Abstract
A self-recombining bacterial artificial chromosome (BAC) containing the 142-kb pseudorabies virus genome was constructed such that the viral genome is rapidly excised from the BAC vector backbone on delivery into mammalian cells. The recombination is mediated by loxP sites in the plasmid and Cre recombinase encoded within the BAC vector. A synthetic intron inserted in the middle of the cre ORF completely inhibits recombination in Escherichia coli, but is spliced out after delivery of the plasmid into mammalian cells. Recombination is efficient, and pure virus lacking the BAC vector backbone is immediately isolated from transfected mammalian cells without the need of serial passage or plaque purification.
Construction of full-length infectious clones of herpesviruses requires the insertion of Escherichia coli vector sequences into the viral genome. The vector sequences used to date are mini F-plasmids (also referred to as bacterial artificial chromosomes; BACs) generally 7 kb or greater in length (1, 2). Transfection of herpesvirus BAC plasmids isolated from E. coli into mammalian cells is sufficient to initiate viral infection, from which a clonal population of recombinant virus can be isolated. However, the resulting recombinant virus is not wild type because of the insertion of the BAC vector sequence in the viral genome. Typically, one or more viral genes are interrupted by the BAC insertion and, as we demonstrate here, the BAC can also disrupt the expression of neighboring genes. This has limited the majority of full-length herpesvirus clones made to date to applications not requiring the production of wild-type virus (3–7).
Applying infectious BAC herpesvirus clones to the study of pathogenesis requires that the virus isolated after delivery of the plasmid DNA into cultured cells is phenotypically indistinguishable from the wild-type parent. Two general approaches to infectious clone design have been applied to address this issue. One was to insert the BAC vector sequences into a viral gene dispensable for viral growth and pathogenesis, for example in the gG gene of pseudorabies virus (PRV) (8). The resulting virus was found to be virulent and spread normally in the vertebrate nervous system, but a subtle growth defect was detected in cultured cells. Although the gG gene of PRV was largely dispensable, the analysis of additional mutations placed in this viral genome are complicated by potential synergistic defects associated with the loss of gG or genes adjacent to gG that may also be affected by the BAC insertion. Furthermore, in this study, the BAC vector sequences were unstable in the viral genome during infection of mammalian cells, resulting in spontaneous deletion of the BAC and surrounding viral sequences. These findings emphasized the need to remove the BAC vector sequence from the viral genome on delivery of the clone into mammalian cells.
Excision of the BAC vector has recently been accomplished by a second approach to herpesvirus BAC clone design that incorporated a duplicated 527-bp viral sequence flanking both sides of the BAC vector insertion (9). On delivery to mammalian cells, homologous recombination between the 527-bp sequences resulted in the loss of the BAC vector insertion and reconstituted a genotypically wild-type virus, which in this case was a mouse cytomegalovirus (MCMV). By regenerating a wild-type genome, this approach avoided stability problems and ensured production of virus identical to the wild-type parental isolate. The conversion of the viral genome to wild type occurred during five serial passages of the virus in culture, which for MCMV takes approximately 5 days.
We present here an alternative general method to produce phenotypically wild-type virus from an E. coli infectious plasmid clone. Like the approach previously described for MCMV, the BAC vector is removed from the viral genome autonomously on delivery of the plasmid DNA into mammalian cells. However, this method offers several additional advantages: (i) The BAC vector is efficiently removed from the virus without the need of serial passages in tissue culture; (ii) The BAC vector is removed by targeted, site-specific, nonhomologous recombination, simplifying mutagenesis of the clone in E. coli; and (iii) The method is amenable to the construction of wild-type infectious clones of other herpesviruses with little or no modification using the constructs described.
Materials and Methods
Virus and Cells.
PRV-Becker, a virulent isolate of PRV, is the parent of all recombinant viruses used in this study (10). All PRV strains were propagated in the PK15 (porcine kidney 15) epithelial cell line. Virus titers were determined in duplicate by plaque assay on PK15 cells. Determination of viral growth rates by single-step growth analysis on PK15 cells was as previously described (8). L2 cells were used for plaque assays in which plaque diameters were measured, based on a previously described method (11). Vero cells were used for the isolation of replicative intermediate viral DNA (see below). PK15, L2 and Vero cells were grown in DMEM supplemented with 10% FBS, whereas viral infections were performed in DMEM supplemented with 2% FBS.
