Abstract
The full-length cDNA of the murine homolog of human nectin1δ (mNectin1δ), also known as human poliovirus receptor related 1 (PRR1) or herpesvirus entry mediator C, was cloned and showed a >90% identity with its human counterpart. mNectin1δ is expressed in some murine cell lines, exemplified by NIH 3T3 and L cells, and in murine tissues. It mediates entry of an extended range of herpes simplex virus (HSV) strains, porcine pseudorabies virus (PrV), and bovine herpesvirus 1. A soluble form of the mediator blocked infectivity in mNectin1δ and human nectin1δ (hNectin1δ)-expressing cells, suggesting a physical interaction of the mediator with virions. The higher concentrations of soluble mNectin1 required to block infectivity relative to soluble hNectin1 suggest that the target of the two molecules is not identical. Entry of HSV, but not PrV, was blocked by soluble mNectin1δ in NIH 3T3 and L cells. Two features were unexpected. First, soluble mNectin1δ failed to physically interact with HSV glycoprotein D (gD) at a detectable level, although it interacted physically with virions. Second, coexpression of mNectin1δ and HSV gD did not restrict HSV or PrV infection, whereas coexpression of hNectin and gD did restrict infection, suggesting that mNectin1δ fails to be sequestered by HSV gD. We conclude that mNectin1δ serves as a species-nonspecific mediator for entry of the human and animal αherpesviruses. This activity, at least for HSV, is independent of a detectable binding to gD.
Herpes simplex virus (HSV) has a broad host range and can infect animals and cultured cells from species other than the natural host. Mouse is the commonly used small animal model in HSV research, including studies on latency, prototypic vaccines, antiviral compounds, and HSV-based vectors. The mouse can be infected by inoculation at peripheral sites, e.g., skin, vagina, or directly into the central nervous system. Infection mimics the infection in humans. Thus, following peripheral inoculation, the virus spreads to nerve endings of the sensory neurons, is transported in anterograde direction to the nuclei of sensitive neurons, where it establishes latency. Virus can be reactivated by exogenous stimuli, and after replication, moves in retrograde direction to peripheral tissues, where it induces lesions. This pattern of infection underscores the existence in mice of receptors for HSV entry into cells, and cell-to-cell spread, with pathways of transmission to tissues analogous to those in humans. Similar to HSV, the porcine αherpesvirus pseudorabies virus (PrV) has a very broad host range in cultured cells, and can infect and cause disease in animal species other than the natural host. Mice have been used for experimental infections and to trace the pattern of virus spread to the nervous system. In contrast, the host range of bovine herpesvirus 1 (BHV-1) appears to be narrower, probably reflecting limited availability also of cellular functions involved in postentry steps. A key question in the validation of the mouse animal model is to what extent the molecular mechanisms of infection and virus spread reflect those in humans.
HSV enters cell cultures by a two-step process. After initial attachment to heparan sulfate glycosaminoglycans, entry occurs through the concerted action of four essential glycoproteins, gD, gB, and the heterodimer gH/gL (1). Their specific roles are not precisely known, except for gD, which represents the glycoprotein with receptor-binding activity (2–5).Thus, cells expressing the gD of HSV, as well as of PrV or BHV-1, become resistant to infection with homologous and heterologous αherpesviruses, by sequestering a cellular molecule that interacts with gD (2, 6–8). This phenomenon has been designated as gD-mediated restriction, or interference, to infection. Moreover, soluble forms of gD, and anti-idiotypic antibodies mimicking gD, bind to cell surfaces in a saturating manner, and prevent infection (9, 10). These findings prompted the search for cellular receptors of HSV, with the expectation that they should interact with gD. The receptors known to date belong to three different molecular families, and all bind gD. Herpesvirus entry mediator A (HveA), a member of the tumor necrosis factor receptor family, has a very narrow distribution (11). 3-O-Sulfation of heparan sulfate enables entry (12). Its HSV receptor activity in human cell lines remains to be determined. The human receptors widespread in a variety of cell lines are members of an Ig family. They are the human nectin1δ [hNectin1δ; also PRR1 or herpesvirus entry mediator C (HveC)] (4, 13), and a splice variant of it, hNectin1α, previously herpesvirus Ig-like receptor (HIgR) (3). hNectin1α and -δ share the ectodomain, made of one V and two C2 Ig domains, and differ in transmembrane and cytoplasmic regions (3). They are expressed in HEp-2, HeLa, and a variety of other cell lines from different lineages, where they serve as HSV receptors (3). hNectin1 interacts physically with gD (5, 14). The family also includes hNectin2α and -δ (also PRR2α/HveB and PRR2δ) (15, 16), that serve as low efficiency receptors for HSV mutants in gD with substitutions at residues 25–27, but not for wild-type virus, and bind mutant gD weakly (17). The third member of the family, the human poliovirus receptor (CD155, or HveD) (18), does not serve as receptor for HSV, but similar to hNectin1, hNectin2, and the mNectin2 (mPRR2, mHveB, or Mph), serves as receptor for PrV (4, 16, 19). Poliovirus receptor/HveD also mediates entry of BHV-1 (4). The two animal αherpesviruses require gD for entry into cells (20–22). hNectin1 and 2 also mediate cell-to-cell spread of HSV (23).
