transitions from viral to cellular sequences were scattered over a range of approximately 1,000 nucleotides downstream of the TRS-RBS motif within AAVS1 (4, 22, 23). These observations are in agreement with the hypothesis that limited cellular DNA replication is involved in the initial steps of the mechanism underlying AAV site-specific integration. This model predicts that Rep-mediated replication is using the RBS and TRS motifs present on AAVS1 as a minimal origin that is potentially unique within the human genome (16).
Several questions remain with respect to the requirements for the chromosomal target sequence. It has not been determined whether active transcription in the vicinity of AAVS1 promotes integration by providing access to the target sequence for the proteins involved in the recombination mechanism. A second question is whether AAV integration could lead to the disruption of an active cellular gene. In most of the previous studies, DNA fragments containing only one viral–cellular junction had been isolated, leaving open the position and characteristics of additional junctions.
In this report, we present our analysis of the region surrounding AAVS1. We show that the target sequence for AAV site-specific integration is closely linked to muscle-specific genes TNNT1 and TNNI3. In support of this finding, we demonstrate that site-specific AAV DNA integration can result in the formation of TNNT1-AAV junctions.
Materials and Methods
Cell Lines.
The 293 cell line, the latently infected KB cell lines M19 (24) and M21 (24), the latently infected HeLa cell line HA16C5 (25), and the latently infected Detroit 6 cell line 7374 (26) were maintained in DMEM supplemented with 10% FBS.
Libraries.
Similar strategies were used to construct the HA16C5 (latently infected) and a human lymphocyte (uninfected) genomic library. The genomic DNA was partially digested with Sau3A and cloned into the BamHI site of the λDASHII vector (Stratagene). The HA16C5 and lymphocyte libraries were screened with an AAV and an AAVS1 probe, respectively. From each library, several phages positive for either AAVS1 (for the uninfected lymphocyte library) or AAV hybridization (for the HA16C5 library) were isolated, digested with NotI, and subcloned into the pBluescript vector. The plasmids were grown at 30°C in SURE bacteria (Stratagene).
Plasmid pHA16–3 was isolated from the HA16C5 library and pS1 from the lymphocyte library. The pS1 plasmid contains a 14-kb insert starting at position 2083 in AAVS1.
Plasmids, Probes, and Cosmids.
pBluescript was used as a cloning vector for the following subcloning steps (exceptions are indicated).
The AAVS1pRVK plasmid (nucleotides 1–3536) was obtained by EcoRI/KpnI digestion of the pRIA plasmid (13). The pS1 plasmid was digested with PstI to generate the AAVS1pPs plasmid (nucleotides 1438 to −2953 relative to AAVS1). AAVS1pPs was digested with DraI/PvuII to generate plasmid AAVS1pPD (nucleotides −1723 to −1970 relative to AAVS1).
The expressed sequence tag (EST) clone AA176454, which is homologous to the TNNT1 mRNA M1 clone (27), was purchased from the American Type Culture Collection (catalog no. 769879) and used to generate the TNNT1 cDNA probes. This clone was digested with XhoI-BamHI to generate the 5′TNNT1 plasmid (nucleotides 56–663) (27) and the 3′TNNT1 plasmid (nucleotides 663–896) (27). The TNNT1ex11–13 insert (nucleotides 562–754) (27) was generated by PCR on the AA176454 clone with the primers RML139 (nucleotides 562–580) (27) and RML140 (nucleotides 733–754) (27), and the PCR product was subsequently cloned into the PCR2.1 vector (Invitrogen). The cosmid c23932 was digested with StuI-BglII to generate the TNNI3ex7 plasmid (nucleotides 4653–4877). All of the inserts of these clones were sequenced.
The cosmids used for this study were generously provided by the Human Genome Center at Lawrence Livermore National Laboratory (LLNL).
The TNNT1- and AAVS1-containing EcoRI fragments of cosmid 31925 were subcloned into pBluescript, and terminal sequences were determined for each fragment by using T3 and T7 primers.