Plasmids.
pGS166 contains the PRV-Becker Us9 gene on a 1.6-kb SphI fragment, and was previously described (12). A green fluorescent protein (GFP) expression cassette flanked by two loxP sites was inserted immediately adjacent to the stop codon of the Us9 gene as follows. Two PCR primers, each containing a loxP site, a NheI site, and either a NsiI or AflII site, were designed with sequence complementary (antisense) to the 3′ Us9 ORF including the stop codon (5′-GGGCTAGCATGCATATAAC-TTCGTATAGCATACATTATACGAAGTTATCTACA-CGTGCCGGGCGATG) or homologous (sense) to the Us9 stop codon and the intergenic sequence immediately downstream the Us9 ORF (5′-GGGCTAGCTTAAGATAACTTCGTATAATGTATGCTATACGAAGTTATGTAGCGAGCGGGTG- GTGG). Each primer was used in a separate PCR with a second primer that resulted in the amplification of a 400- to 800-bp fragment. Both fragments were cut internally with either EagI (Us9 containing fragment) or NcoI (downstream Us9 fragment) and NheI, and ligated to each other at the NheI sites and pGS166 digested with EagI and NcoI in a triple ligation. The resulting clone, pGS181, contained a unique NsiI and AflII site together flanked by two loxP sites in direct orientation with each other, all collectively positioned immediately downstream the Us9 ORF. A GFP expression cassette was inserted into the NsiI and AflII sites from pEGFP-N1 (CLONTECH), resulting in pGS187.
The pBluelox BAC vector was derived from pMBO131 (1). A synthetic adapter was cloned into the SalI site of pMBO131, resulting in pGS213. The adapter was composed of two oligonucleotides (5′-TCGAGCTAGCATGCATAACTTCGTATAATG-TATGCTATACGAAGTTATGCGGCCGCGGGAAGCTT-GGATCCGGGGG and 5′-TCGACCCCCGGATCCAAGCTT-CCCGCGGCCGCATAACTTCGTATAGCATACATTATA-CGAAGTTATGCATGCTAGC) that together encoded a loxP site and several restriction sites: BamHI, HindIII, NheI, NotI, NsiI, and SphI. The pSVβ lacZ expression cassette (CLONTECH), from which the intron had been first deleted, was subcloned into the BamHI and remaining SalI sites resulting in pGS271. PCR was used to amplify oriT adjacent to a PacI site from pCTC1 with primers 5′-CCCCGTCGACCGGCCAGCCTCGCAGAGCAGGAT and 5′-GGGGGTCGACGAATTCTTAATTAAGAATTCCG-GGCAGGATAGGTGAAGTAGGCC (13). The product was ligated into pGS271 cut with SalI using SalI-generated DNA ends, resulting in pBluelox. A SphI/PstI fragment from pBluelox, containing the PacI site, was subcloned into the allelic exchange vector, pGS284, to provide flanking sequence for recombination into infectious clones derived from pBluelox (see below), resulting in pGS382 (8).
Plasmid pOG231 encodes a Cre-expression cassette fused to a nuclear localization signal, and has previously been described (14). A 5′ intron was removed from pOG231 by digestion with SmaI and BglII, filling in of the BglII end by Klenow, and religation resulting in pGS396. An intron from vector pCI (Promega) was PCR amplified with primers: 5′-GGGTACGTAAGTATCAAGGTTACAAG and 5′-GGGCAGCTGTGGAGAGAAAGGC. The product was ligated into pGS396 cut with EcoRV using SnaBI and PvuII-generated DNA ends, resulting in pGS403. The Cre cassette from pGS403 was PCR amplified with primers: 5′-GGGTTAATTAACCCTCACTAAAGGG and 5′-GGGTTAATTAATACGACTCACTATAGGG. The product was ligated into pGS382 cut with PacI using PacI-generated DNA ends, resulting in the allelic exchange plasmid, pCREin. Recombination of pCREin with pBecker2 (see below) resulted in pBecker3, and was achieved by the method of allelic exchange as previously described (8). The pBecker3 construct contains the Cre expression cassette with intron in the BAC vector backbone, but does not contain any additional pCREin vector sequence.