In the present studies, we identified the murine homolog of hNectin1δ (mNectin1δ), with the objective to determine whether it serves as mediator of αherpesviruses entry into murine cells, and investigate whether the process of entry through mNectin1δ involves the same molecular mechanisms as entry through hNectin1.
Materials and Methods
Cells and Viruses.
Cells were grown in Dulbecco–Vogt-modified Eagle's medium and 5% FCS. The HSV strains were as mentioned (3). R8102, R7032, and a PrV strain carrying LacZ (PrV-LacZ) in place of the gH gene and grown on a gH-expressing complementing cell line were described (3, 24, 25). Detection of infectivity by β-galactosidase (β-gal) was described (3, 11). J cells harboring α27p-LacZ were constructed by transfection with pCEPα27p-LacZ (17), and selection was with 200 μg/ml hygromycin.
Cloning of mNectin1δ.
Sequence no. AA277558 in murine Expressed Sequence Tags is homologous to the 5′ end of hNectin1. The entire ectodomain (aa 1–334) was amplified from retrotranscribed L-cell mRNA with primer muR15n (5′-GAT GGC TCG GAT GGG GCT TGC GGG-3′) and the human primer CFLPRR13 (5′-CCG ATC GGC CGA TGT GAT ATT GAC CTC CAC-3′). The 3′ portion of the molecule was amplified from a cDNA library of 3T3 cells (Invitrogen) with primer 5′muPPR1 designed on the sequence of the ectodomain (5′-GAA CCT ACA TCT GTG AGG CCA CCA A-3′), and a 3′ primer annealing to the vector, R3-pcDNA31, (5′-CAG ATG GCT GGC AAC TAG AAG GC-3′). The full-length cDNA (GenBank accession no. AF239762) amplified from the murine library was cloned in pGemTeasy (Promega), generating pEA6. The entire mNectin1 cDNA, excised with NotI, was transferred to pcDNA3.1(−) (Invitrogen), generating pEA6.6. Transfections were carried out by means of LipofectAmine (GIBCO).
Reverse Transcription–PCR.
Total RNA (2 μg) extracted with TRIzol (GIBCO/BRL), was retrotranscribed and amplified with primers muR15n (5′-GAT GGC TCG GAT GGG GCT TGC GGG-3′) and mu12L (5′-CTG CCA TGT GAC CTG GGT GAT TTT CAC-3′).
Northern Blot Analysis.
Murine multiple-tissue Northern membrane (CLONTECH) was hybridized with the 1,002-bp EcoRI/EagI ectodomain probe labeled with [32P]dCTP, and with a β-actin probe.
Soluble mNectin1-Fc.
The ectodomain of mNectin1 (aa 1–334) was amplified from L-cell mRNA with primers muR15n and CFLPRR13. The PCR product, cloned in the pGEMTeasy, was transferred to the CosFcLink vector (SmithKline Beecham) and transfected in COS 1 cells (14), kept in synthetic CHOSFMII medium (Life Technologies, Grand Island, NY). Purification was by affinity chromatography to ProteinA-Affigel. Where indicated, the medium of transfected COS cells was concentrated. Soluble hNectin1-Fc was described (14).
gD Expression.
The coding sequence of gD, containing 23 bp upstream the start codon and 35 bp downstream the stop codon, was amplified by PCR with primers 5′-TAT CCT TAA GGG ATC CTT TGT GTG GTG CG-3′ and 5′-GGT CTG CGG GGT AAG CTT GGG ACC TTA-3′, and cloned in pcDNA 3.1(−) (Invitrogen) to obtain pEA99.
ELISA.
gD was purified to homogeneity from HSV-1-infected BHK cell lysates by affinity chromatography to monoclonal antibody no. 30 (26), and immobilized to Affigel. Microwell plate coating was performed in 100 μl of bicarbonate buffer (pH 9.5) with either 16 nM gD, or fetuin, or 107 pfu of R7032 virions. Coated wells were reacted with mNectin1-Fc or hNectin1-Fc for 2 h at 37°C, after blocking unspecific binding sites with 2% BSA. Binding was revealed with anti-human IgG peroxidase (Dako; 1:6000), by developing with 0.5 mg/ml o-phenylenediamine (Sigma) in 2.5 mM citric acid/5 mM Na2HPO4/0.009% H2O2; blocking with 1:6 H2SO4; and monitoring at 490 nm. In the second ELISA, serial dilutions of mNectin1-Fc or hNectin1-Fc were bound to microwells in bicarbonate buffer, and reacted with biotinylated soluble gD (gD Δ290–299t) (27) for 2 h at 37°C after blocking unspecific binding with 2% BSA. Binding was revealed with streptavidin-conjugated peroxidase (ABC Kit, Vector Laboratories) and developing was as above. gD biotinylation was described (3).
Fluorescence.
Cells grown on cover slips were transfected with pEA6.6, pCF18, or pcDNA3.1(−), and reacted with biotinylated soluble gD (gD Δ290–299t) (27), and extravidin-tetramethylrhodamine B isothiocyanate.
Results
Cloning of the Full-Length cDNA of mNectin1δ.