Southern Analysis.
Human genomic DNA was extracted from peripheral blood by using the QIAmp blood DNA kit (Qiagen, Chatsworth, CA). Genomic DNA from cell lines was isolated by standard procedures. Genomic DNA (10 μg) was digested, separated by gel electrophoresis on a 0.8% agarose gel, transferred onto a nylon membrane, and probed with a radiolabeled [α-32P]dCTP probe. Probes are indicated in the figure legends. The membranes were hybridized overnight at 65°C in a 0.75× nylon buffer (1× = 14% SDS/80 mM Na2HPO4/14 mM Na2H2EDTA⋅2H2O) and washed for 20 min each at 65°C in 0.5×, 0.1×, and 0.05× nylon buffer.
GenBank Accession Numbers.
AAV2, AF043303; AAVS1, S51329; cosmid 31855, AC005782; TNNT1 M1 mRNA clone, M19308; TNNT1 gene exons 1–11, AJ011712; TNNT1 gene exons 12–14, AJ011713; TNNI3 gene, X90780.1.
Results and Discussion
To determine whether AAV integration can lead to the disruption of a functional gene closely linked to AAVS1, a genomic library of HeLa cells latently infected with AAV (HA16C5) was screened with an AAV probe, and junction sequences were analyzed. All of the plasmids positive for an AAV-specific probe contained AAVS1 junction sequences (data not shown). One plasmid contained a fragment with a second junction between AAV and cellular DNA in addition to the AAV-AAVS1 junction. Sequence analysis revealed that this cellular fragment was derived from the TNNT1 gene, suggesting the possibility that AAVS1 and TNNT1 are linked. This possibility is in agreement with the previously mapped position of TNNT1 at 19q13.4 (28), which is included within the region reported for AAVS1 (3, 4).
Characterization of the Region Surrounding AAVS1.
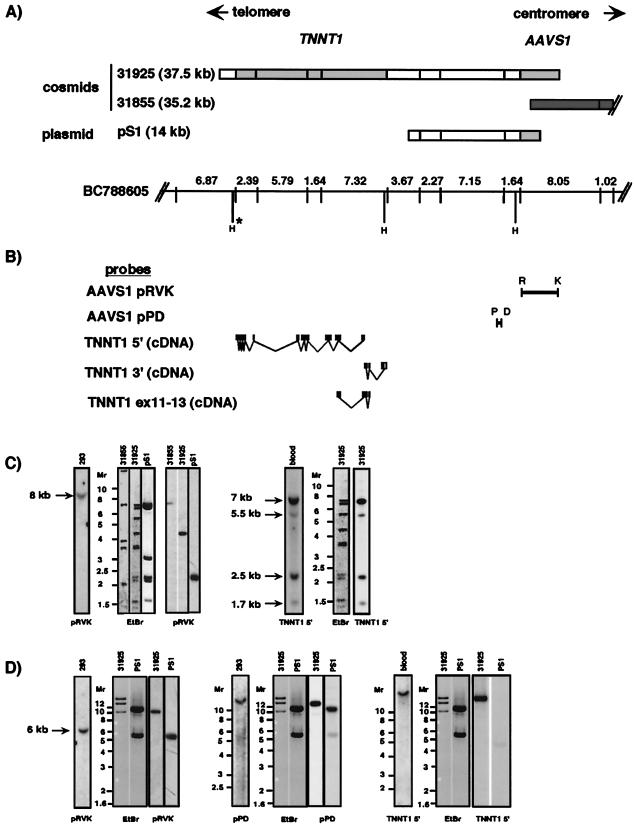
Although the presence of AAV-AAVS1 and AAV-TNNT1 junctions within the same fragment was indicative for colocalization of AAVS1 with TNNT1, the possibilities could not be excluded that both sequences were brought to close vicinity either by cellular rearrangements inherent to the transformed HeLa cell line or by rearrangements induced by AAV integration. To test the hypothesis that AAVS1 and TNNT1 are linked, several AAVS1 probes and TNNT1 cDNA probes (Fig. 1) were used for Southern analyses of nine cosmids that had been preselected for the presence of AAVS1. These cosmids were derived from a library from flow-sorted chromosomes of a human–hamster hybrid, which carries chromosome 19 as its only human chromosome (constructed by the Human Genome Center at LLNL). In addition, the plasmid pS1, which was derived from a different, independently generated genomic library was used in these analyses. The cosmids and pS1 DNA were digested with EcoRI, and the resulting fragments were hybridized to the probes indicated in Fig. 1B. The resulting hybridization patterns were compared with those derived from genomic DNA isolated from both human peripheral blood and HEK293 cells. The analysis of genomic DNA was added to exclude rearrangements of cosmid sequences. Cosmid 31855 (insert size 35.2 kb) (Fig. 1A) extended from AAVS1 into the centromeric region. During the course of our studies, the complete sequence of cosmid 31855 was determined by the LLNL. Sequence comparison of AAVS1 with cosmid 31855 clearly linked AAVS1 