Virus Construction.
PRV252 was isolated from PK15 cells cotransfected with pGS187 linearized with SphI and PRV-Becker nucleocapsid DNA. The desired recombinant virus was identified and plaque-purified based on GFP expression. PRV253 was isolated from PK15 cells cotransfected with uncut pOG231 and PRV252 nucleocapsid DNA, and virus lacking GFP expression was plaque-purified. PRV257 was isolated from PK15 cells cotransfected with uncut pOG231, uncut pBluelox, and PRV253 nucleocapsid DNA, and the desired recombinant virus was identified and plaque-purified by a β-galactosidase (β-gal) expression assay as previously described (Fig. 1) (15). PRV161 harbors a deletion in the Us9 gene, and was previously described (16).
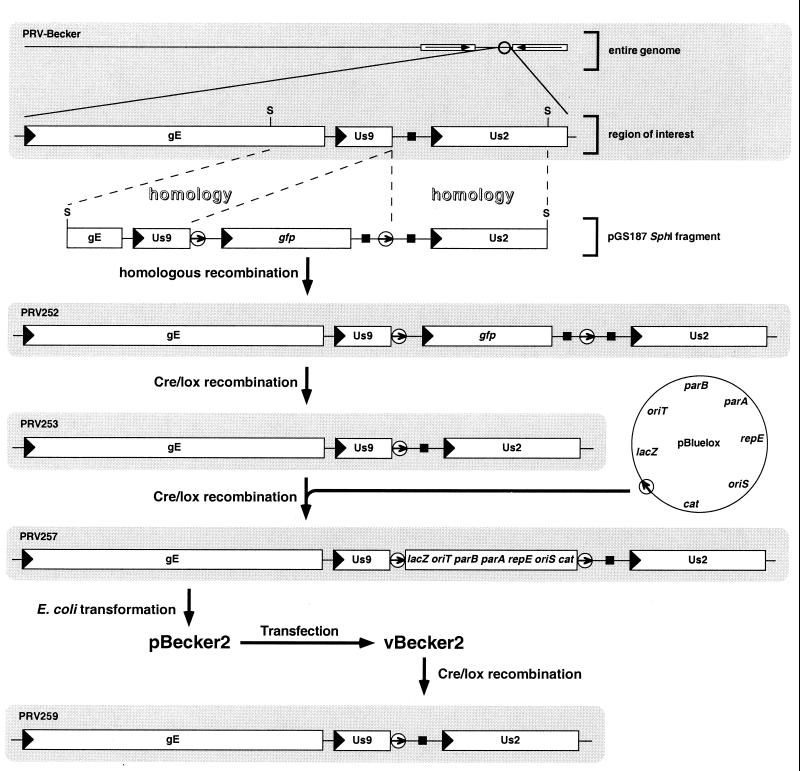
Figure 1.
Construction of the loxP recombinant virus and insertion of the pBluelox vector into the viral genome are illustrated. The PRV-Becker 142-kb genome is diagrammed at top, with a portion of the right end of the genome expanded to show the gE, Us9, and Us2 genes. For clarity, only this region of all subsequent viral genomes is illustrated. All recombinations were carried out in transfected PK15 cells. Transformation of PRV257 DNA into E. coli yielded the full-length clone, pBecker2. Transfection of pBecker2 back into PK15 cells yielded vBecker2, the latter of which is indistinguishable from PRV257. Viral genes are represented as open rectangles with an arrowhead denoting direction of transcription. Open circles represent loxP sites with the enclosed arrow indicating relative orientation. Polyadenylation sequences are denoted by solid squares, and viral inverted repeats are shown as open rectangles with enclosed arrows (additional notations: lacZ, β-galactosidase gene; cat, chloramphenicol-resistance gene; repE, parA, and parB, replication and partitioning genes; oriS, BAC origin of replication; oriT, origin of transfer; S, SphI site).
Viral DNA Isolation, Transfection, Transformation, and Electrophoresis.