The first portion of the murine homolog of hNectin1δ was identified by analysis of the murine Expressed Sequence Tags database. Sequence no. AA277558 starts at −149 from the first ATG (+1), up to +297. The ectodomain of mNectin1δ (aa 1–334) was then amplified from retrotranscribed L-cell mRNA, with a 5′ primer designed on the Expressed Tag Sequence and a 3′ primer designed on the sequence of the hNectin1δ. The C-terminal portion of the molecule was subsequently amplified from a cDNA library of NIH 3T3 cells with a 5′ primer designed on the sequence of the ectodomain, and a 3′ primer annealing to the vector. The nucleotide sequence of the full-length cDNA, cloned from the murine library, revealed that mNectin1δ is 94% identical at the aa level to its human homolog (Fig. 1). Characteristic features are (i) a predicted signal sequence at the N terminus. The Ser at position +30, which is supposed to serve for cleavage of the signal sequence in hNectin1δ, is substituted with Thr in mNectin1δ; (ii) an ectodomain with six conserved Cys, which define three domains, one N-terminal V, and two C2 domains; (iii) a transmembrane region from +354 to +377; and (iv) a cytoplasmic tail terminating with a EWYV motif. In hNectin1δ and hNectin2, the conserved A/ExYV motif binds the PDZ domain of afadin (28).
Figure 1.
Amino acid sequence of mNectin1δ. The six conserved Cys that define the V and C2 domains are boxed. The predicted signal and transmembrane sequences are shaded. Amino acids in hNectin1δ that differ from the corresponding in mNectin1δ are marked.
Expression of mNectin1 in Murine Cell Lines and Mouse Tissues.
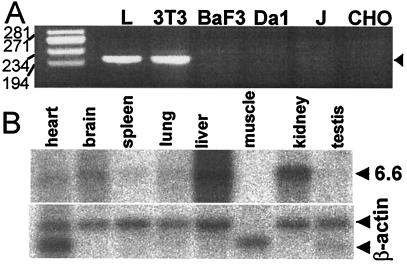
Expression of mNectin1 was assessed by reverse transcription–PCR in a number of murine cell lines, NIH 3T3, L (fibroblastic), BaF3, and Da1 (hematopoietic). An amplification product of expected size (204 bp) was clearly visible with cDNA of NIH 3T3 and L, but not BaF3 and Da1 cells (Fig. 2A), indicating that some murine cell lines express mNectin1δ. The expression in murine tissues was assessed by Northern blot analysis of murine multiple-tissue Northern membranes (CLONTECH), probed with the 1,002-bp EcoRI/EagI fragment encompassing the ectodomain. The results in Fig. 2B show that a unique 6.6-kbp band was visible in several tissues, with the highest level of expression in the liver, brain, and kidney. These organs are targets of HSV infection. The brain and spinal cord showed the highest levels of expression of hNectin1δ in human tissues (3). In contrast, kidney was positive in murine, but negative in human tissues (3). In addition, whereas the Northern blot of human tissues showed two bands, reflecting the mRNAs for the α and δ isoforms of hNectin1, only one band was visible with murine tissues.
Figure 2.
Expression of mNectin1δ in (A) murine cells and (B) mouse tissues. (A) Reverse transcription–PCR detection in murine, J, and CHO cells. The amplification product was 204 bp (arrow). Numbers on the left represent molecular weight markers. (B) Tissue distribution of mNectin1δ determined by Northern blot analysis of murine multiple-tissue Northern membrane hybridized with a probe encompassing the ectodomain of mNectin1δ. Lower panel, hybridization with β-actin probe.
mNectin1δ Mediates Entry of an Extended Range of HSV Strains.
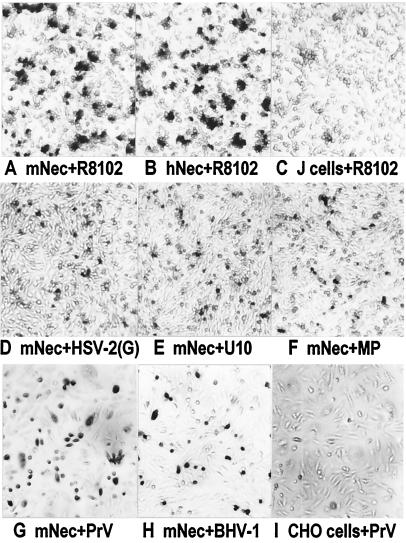
To assess whether mNectin1δ mediates HSV entry, J cells, resistant to HSV infection because of failure to express suitable entry receptors (3), were transfected with the mNectin1δ cDNA, cloned into pcDNA3.1(−), under the cytomegolovirus IE promoter (pEA6.6). The transfected cells were inoculated with R8102, a HSV recombinant that carries a LacZ gene under the α27 promoter. Entry was monitored as αgene (β-gal) expression (11). Fig. 3 shows that mNectin1δ enabled infection of J cells with R8102. The number of cells acquiring infectivity resembled that in a parallel culture transfected with pCF18, which carries the cDNA of hNectin1α (3). To investigate whether mNectin1δ enabled entry of a broad spectrum of HSV strains, a derivative of J cells harboring a LacZ gene driven by the α27 promoter, designated as J-αLacZ, was derived. The J-αLacZ cells expressing in transient mNectin1δ were infected with HSV-1(U10) and HSV-1(U21), two gD-unrestricted mutant strains which can enter cells via either hNectin1 or hNectin2 (17, 26); HSV-1(MP) and HFEM, two syncytial strains; and HSV-2(G). Infection was monitored as β-gal expression from the cell-resident LacZ gene. mNectin1δ mediated entry of all HSV strains tested (representative examples in Fig. 3).
Figure 3.