to the EcoRI map graphically displayed in Fig. 1A. Note that, to date, within the 35.2 kb contained within this cosmid, no known cellular gene is found. Of eight cosmids extending into the telomeric direction of AAVS1, two cosmids hybridized to TNNT1- and AAVS1-specific probes (cosmids 31925, insert size: 37.5 kb; and 28847, insert size: 37 kb). However, cosmid 28847 had undergone rearrangements as determined by restriction patterns inconsistent with the respective EcoRI map (Fig. 1A, BC788605) provided by the LLNL and was therefore omitted (data not shown). EcoRI fragments of the inserts from cosmid 31925 and plasmid pS1 were subcloned, and terminal sequences were determined. These data allowed positioning of the inserts with respect to the previously published AAVS1 sequence (data not shown) (13). Consistent hybridization patterns were observed with cosmid 31925 and genomic DNA with the AAVS1 and TNNT1 probes (Fig. 1C). Note that the hybridization pattern of cosmid 31925 and plasmid pS1 with the AAVS1 probe is different from genomic DNA. As seen in Fig. 1A, cosmid 31925 contains only 4.5 kb of the 8-kb EcoRI fragment of AAVS1 in addition to the full TNNT1 gene and pS1 contains ca. 2 kb of the AAVS1 EcoRI fragment. In summary, our data support a close linkage between AAVS1 and TNNT1.
Figure 1.
AAVS1 and TNNT1 are linked. (A) EcoRI maps of two cosmids (31925 and 31855), a plasmid isolated from a genomic library (pS1), and a bacterial artificial chromosome (BC788605) are shown. The cosmids were preselected for containing AAVS1. pS1 was selected based on hybridization to AAVS1 probes. Gray boxes indicate EcoRI fragments for which independent DNA sequences are available [AAVS1, accession no. S51329, Kotin et al. (13); TNNT1, accession nos. AJ011712 and AJ011713]. The dark gray box indicates cosmid 31855. The DNA sequence of this cosmid has been made available by the human genome center at LLNL (AC005782) and is in agreement with the published AAVS1 sequence (S51329). The first 8 kb of this cosmid are graphically displayed. The sequence starts in AAVS1 at nucleotide 991 and extends for 35 kb into the centromeric region. The cosmids were digested with EcoRI and hybridized to probes indicated in the bottom part. The EcoRI fragments were aligned with help of the map of BC788605 (URL: http://www-bio.llnl.gov). In BC788605, the EcoRI sites are indicated by vertical lines, and the distances between them are shown in kb. In addition, HindIII (H) sites are shown. The positions of the HindIII sites are deduced from published sequences (for TNNT1 and AAVS1) and from sequence data generated for this study (for AAVS1). H*: this HindIII site was deduced from the published cardiac troponin I gene (TNNI3) sequences (30, 34). The HindIII site within TNNI3 was included based on the recent observation by Barton et al. (30) that the TNNT1 gene is closely linked to the TNNI3 gene. (B) Hybridization probes used for this study. AAVS1 probes: AAVS1pRVK: EcoRI-KpnI (nucleotides 1–3536 in S51329) and AAVS1pPD: DraI-PvuII (nucleotides −1723 to −1970 relative to S51329). TNNT1 probes: TNNT1 5′, nucleotides 56–663 (27); TNNT1 3′, nucleotides 663–896 (27); TNNT1ex11–13, nucleotides 562–754 (27). (C and D) Genomic