The source of linear viral DNA used for cotransfections was nucleocapsids harvested from infected PK15 cells as previously described (8). Transfection of nucleocapsid DNA was by the calcium phosphate precipitation method as previously described (17). For cotransfections with the Cre-expression plasmid, ≈5 μg of pOG231 was included in the precipitation. For the cloning of pBecker2, replicative intermediate (covalently closed circular) PRV257 DNA was isolated and transformed into E. coli as previously described, except that Vero cells were infected and harvested 2 h postinfection (8). Examination of DNA by pulse-field electrophoresis was as previously described (8).
Western Blot Analysis.
Cell lysates from PK15 cells infected for 12 h were collected in TNX buffer [10 mM Tris (pH 7.4)/150 mM NaCl/1% Triton X-100]. Insoluble cell debris was removed by a 10-min microcentrifugation at 4°C. A mixture of 1.5 μl of supernatant (approximately equivalent to the lysate from 3,000 infected cells) and 1.5 μl of 2× final sample buffer [2% SDS/0.06 M Tris (pH 6.8)/10% glycerol/0.05% bromophenol blue/10% 2-mercaptoethanol) was boiled for 5 min, electrophoresed through a 12.5% polyacrylamide gel, and transferred to Hybond ECL membrane (Amersham Pharmacia). Western blot analysis was as previously described, except that a ECL + Plus detection kit was used (Amersham Pharmacia) (12). Signal intensities were quantitated with a Storm 860 FluorImager (Molecular Dynamics). The Us9 signal from PRV161 was subtracted from the Us9 signals of the other samples, correcting for background. The ratio of the corrected Us9 signal relative to the gI signal was expressed as percentages relative to PRV-Becker. Mouse monoclonal antibody 1A8 was isolated as a sibling clone to the previously described mouse monoclonal 2A2, both of which were raised to the PRV Us9 protein in a single BALB/c mouse (18). Rabbit antiserum RB1544, raised to the PRV gI protein, was described previously (19).
Results
Construction of a Full-Length PRV Clone by Cre/lox Recombination.
Establishing a full-length herpesvirus clone in E. coli first requires the insertion of a BAC/miniF vector into the viral genome. This is typically accomplished by targeted homologous recombination during infection in cell culture (3–5, 8, 20). We instead used Cre/lox site-specific recombination, which would allow for the BAC vector sequence to be removed by an additional Cre/lox reaction at a later time. To accomplish this, we first made a derivative of the wild-type PRV-Becker isolate with a single 34-bp loxP insertion in its genome. We chose to place the loxP site in a large intergenic region in the viral genome, with the intention that the insertion would not interfere with viral gene expression. However, as there was no direct method to identify the desired loxP recombinant virus during plaque purification, we began by inserting a GFP expression cassette flanked by two loxP sites (Fig. 1). The resulting virus, PRV252, was plaque-purified based on expression of GFP. The loxP sites in the PRV252 DNA were then recombined by cotransfection of the viral DNA with a Cre recombinase expression plasmid (see Materials and Methods), resulting in excision of the GFP cassette. One GFP-minus virus (PRV253) was plaque-purified, and the region around the insertion was sequenced. As expected, the gfp cassette was excised by Cre recombination, leaving behind a single loxP site immediately downstream the Us9 ORF (Fig. 2).
Figure 2.
Sequence of the loxP insertion from PRV253. A portion of the Us9 ORF is shown with translation immediately upstream the loxP site, and the codon encoding the start methionine of Us2 is included at the end of the sequence. The loxP site is underlined, and the polyadenylation consensus for the Us9 gene is double underlined.
A BAC vector, pBluelox, containing a single loxP site and a β-gal expression cassette was recombined into the loxP site of PRV253 by cotransfection into PK15 cells with a Cre-expression plasmid (Fig. 1). Approximately 10% of the resulting virus yield expressed β-gal, and one such virus was plaque-purified (PRV257). Circular viral DNA from PRV257-infected cells was isolated and used to transform E. coli strain DH10B (Research Genetics, Huntsville, AL). Of 18 chloramphenicol-resistant isolates screened by restriction digests and pulse-field electrophoresis, 17 carried the entire 142-kb PRV genome (data not shown). One clone, pBecker2, was examined further.
Transfection of pBecker2 and Cre Recombination.