Susceptibility of J and CHO cells expressing mNectin1δ to infection with HSV, PrV, and BHV-1. Micrographs showing J cells expressing mNectin1δ (A and D–F), hNectin1α (B), or J cells transfected with pcDNA3.1(−) (C), infected with indicated viruses. CHO cells transfected with (G and H) mNectin1δ, or (I) pcDNA3.1(−), infected with (G) PrV or (H) BHV-1. Infection was detected as β-gal (A–F), or by immunostaining (G–I). Differences in intensities between A and B compared with D–F are because of the fact that in A and B, the LacZ gene is expressed from the viral genome, and supposedly in higher copy number, whereas in D–F, the LacZ is expressed from a gene resident in the cell.
mNectin1δ Mediates Entry of PrV and BHV-1.
Chinese hamster ovary (CHO) cells, resistant to HSV, PrV, and BHV-1 infection (11), transfected with pEA6.6, were exposed to wild-type PrV, PrV-LacZ (25), or BHV-1. mNectin1δ enabled entry of all three viruses (typical examples in Fig. 3). Thus, mNectin1δ had a broad promiscuous mediator activity, and could not be differentiated from hNectin1δ with respect to the range of αherpesviruses for which it served as an entry mediator.
Soluble mNectin1 Blocks HSV Entry Through mNectin1δ and hNectin1δ.
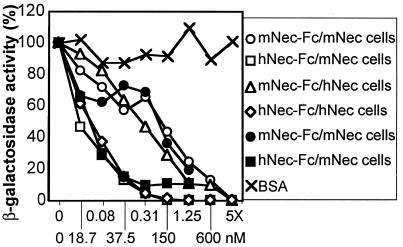
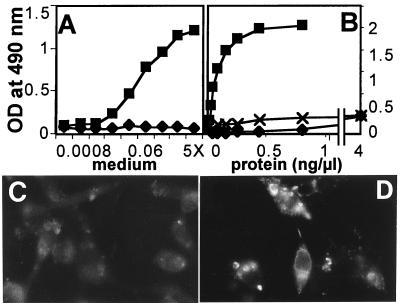
If the entry of HSV through mNectin1δ depends on direct interaction of a HSV envelope protein with the mediator, a soluble form of mNectin1 could bind to virus and compete with cell-bound molecule, and thereby reduce virus entry. Moreover, if the binding of mNectin1δ to its target influences the binding of hNectin1δ to gD, or if the target of mNectin1δ in HSV virions overlaps, at least in part, with the target of hNectin1δ, the soluble form of mNectin could compete also with the entry through hNectin1. A soluble form of mNectin1 was constructed by fusion of the ectodomain with the Fc portion of human IgG (mNectin1-Fc). Previously, a soluble form of hNectin1 was constructed in the same manner (hNectin1-Fc) (14). R8102 was preincubated with increasing concentrations of mNectin1-Fc, or hNectin1-Fc, and allowed to infect J cells expressing mNectin1δ or hNectin1α. For each soluble mediator, we used two batches. In one instance, the soluble mediators were purified to homogeneity by affinity chromatography to Affigel-Protein A. In the other instance, the concentrated media from COS cells transfected with the corresponding Fc-link vectors were produced, processed, and used in parallel for a direct comparison. Results with the two batches were very similar (Fig. 4), and showed that soluble mNectin1-Fc, but not BSA, reduced in a dose-dependent manner the HSV infectivity in cells expressing mNectin1δ or hNectin1α. Similarly, soluble hNectin1-Fc inhibited entry in both cell types. Unexpectedly, the concentrations required for 50% inhibition were constantly higher for mNectin1-Fc than for hNectin1-Fc. The inhibitory effect of soluble mNectin1-Fc implies a direct interaction of mNectin1δ with HSV. The ability of soluble mediators to compete with entry in cells expressing the heterologous mediator suggests that either the binding of mNectin1δ to its target affects the ability of hNectin1 to bind gD, or that the target of mNectin1δ coincides in part with that of hNectin1α. The unexpected finding that higher concentrations were required for soluble mNectin1-Fc suggests that the target of the two molecules is not identical, or that the binding of mNectin1δ to its target may be weaker than the binding of hNectin1 to gD. mNectin1-Fc and hNectin-Fc inhibited PrV entry in mNectin1δ-expressing cells with a similar pattern (data not shown).
Figure 4.
Soluble mNectin1-Fc or hNectin1-Fc block R8102 entry in cells expressing mNectin1δ or hNectin1α. R8102 was preincubated with increasing dilutions of mNectin-Fc (mNec-Fc), hNectin1-Fc (hNec-Fc), or BSA, and allowed to infect cells expressing mNectin1δ (mNec) or hNectin1δ (hNec). Soluble mediators were either purified by affinity chromatography (closed symbols), or concentrated media of transfected COS cells (open symbols). Cells were grown in 96-well trays. Infection was monitored as β-gal expression. Each point represents the average of triplicates.
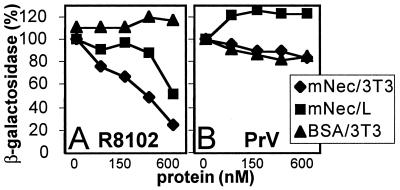
Entry of HSV, but Not of PrV, Is Blocked by Soluble mNectin1δ in NIH 3T3 and L Cells.