DNA isolated from blood and from HEK293 cells (293) and cosmid DNA (cosmids 31925, 31855) as well as plasmid pS1 DNA was digested with either EcoRI (Fig. 1C) or HindIII (D) and separated by gel electrophoresis. Southern analysis was performed by using AAVS1 (pRVK and pPD) and TNNT1 (TNNT1 5′) probes. Ethidium bromide (EtBr) stainings of the gels before hybridization are shown where indicated. Molecular weights (Mr) are indicated in kb. Note, the genomic and cosmid DNA samples were run on separate gels to avoid cross-contaminations. The sizes of the genomic bands were determined from these independent Southern blots and are indicated in kb. The hybridization patterns were consistent with the EcoRI and HindIII maps shown in A.
The LLNL EcoRI map (Fig. 1A; BC788605) was used to organize the fragments derived from our restriction and Southern analyses of the cosmids, pS1, and genomic DNA, resulting in an estimated distance between AAVS1 and TNNT1 of 14.7 kb. To confirm this distance between TNNT1 and AAVS1, cosmid 31925, plasmid pS1, and genomic DNA were digested with HindIII and hybridized to TNNT1 and to AAVS1 probes (Fig. 1D). As evident from the hybridization patterns shown in Fig. 1D, the distances deduced from the HindIII digests are consistent with those from the EcoRI digests, and we therefore conclude that AAVS1 and TNNT1 are separated by 14.7 kb.
TNNT1 Rearrangements in Latently Infected Cells.
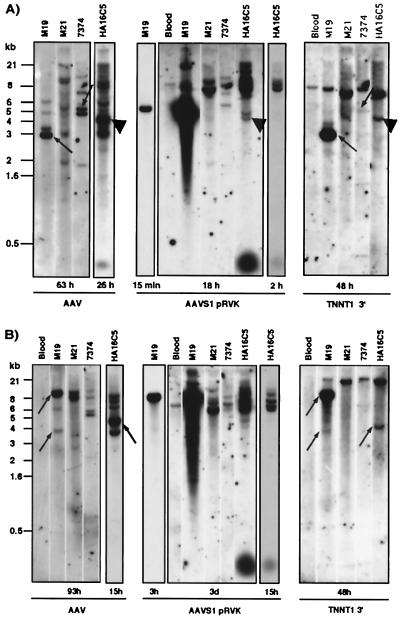
The proximity of AAVS1 and TNNT1 led us to examine several cell lines for evidence that site-specific integration of AAV DNA into AAVS1 could lead to the disruption of the TNNT1 gene. Southern blots were performed on genomic DNA isolated from several latently infected cell lines, the Detroit 6 (D6) line 7374 (26), the KB lines M19 (24), M21 (24), and the HeLa line HA16C5 (25). These cell lines have previously been characterized as containing at least one copy of AAV DNA integrated into AAVS1 (refs. 23 and 29, and unpublished observations). As shown in Fig. 2, the 8-kb and 6-kb bands were detected after EcoRI and HindIII digestion, respectively, followed by hybridization to the pRVK probe. These bands correspond to the undisrupted AAVS1 locus, apparently present in all of the latently infected cells as well as in the control DNA isolated from peripheral blood (4, 13, 23, 29) (Fig. 2 A and B). As previously shown, only the infected cells showed supernumerous bands with an AAVS1 probe (4, 23, 29); some of these bands cohybridized with an AAV probe, indicating a possible AAV-AAVS1 junction fragment (Fig. 2 A and B). Hybridization of the same DNA to a 3′TNNT1 probe resulted in the detection of the undisrupted 7-kb (EcoRI, Fig. 2A) and 23-kb (HindIII, Fig. 2B) fragments (27, 30). After an EcoRI and a HindIII digestion, the M19 and HA16C5 cells showed additional bands, indicating a possible disruption of the TNNT1 gene (Fig. 2 A and B); some of these bands cohybridize with an AAV probe, suggesting linkage between AAV DNA and the TNNT1 gene. Moreover, in the HA16C5 DNA, one band cohybridized with the AAV, AAVS1, and 3′TNNT1 probes (Fig. 2A). These data indicate that integration of AAV into the AAVS1 locus can lead to rearrangements of the TNNT1 gene.