Transfection of pBecker2 plasmid into PK15 cells resulted in productive viral infection, and 100% of the virus expressed β-gal (data not shown). Together, this indicated that pBecker2 was an infectious clone, and the pBluelox sequence was intact in the recombinant viral genome. Virus isolated from pBecker2 transfection was named vBecker2. Nucleocapsid DNA isolated from vBecker2 was digested with either HindIII or EcoRI and compared with digestions of PRV-Becker nucleocapsid DNA and pBecker2 by pulse-field gel electrophoresis (Fig. 3). The presence of 4.2-kb and 5.8-kb BAC-derived EcoRI fragments, and the replacement of the PRV-Becker 24-kb HindIII fragment with two smaller fragments confirmed vBecker2 to be derived from the pBecker2 clone. Furthermore, there was no evidence of vBecker2 isoforms that lacked any of the BAC restriction sites, unlike our previous observation for vBecker1, indicating the BAC vector was stable in the virus (8). The stability of the vBecker2 genome was confirmed by assaying vBecker2 serial passaged four rounds in PK15 cells for β-gal expression. Of ≈200 viral plaques examined, 100% expressed β-gal.
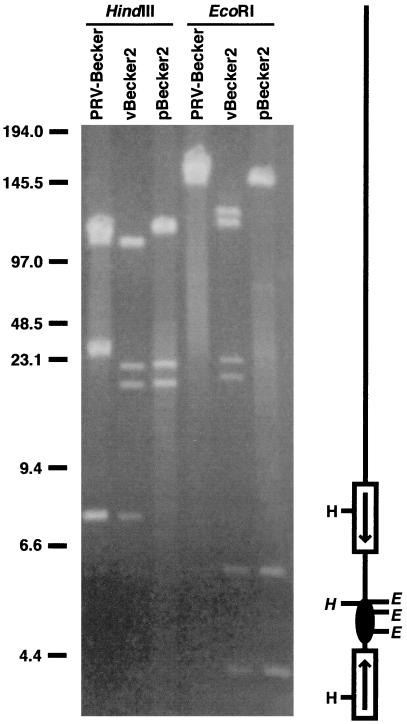
Figure 3.
Pulse-field gel electrophoresis of HindIII- and EcoRI-digested viral nucleocapsid (PRV-Becker and vBecker2) and E. coli plasmid (pBecker2) DNA. To the right of the gel is a diagram of the viral DNA, indicating positions of HindIII sites at left and EcoRI sites at right. The BAC vector is shown as a solid ellipse. Restriction sites within the BAC vector insertion are shown in italics, which includes one HindIII site and all three EcoRI sites, and are absent from PRV-Becker nucleocapsid DNA. The viral inverted repeats (shown as open rectangles with enclosed arrows) isomerize during viral replication in mammalian cells, as evident in the EcoRI digestion of vBecker2 (reviewed in ref. 23). The ends of the viral DNA are ligated together in the pBecker2 plasmid from E. coli. Size standards are indicated in kb.
Cotransfection of PK15 cells with pBecker2 with a Cre-expression plasmid resulted in a virus yield with 85–90% of viruses lacking β-gal activity because of excision of the DNA between the loxP sites. One such virus was plaque-purified and named PRV259 (Fig. 1).
Viral Gene Expression from vBecker2.
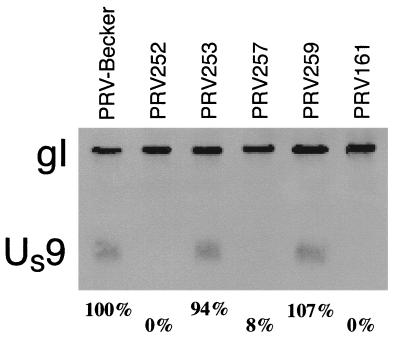
We were concerned that the loxP insertions may affect expression of the upstream Us9 gene. Expression of the Us9 gene was examined by Western blot analysis of infected PK15 cell lysates (Fig. 4). Expression of Us9 from either PRV253 and PRV259, both of which contain a single loxP site downstream Us9, was indistinguishable from that found after infection by the parental PRV-Becker strain. Us9 protein could generally not be detected from PRV252 or vBecker2 infections, which likely do not make stable Us9 transcripts because of their large insertions between the Us9 ORF and its downstream polyadenylation signal. Expression of the two genes immediately upstream Us9, gE, and gI, as well as the downstream Us2 gene was unaffected by a single loxP insertion (data not shown).