The two murine cell lines that expressed mNectin1δ at high levels were NIH 3T3 and L cells. These cells are susceptible to HSV and PrV infection. If entry is mediated by mNectin1δ, or by a mediator that recognizes in HSV, or PrV, virions the same target as mNectin1δ, and if this is the principal available mediator, the soluble mNectin1δ could compete with, and reduce, virus entry. HSV, or PrV, were preincubated with soluble mNectin1-Fc, and then allowed to infect NIH 3T3 or L cells. Fig. 5A shows that HSV infection was reduced in NIH 3T3 cells, and to a somewhat lower extent in L cells, by mNectin1-Fc, but not by BSA, in a dose-dependent manner. The extent of inhibition in NIH 3T3 cells was about 80% at the highest concentration used. In contrast with HSV, PrV entry was not reduced either in NIH 3T3, or L cells (Fig. 5B). The results indicate that the major entry mediator for HSV in NIH 3T3 cells is a molecule that recognizes in the virions the target recognized by mNectin1δ. Given that the cells express mNectin1δ, the mediator is very likely mNectin1δ itself. Lack of inhibition on PrV entry indicates the presence of alternative mediators that recognize in PrV virions a target different from that recognized by mNectin1δ.
Figure 5.
Soluble mNectin1-Fc reduces entry of R8102 (A), but not PrV (B) in NIH 3T3 and L cells. R8102, or PrV, were preincubated with increasing dilutions of affinity-purified mNectin-Fc (mNec), or BSA, and allowed to infect NIH 3T3 (⧫), or L (■) cells. Each point represents the average of triplicates.
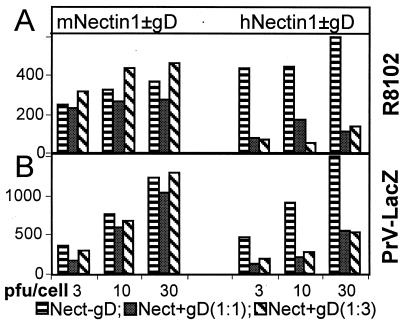
Coexpression of HSV gD and mNectin1δ Does Not Restrict HSV and PrV Infection.
To ascertain whether HSV and PrV infection in mNectin1δ-expressing cells is sensitive to gD-mediated restriction, J cells, resistant to infection with HSV and PrV-LacZ, were cotransfected with a mixture of mNectin1δ cDNA and HSV gD expression plasmids, in 1:1 and 1:3 ratios, and then exposed to R8102, or PrV-LacZ (29). In parallel experiments, the hNectin1α, cloned in the same expression vector pcDNA3.1 as mNectin1δ (pCF18, ref. 3), was also cotransfected with gD expression vector (1:1 and 1:3). Surprisingly, coexpression of HSV gD and mNectin1δ failed to restrict HSV, as well as PrV, infection (Fig. 6). In sharp contrast, under the same conditions, coexpression of hNectin1α and gD did restrict infection of both HSV and PrV-LacZ. These results were unexpected. They imply that mNectin1δ fails to be sequestered by gD, and suggest that either the binding of mNectin1δ to gD is very weak, or that the target of mNectin1δ is not the same as that of hNectin1α.
Figure 6.
Coexpression of mNectin1δ and gD (Left), but not hNectin1α and gD (Right), fails to restrict infection with R8102 (A), or PrV (B). J cells were transfected with mNectin1δ, or hNectin1α (Nect-gD, horizontal hatching), or cotransfected with mixtures of mNectin1δ, or hNectin1α, plus gD expression vectors at ratios of 1:1 [Nect + gD (1:1), full hatching)] or 1:3 [Nect + gD (1:3), slanted hatching]. At 24 h after transfection, cells were replated in 96-well trays, and 24 h later exposed to R8102 (A), or PrV-LacZ (B), at multiplicities of infection of 3, 10, or 30 pfu/cell. Infection was monitored as β-gal activity.
Soluble mNectin1δ Fails to Interact with HSV gD at a Detectable Level.
To test whether mNectin1δ interacts physically with gD, three series of experiments were performed, two ELISAs and one assay based on a gD-fluorescent probe. In the two ELISAs, three batches were tested for each mediator. One batch each consisted of soluble mediators purified to homogeneity by affinity chromatography. Two batches consisted of concentrated media from transfected COS cells. In the first ELISA, gD, purified to homogeneity from infected cells, was immobilized to microwells and reacted with mNectin1-Fc, followed by peroxidase-coupled anti-human IgG. Fig. 7A represents an experiment with batches of mNectin1-Fc and hNectin-Fc prepared in parallel, and shows that mNectin-Fc failed to react with gD, whereas hNectin1-Fc did react. Fig. 7B represents results with purified molecules. At concentrations of mNectin1-Fc 10-fold higher than that required for maximum binding of hNectin1-Fc, there was a barely detectable reactivity to gD. This was the only case in which we observed a very low reactivity. However, parallel reactivity to a control unrelated glycoprotein, human fetuin, was of the same order of magnitude. We interpret this as an unspecific binding. In the second ELISA, the soluble mediators were immobilized onto microwells and then reacted with a biotinylated recombinant soluble gD (gD Δ290–299t) (27). Binding was revealed by streptavidin-conjugated peroxidase. In both assays, mNectin1-Fc, but not hNectin1-Fc, failed to show a detectable binding to gD (data not shown). Finally, a third series of experiments did not make use of soluble forms of the mediator. Cells transfected with mNectin1δ cDNA were reacted with biotinylated soluble gD (gD Δ290–299t) (27), followed by extravidin-tetramethylrhodamine B isothiocyanate. Binding of cells expressing mNectin1δ with gD could not be detected, whereas binding of cells expressing hNectin1α with gD was positive (Fig. 7 C and D). All results consistently indicated that mNectin1δ fails to interact with HSV gD at detectable level.