Figure 2.
wtAAV2 DNA integration can lead to the disruption of both AAVS1 and TNNT1. Southern blots of cellular DNA (ca. 10 μg) isolated from blood (negative control) and from cells latently infected with AAV (M19, M21, 7374, HA16) are shown. The genomic DNA was digested with either EcoRI (A) or HindIII (B). The blot was first hybridized to the 3′TNNT1 cDNA probe followed by hybridization to an AAV probe and subsequently to AAVS1pRVK. Between hybridizations, the blot was stripped (50% formamide, 2 h, 65°C) and the membrane was exposed to a PhosphorImager screen for 12 h to quantify remaining label. The exposure times are indicated for each blot. Cohybridization of AAV and TNNT1 is indicated by arrows (A, M19; B, M19, HA16) and of AAV, AAVS1, and TNNT1 by ▴ (A, HA16). Potential cohybridization of AAV and TNNT1 in 7374 cells was seen after EcoRI digestion (A); however, no TNNT1 disruption was apparent after HindIII digestion (B). Note, the variations in hybridization intensities with AAV as well as with AAVS1 probes are consistent with previous observations (see for example ref. 29)
Recently, Barton et al. (30) found that the TNNT1 gene is closely linked to the cardiac troponin I gene (TNNI3), which had previously been mapped to 19q13.4 (31). The authors concluded that the distance between TNNT1 and TNNI3 is only 2.6 kb. Based on these findings, we performed Southern analysis on genomic DNA of the four latently infected cell lines. However, when the TNNI3 probe indicated in Materials and Methods was used, no evidence for disruption was found (data not shown).
Sequence Analysis of an AAV-TNNT1 Junction Fragment.
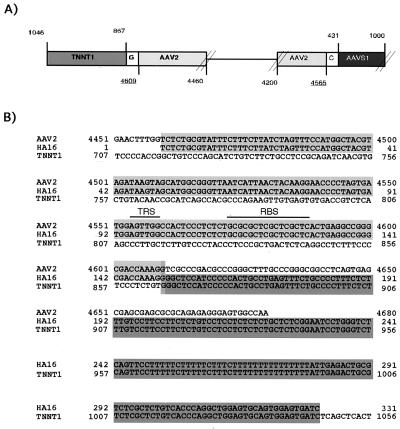
The junction sequences isolated from the HA16 library (and cloned to give plasmid pHA16-3) provide direct support for the hypothesis that AAV integration was the cause for the observed TNNT1 rearrangements (Fig. 3 A and B). As indicated above, this fragment contains two junctions, one between the right AAV ITR and AAVS1 (near the RBS) and a second between another right AAV ITR and the last intron (intron 13) of the TNNT1 gene. This sequence analysis demonstrates that integration of AAV into AAVS1 can indeed disrupt the TNNT1 gene. The junction fragment showed characteristics similar to those previously described for AAV-AAVS1 junctions (4, 13, 32): the ITR was partially deleted, the immediate junction consisted of nucleotides common to both donor and acceptor DNA, and the AAV RBS was found at the viral–cellular junctions.