Figure 7.
(A and B) ELISA binding of mNectin1δ to HSV gD. gD was immobilized to microwells, and then allowed to react with increasing concentrations of mNectin1-Fc (⧫) and hNectin1-Fc (■). Soluble mediators were in concentrated media of transfected COS (A) or purified by affinity chromatography (B). In B, crosses (✖) indicate binding of mNectin1-Fc to fetuin. (C and D) Fluorescence of J cells expressing mNectin1δ (C) or hNectin1α (D) reacted with biotinylated gD (Δ290–299t) and streptavidin-tetramethylrhodamine B isothiocyanate.
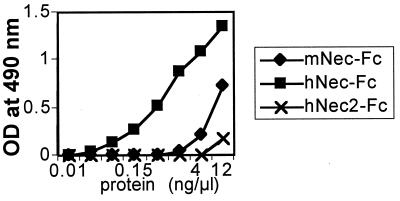
mNectin1δ Interacts with HSV Virions.
To prove an interaction of mNectin1δ with virions more directly, virions of R8102 or a gE-gI− HSV recombinant, R7032 (24), were immobilized to microwells and reacted with mNectin1-Fc or hNectin1-Fc. Binding was revealed by anti-human IgG. Binding of virions to soluble hNectin2-Fc, which is known not to interact with gD of wild-type virus, represented the negative control (17). Fig. 8 shows results with R7032. mNectin1-Fc bound HSV virions to a significantly higher level than soluble hNectin2-Fc. The reactivity was lower than that to hNectin1-Fc. This is in accordance with the lower inhibitory activity of soluble mNectin1-Fc, and probably reflects the absence of a binding activity to gD, which represents a strong ligand for hNectin1.
Figure 8.
Soluble mNectin1-Fc binds to HSV virions. R7032 virions (107 pfu) were immobilized to microwells, and allowed to react with increasing concentrations of purified mNectin1-Fc (mNec-Fc, ⧫), hNectin1-Fc (hNec-Fc, ■), and hNectin2-Fc (hNec2-Fc, ✖).
Discussion
The mouse genome encodes a homolog of hNectin1δ that serves as a species nonspecific mediator for entry of the human, porcine, and bovine αherpesviruses. Entry of HSV is reduced by soluble mNectin1δ in NIH 3T3 and L cells, suggesting that this molecule represents the major mediator of HSV entry in some murine cells. The unexpected finding reported here is that mNectin1δ binds HSV virions but fails to bind gD at detectable levels.
The Mouse Genome Encodes a Murine Homolog of Nectin1δ.
Based on a partial sequence present in the murine Expressed Sequence Tags database, we cloned the full-length cDNA of mNectin1δ. The degree of identity with the human counterpart is 94% at the aa level. Both murine and human nectin1δ carry at the C terminus the A/ExYV motif that binds hNectin1δ to the PDZ domain of afadin, and recruits it to adherens junctions (28). Conservation of the motif predicts a localization at adherens junctions also for mNectin1δ. In addition to mNectin1δ, the mouse encodes a second related member of the Ig family, mNectin2, of which two variants, α and δ, are known (30, 31). No murine homolog of hNectin1α and poliovirus receptor/HveD is known to date. Northern blot analysis of mouse tissues with a probe covering the entire ectodomain revealed only one band (Fig. 2B), in contrast with two bands seen in Northern blot analysis of human tissues (3). Although a systematic study at the genome level has not been performed, only some of the human members of the Ig family appear to have a homolog in mice.
mNectin1δ Serves as a Species Nonspecific Mediator for Entry of the Human, Porcine, and Bovine αherpesviruses, and Entry of HSV Is Reduced by Soluble mNectin1δ in NIH 3T3 and L Cells.
Receptor-negative cells, transfected with mNectin1δ cDNA, acquired susceptibility to infection with an extended range of HSV strains, PrV and BHV-1. The range of viruses entering cells via mNectin1δ closely paralleled the range of viruses entering cells via hNectin1. A promiscuous entry mediator activity seems therefore to be a property common to the mouse and human nectin1 (3, 4). In contrast, other members of the Ig family serve a more restricted range of viruses. Thus, hNectin2 serves for gD-unrestricted HSV-1 mutants and PrV, mNectin2 serves for PrV only, and poliovirus receptor/HveD serves for PrV and BHV-1 (3, 4, 16, 17, 19). A central question addressed by these studies was whether mNectin1δ is expressed in murine cells susceptible to HSV and PrV infection, and in murine tissues target of the infection. The positive cells are exemplified by NIH 3T3 and L cells, both susceptible to HSV and PrV infection. Entry of HSV, but not PrV, was reduced by soluble mNectin1δ in NIH 3T3 and L cells, providing evidence that in these cells entry is mediated by a molecule that has the same target in HSV virions as mNectin1δ. Lack of inhibition on PrV infectivity suggests that alternative mediators may be functional for PrV entry into the murine cells analyzed here. The properties of mNectin1δ provide a molecular basis for the broad susceptibility of mouse cells to infection with HSV, PrV, and BHV-1. mNectinδ is expressed in mouse tissues target of HSV and PrV infection, and therefore has the potential to act as entry mediator into murine tissues. The extent to which this molecule is functional in mouse tissues could be explored directly by knocking out the gene, a procedure made possible by current studies.
mNectin1δ Binds HSV Virions but Fails to Bind gD at a Detectable Level.
The novel finding was the failure to reveal a detectable binding between mNectin1 and HSV gD. Under the same experimental conditions, binding of hNectin1 to gD, and also of hNectin2 to mutant gD was readily detectable (14, 17, 32). This, in conjunction with the inability of HSV gD to restrict HSV infection, when coexpressed with mNectin1δ (Fig. 6), indicates that mNectin1δ does not significantly bind gD in the micromolar concentration range used in current assays. The situation with PrV may be similar, because cotransfection of mNectin1δ and HSV gD failed to restrict PrV. The functional domain of hNectin1 in HSV entry, and in binding to gD, is located in the V domain (14, 32). In this region, only 10 aa are substituted in mNectin1δ relative to hNectin1. These substitutions seem to be sufficient to substantially modify the binding activity to gD. The possibility that binding to gD is so weak that it cannot be detected under the measurement conditions used in this study, but nonetheless sufficient to mediate entry cannot be ruled out formally. Against it argues the inability of mNectin1δ to restrict HSV infection when coexpressed with HSV gD. If the binding to gD were too low to be detected, but sufficient to mediate entry, it should be sufficient also to sequester mNectin1δ and restrict infection. mNectin1δ constitutes the first known example of a cellular mediator of HSV entry that does not show a detectable binding to gD in the micromolar concentration range. It is surprising that, not only has HSV evolved the ability to exploit structurally and functionally unrelated molecules to mediate entry, but in some murine cells, the pathway of entry may be independent of a detectable binding to gD. At present, it is not known if gD participates in the process of HSV entry into human cells merely as a receptor-binding protein, or with two activities, i.e., as a receptor-binding as well as a fusogenic glycoprotein. The ability of soluble mNectin to interfere with entry via hNectin1 is consistent with a dual role for gD. mNectin1δ may represent a useful tool to dissect these activities.
Two lines of evidence indicate that mNectin1δ interacts with HSV virions, despite the fact that it does not significantly bind gD. First, preincubation of virions with soluble mNectin1-Fc inhibited virus infectivity. Second, a physical binding was detectable in ELISA. The HSV entry mediator activity is likely to correlate with the binding activity to a virion component. The target of mNectin1δ in HSV virions still has to be identified. The ability of soluble mNectin1 to block entry in cells expressing hNectin1, and vice versa, suggests that the binding of mNectin1δ to its target affects the ability of hNectin1 to bind gD.
The basic objective of the present study was to ascertain whether the molecular mechanism of HSV, and PrV entry into murine cells, and supposedly of HSV and PrV infection of mice, is homologous to that functional in human cell lines. The present results demonstrate that, although the murine homolog of hNectin1δ can mediate HSV and PrV entry into cells, the molecular mechanisms underlying these activities are different. This difference may be relevant to studies dealing with HSV transmission to tissues, or the protective effect of vaccines that elicit a neutralizing response to HSV gD.
Acknowledgments
We thank Drs. M. Ackermann (University of Zurich), T. Mettenleiter (Federal Veterinary Research Institute, Insel Reims, Germany), and G. H. Cohen (University of Pennsylvania) for gifts of mAb to BHV-1 gB, PrV-gH-complemented, mAb to PrV, and soluble gD. The studies at the University of Bologna were aided by Telethon Grant A141, grants from the Progetto Finalizzato Biotecnologie and Ministry of Education and Research (40%). Studies in Marseille were aided by the Institut National de la Santé et de la Recherche Médicale U119, the Association pour la Récherche sur le Cancer, and the Ligue Nationale Française Contre le Cancer.
Abbreviations
- HSV
herpes simplex virus
- PRR1
poliovirus receptor related 1
- Hve
herpesvirus entry mediator
- gD
glycoprotein D
- PrV
pseudorabies virus
- BHV-1
bovine herpesvirus 1
- β-gal
β-galactosidase
- hNectin
human nectin
- mNectin
murine homolog of human nectin
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF239762).