Figure 3.
The plasmid pHA16–3 contains viral–cellular junctions involving both AAVS1 and TNNT1 sequences. (A) A schematic representation of viral-cellular junctions in the plasmid pHA16-3 is shown. The plasmid pHA16-3 was isolated from a genomic library of latently infected HeLa cells (HA16C5) by selection for hybridization to an AAV probe. Subsequently, the viral–cellular junctions were sequenced. pHA16-3 contains a 14-kb insert with one junction between AAV2 (light gray) and AAVS1 (dark gray) and a second one between AAV (light gray) and TNNT1 (intermediate gray). At each junction, one nucleotide (white box) is common to both sequences. Underlined nucleotide numbers correspond to sequences at the recombination junctions (AAV2, accession no. AF043303; TNNT1, accession no. AJ011713; AAVS1, accession no. S51329). Nucleotide 1046 in TNNT1 corresponds to the cloning site. The DNA sequences between the junctions have not been determined (thin line). (B) DNA sequences at the AAV-TNNT1 junction are shown. The AAV RBS and TRS sites are indicated.
To rule out the possibility that the junction analyzed (and presented in Fig. 3) might be the result of recombination in Escherichia coli, PCR analysis was performed on genomic DNA from HA16 cells. PCR using a TNNT1 (derived from exon 14) and an AAV-specific primer (derived from the cap gene) resulted in one band of the expected size, the identity of which was confirmed by restriction analysis (data not shown).
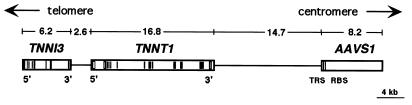
In summary, we conclude that AAVS1 is located 14.7 kb and 34.1 kb centromeric to TNNT1 and TNNI3 (Fig. 4), respectively, at position 19q13.4. Southern analysis of latently infected cells and sequence analysis of a viral-cellular junction showed that the site-specific integration of AAV DNA at AAVS1 led to rearrangements of the TNNT1 gene in two of the four cell lines studied.
Figure 4.
The organization of the troponin genes is shown relative to AAVS1. Boxes within TNNI3 and TNNT1 indicate the positions of the exons (30). Dark boxes indicate coding regions within exons, and open boxes indicate noncoding regions. The positions of TRS and RBS are indicated as lines within AAVS1. The sizes are shown in kb.
Troponin and tropomyosin constitute the Ca2+-sensitive complex regulating the contraction of muscle fibers. The troponin complex consists of three subunits, troponin C (Ca2+ binding), troponin I (inhibitory subunit), and troponin T (tropomyosin binding subunit). There are three genes encoding troponin I isoforms, TNNI1–3, and three genes encoding troponin T isoforms, TNNT1–3. The three respective isoforms represent the slow (TNNT1, TNNI1), fast (TNNT2, TNNI2), and cardiac troponins (TNNT3, TNNI3). The expression of the troponin genes is tightly regulated in a tissue-dependent manner, as well as during development. In adults, TNNT1 is exclusively expressed in skeletal muscle, whereas TNNI3 is exclusively expressed in the heart. It is of note that TNNT1 expression was detected by Northern blot analysis in two latently infected cell lines, Detroit 6 (7374) and HA16 (data not shown). It remains to be determined whether induction of TNNT1 expression occurred as a result of AAV DNA integration or whether the parental, uninfected cells expressed TNNT1 before AAV infection.
The site-specific integration of AAV DNA has attracted considerable attention with respect to its potential for a targeted gene transfer that would minimize the risk for insertional mutagenesis. Our report highlights that the TNNT1 gene can be disrupted as a result of wild-type AAV DNA integration, raising several issues. Studies of AAV integration have predominantly used immortalized or transformed cells that have lost chromosome stability. The extent of AAV-induced rearrangements, including the disruption of TNNT1 observed in our study, may be related to loss of these, as yet unknown, control mechanisms. Furthermore, the troponin genes are telomeric with respect to AAVS1. Most of the previously characterized junctions (4, 23, 29, 32), however, were found centromeric (“downstream”) of the AAVS1 TRS (see also Fig. 3A). A model for the mechanism of AAV integration must incorporate the potential for “upstream” rearrangements.