References
- 1.Spear P G. Semin Virol. 1993;4:167–180. [Google Scholar]
- 2.Campadelli-Fiume G, Arsenakis M, Farabegoli F, Roizman B. J Virol. 1988;62:159–167. doi: 10.1128/jvi.62.1.159-167.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cocchi F, Menotti L, Mirandola P, Lopez M, Campadelli-Fiume G. J Virol. 1998;72:9992–10002. doi: 10.1128/jvi.72.12.9992-10002.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Geraghty R J, Krummenacher C, Cohen G H, Eisenberg R J, Spear P G. Science. 1998;280:1618–1620. doi: 10.1126/science.280.5369.1618. [DOI] [PubMed] [Google Scholar]
- 5.Krummenacher C, Nicola A V, Whitbeck J C, Lou H, Hou W, Lambris J D, Geraghty R J, Spear P G, Cohen G H, Eisenberg R J. J Virol. 1998;72:7064–7074. doi: 10.1128/jvi.72.9.7064-7074.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson R M, Spear P G. J Virol. 1989;63:819–827. doi: 10.1128/jvi.63.2.819-827.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chase C C, Lohff C, Letchworth G J D. Virology. 1993;194:365–369. doi: 10.1006/viro.1993.1269. [DOI] [PubMed] [Google Scholar]
- 8.Petrovskis E A, Meyer A L, Post L E. J Virol. 1988;62:2196–2199. doi: 10.1128/jvi.62.6.2196-2199.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson D C, Burke R L, Gregory T. J Virol. 1990;64:2569–2576. doi: 10.1128/jvi.64.6.2569-2576.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang T, Campadelli Fiume G. Proc Natl Acad Sci USA. 1996;93:1836–1840. doi: 10.1073/pnas.93.5.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery R I, Warner M S, Lum B J, Spear P G. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 12.Shukla D, Liu J, Blaiklock P, Shworak N W, Bai X, Esko J D, Cohen G H, Eisenberg R J, Rosenberg R D, Spear P G. Cell. 1999;99:13–22. doi: 10.1016/s0092-8674(00)80058-6. [DOI] [PubMed] [Google Scholar]
- 13.Lopez M, Eberlé F, Mattei M G, Gabert J, Birg F, Bardin F, Maroc C, Dubreuil P. Gene. 1995;155:261–265. doi: 10.1016/0378-1119(94)00842-g. [DOI] [PubMed] [Google Scholar]
- 14.Cocchi F, Lopez M, Menotti L, Aoubala M, Dubreuil P, Campadelli-Fiume G. Proc Natl Acad Sci USA. 1998;95:15700–15705. doi: 10.1073/pnas.95.26.15700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eberlé F, Dubreuil P, Mattei M G, Devilard E, Lopez M. Gene. 1995;159:267–272. doi: 10.1016/0378-1119(95)00180-e. [DOI] [PubMed] [Google Scholar]
- 16.Warner M S, Geraghty R J, Martinez W M, Montgomery R I, Whitbeck J C, Xu R, Eisenberg R J, Cohen G H, Spear P G. Virology. 1998;246:179–189. doi: 10.1006/viro.1998.9218. [DOI] [PubMed] [Google Scholar]
- 17.Lopez M, Cocchi F, Menotti L, Avitabile E, Dubreuil P, Campadelli-Fiume G. J Virol. 2000;74:1267–1274. doi: 10.1128/jvi.74.3.1267-1274.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mendelsohn C L, Wimmer E, Racaniello V R. Cell. 1989;56:855–865. doi: 10.1016/0092-8674(89)90690-9. [DOI] [PubMed] [Google Scholar]
- 19.Shukla D, Rowe C L, Dong Y, Racaniello V R, Spear P G. J Virol. 1999;73:4493–4497. doi: 10.1128/jvi.73.5.4493-4497.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karger A, Mettenleiter T C. Virology. 1993;194:654–664. doi: 10.1006/viro.1993.1305. [DOI] [PubMed] [Google Scholar]
- 21.Rauh I, Mettenleiter T C. J Virol. 1991;65:5348–5356. doi: 10.1128/jvi.65.10.5348-5356.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fehler F, Herrmann J M, Saalmuller A, Mettenleiter T C, Keil G M. J Virol. 1992;66:831–839. doi: 10.1128/jvi.66.2.831-839.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cocchi F, Menotti L, Dubreuil P, Lopez M, Campadelli Fiume G. J Virol. 2000;74:3909–3917. doi: 10.1128/jvi.74.8.3909-3917.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meignier B, Longnecker R, Mavromara-Nazos P, Sears A E, Roizman B. Virology. 1988;162:251–254. doi: 10.1016/0042-6822(88)90417-5. [DOI] [PubMed] [Google Scholar]
- 25.Babic N, Klupp B G, Makoschey B, Karger A, Flamand A, Mettenleiter T C. J Gen Virol. 1996;77:2277–2285. doi: 10.1099/0022-1317-77-9-2277. [DOI] [PubMed] [Google Scholar]
- 26.Brandimarti R, Huang T, Roizman B, Campadelli Fiume G. Proc Natl Acad Sci USA. 1994;91:5406–5410. doi: 10.1073/pnas.91.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicola A V, Willis S H, Naidoo N N, Eisenberg R J, Cohen G H. J Virol. 1996;70:3815–3822. doi: 10.1128/jvi.70.6.3815-3822.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takahashi K, Nakanishi H, Miyahara M, Mandai K, Satoh K, Satoh A, Nishioka H, Aoki J, Nomoto A, Mizoguchi A, et al. J Cell Biol. 1999;145:539–549. doi: 10.1083/jcb.145.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nixdorf R, Schmidt J, Karger A, Mettenleiter T C. J Virol. 1999;73:8019–8026. doi: 10.1128/jvi.73.10.8019-8026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morrison M E, Racaniello V R. J Virol. 1992;66:2807–2813. doi: 10.1128/jvi.66.5.2807-2813.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki J, Koike S, Ise I, Sato-Yoshida Y, Nomoto A. J Biol Chem. 1994;269:8431–8438. [PubMed] [Google Scholar]
- 32.Krummenacher C, Rux A H, Whitbeck J C, Ponce-de-Leon M, Lou H, Baribaud I, Hou W, Zou C, Geraghty R J, Spear P G, et al. J Virol. 1999;73:8127–8137. doi: 10.1128/jvi.73.10.8127-8137.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]