It is noteworthy that AAV has evolved to integrate close to, and potentially disrupt, the highly muscle-specific gene, TNNT1. Based on the idea that, in the adult, TNNT1 is exclusively expressed in skeletal muscle tissues (33), the question arises whether muscle represents a natural target tissue for latent AAV infection. In support of this possibility, (E. P. Hoffman and coworkers, personal communication) find AAV DNA in 17% of normal human muscle biopsies tested.
It is possible that integration of AAV into or close to TNNT1 might change the level of expression of either the TNNT1 or the closely linked TNNI3 genes. It can now also be asked whether altered mRNA species and potentially proteins are generated in human muscle cells as a result of AAV integration. However, skeletal muscle fiber cells are multinucleated, so the disruption of a subset of TNNT1 alleles might not lead to phenotypic changes in these cells. Hence, AAV site-specific integration into this locus might represent yet another strategy by the virus to optimize the virus-host relationship.
Acknowledgments
We thank Agnes Begue for her help in constructing the genomic library HA16C5 and for providing the lymphocyte library. Furthermore, we thank Dr. Linda Ashworth for helpful discussions and for providing the cosmids, and Dr. Jörg R. Schlehofer for providing the parental HA16 cells from which HA16C5 was subcloned. We thank Drs. Mary Klotman, Nuria Morral, Nick Muzyzcka, and Peter Ward for critical reading of the manuscript. This work was supported in part by Grant DK55609 (to R.M.L.) from the National Institutes of Health.
Abbreviations
- AAV
adeno-associated virus type 2
- ITR
inverted terminal repeat
- RBS
Rep binding site
- TRS
terminal resolution site
- LLNL
Lawrence Livermore National Laboratory
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080079397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080079397
References
- 1.Berns K I, Linden R M. BioEssays. 1995;17:237–245. doi: 10.1002/bies.950170310. [DOI] [PubMed] [Google Scholar]
- 2.Berns K I. In: Fields Virology. Fields B N, Knipe D M, Howley P M, editors. Vol. 2. Philadelphia: Lippincott–Raven; 1996. pp. 2173–2197. [Google Scholar]
- 3.Kotin R M, Menninger J C, Ward D C, Berns K I. Genomics. 1991;10:831–834. doi: 10.1016/0888-7543(91)90470-y. [DOI] [PubMed] [Google Scholar]
- 4.Samulski R J, Zhu X, Xiao X, Brook J D, Housman D E, Epstein N, Hunter L A. EMBO J. 1991;10:3941–3950. doi: 10.1002/j.1460-2075.1991.tb04964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava A, Lusby E W, Berns K I. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarty D M, Pereira D J, Zolotukhin I, Zhou X, Ryan J H, Muzyczka N. J Virol. 1994;68:4988–4997. doi: 10.1128/jvi.68.8.4988-4997.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ryan J H, Zolotukhin S, Muzyczka N. J Virol. 1996;70:1542–1553. doi: 10.1128/jvi.70.3.1542-1553.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chiorini J A, Yang L, Safer B, Kotin R M. J Virol. 1995;69:7334–7338. doi: 10.1128/jvi.69.11.7334-7338.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Im D S, Muzyczka N. Cell. 1990;61:447–457. doi: 10.1016/0092-8674(90)90526-k. [DOI] [PubMed] [Google Scholar]
- 10.McCarty D M, Ryan J H, Zolotukhin S, Zhou X, Muzyczka N. J Virol. 1994;68:4998–5006. doi: 10.1128/jvi.68.8.4998-5006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brister J R, Muzyczka N. J Virol. 1999;73:9325–9336. doi: 10.1128/jvi.73.11.9325-9336.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith D H, Ward P, Linden R M. J Virol. 1999;73:2930–2937. doi: 10.1128/jvi.73.4.2930-2937.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kotin R M, Linden R M, Berns K I. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Giraud C, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1994;91:10039–10043. doi: 10.1073/pnas.91.21.10039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linden R M, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1996;93:7966–7972. doi: 10.1073/pnas.93.15.7966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Linden R M, Ward P, Giraud C, Winocour E, Berns K I. Proc Natl Acad Sci USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balague C, Kalla M, Zhang W W. J Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Surosky R T, Urabe M, Godwin S G, McQuiston S A, Kurtzman G J, Ozawa K, Natsoulis G. J Virol. 1997;71:7951–7959. doi: 10.1128/jvi.71.10.7951-7959.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzman M D, Kyostio S R, Kotin R M, Owens R A. Proc Natl Acad Sci USA. 1994;91:5808–5812. doi: 10.1073/pnas.91.13.5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiorini J A, Weitzman M D, Owens R A, Urcelay E, Safer B, Kotin R M. J Virol. 1994;68:797–804. doi: 10.1128/jvi.68.2.797-804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kotin R M. Hum Gene Ther. 1994;5:793–801. doi: 10.1089/hum.1994.5.7-793. [DOI] [PubMed] [Google Scholar]
- 22.Zhu X. Ph.D. thesis. Pittsburgh: Univ. of Pittsburgh; 1993. [Google Scholar]
- 23.Kotin R M, Berns K I. Virology. 1989;170:460–467. doi: 10.1016/0042-6822(89)90437-6. [DOI] [PubMed] [Google Scholar]
- 24.Laughlin C A, Cardellichio C B, Coon H C. J Virol. 1986;60:515–524. doi: 10.1128/jvi.60.2.515-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walz C, Schlehofer J R. J Virol. 1992;66:2990–3002. doi: 10.1128/jvi.66.5.2990-3002.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berns K I, Pinkerton T C, Thomas G F, Hoggan M D. Virology. 1975;68:556–560. doi: 10.1016/0042-6822(75)90298-6. [DOI] [PubMed] [Google Scholar]
- 27.Gahlmann R, Troutt A B, Wade R P, Gunning P, Kedes L. J Biol Chem. 1987;262:16122–16126. [PubMed] [Google Scholar]
- 28.Samson F, de Jong P J, Trask B J, Koza-Taylor P, Speer M C, Potter T, Roses A D, Gilbert J R. Genomics. 1992;13:1374–1375. doi: 10.1016/0888-7543(92)90077-6. [DOI] [PubMed] [Google Scholar]
- 29.Kotin R M, Siniscalco M, Samulski R J, Zhu X D, Hunter L, Laughlin C A, McLaughlin S, Muzyczka N, Rocchi M, Berns K I. Proc Natl Acad Sci USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barton P J, Cullen M E, Townsend P J, Brand N J, Mullen A J, Norman D A, Bhavsar P K, Yacoub M H. Genomics. 1999;57:102–109. doi: 10.1006/geno.1998.5702. [DOI] [PubMed] [Google Scholar]
- 31.Mogensen J, Kruse T A, Borglum A D. Cytogenet Cell Genet. 1997;79:272–273. doi: 10.1159/000134740. [DOI] [PubMed] [Google Scholar]
- 32.Giraud C, Winocour E, Berns K I. J Virol. 1995;69:6917–6924. doi: 10.1128/jvi.69.11.6917-6924.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabry M A, Dhoot G K. J Muscle Res Cell Motil. 1991;12:262–270. doi: 10.1007/BF01745116. [DOI] [PubMed] [Google Scholar]
- 34.Bhavsar P K, Brand N J, Yacoub M H, Barton P J R. Genomics. 1996;35:11–23. doi: 10.1006/geno.1996.0317. [DOI] [PubMed] [Google Scholar]