Figure 4.
Detection of Us9 expression in infected PK15 cells by Western blot analysis. Lysates were probed with an anti-Us9 monoclonal antibody, and reprobed with an anti-gI polyclonal antiserum. The latter provided a gel loading control. Percentages listed at bottom are quantitation of Us9 signal relative to gI signal, as described in Materials and Methods. The quantitation was an average of two gels, only one of which is shown. PRV161 harbors a deletion in the Us9 gene.
Plaque Formation by vBecker2.
Growth and spread of the recombinant viruses was assayed by plaque formation in confluent L2 cells (Table 1). Viruses containing a single loxP site formed normal-sized plaques. However, insertion of either the GFP cassette (PRV252) or BAC vector (PRV257) resulted in a notable decrease in plaque size.
Table 1.
Virus plaque size in L2 cells
| Virus | Average plaque diameter, %* |
|---|---|
| PRV-Becker | 100 (n = 145) |
| PRV252 | 72 (n = 103) |
| PRV253 | 92 (n = 53) |
| PRV257 | 84 (n = 42) |
| PRV259 | 100 (n = 43) |
| vBecker2 | 80 (n = 135) |
Diameters are expressed as a percentage relative to wild-type PRV-Becker, which is assigned a value of 100%. Number of plaques measured is listed in parentheses.
Insertion of a Cre-Expression Cassette into pBecker2.
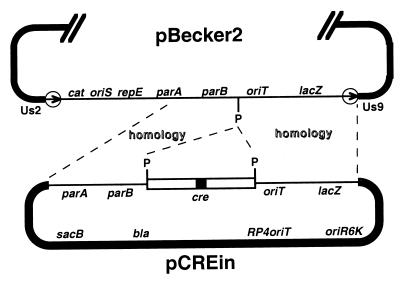
Lack of detectable Us9 expression and decreased plaque size confirmed that insertion of BAC vector sequences into a herpesvirus genome is detrimental to viral function (8). With the pBecker2 construct, these defects can be overcome by cotransfecting the clone with a Cre-expression plasmid, and subsequent excision of the BAC vector from the viral genome. However, cotransfection is not an ideal method as the recombinant virus lacking the BAC sequences must be plaque-purified over the course of several days. We therefore inserted a Cre-expression cassette into the BAC vector backbone of pBecker2 by homologous recombination in E. coli (Fig. 5). The BAC vector backbone provides a location for the insertion of foreign genes that will be delivered to mammalian cells with the viral genome, but will not affect viral function once removed by Cre-mediated recombination. Additionally, by expressing Cre recombinase from the infectious clone, excision of the BAC vector from the viral genome is forced to go to completion as any viral genome carrying the BAC vector insertion will also express the Cre recombinase. Unfortunately, loxP recombination occurred in E. coli when the cre ORF was recombined into pBecker2, indicating Cre protein was produced in the absence of any known prokaryotic promoter upstream the cre ORF (data not shown). To completely block Cre expression in E. coli, a synthetic 133-bp intron encoding four nonsense codons in-frame with the cre ORF was inserted in the middle of the cre ORF. The intron splice acceptor and donor sites were positioned such that splicing of the intron would reconstitute an in-frame cre ORF in eukaryotic cells. This Cre construct was crossed into the pBecker2 plasmid without any detectable recombination between the loxP sites, and one isolate containing the Cre-expression cassette was named pBecker3.
Figure 5.
Construction of pBecker3 by insertion of the intron-containing Cre-expression cassette into pBecker2 is illustrated. A portion of the 156-kb pBecker2 plasmid is shown above, with the BAC vector region and loxP sites (circled arrows) oriented down. The pCREin vector is shown below with the Cre cassette and homologous flanking regions, the latter derived from pBluelox, oriented up. Homologous recombination in E. coli between the two plasmids yielded pBecker3. The pGS284 allelic exchange vector, which is the source of the pCREin backbone, and the E. coli allelic exchange method are described in ref. 8 (notations are as in Fig. 1, with the addition of: sacB, levansucrase gene; bla, ampicillin-resistance gene; RP4oriT, origin of transfer; oriR6K, conditional origin of replication; P, PacI site).
Transfection of pBecker3 and vBecker3 Characterization.
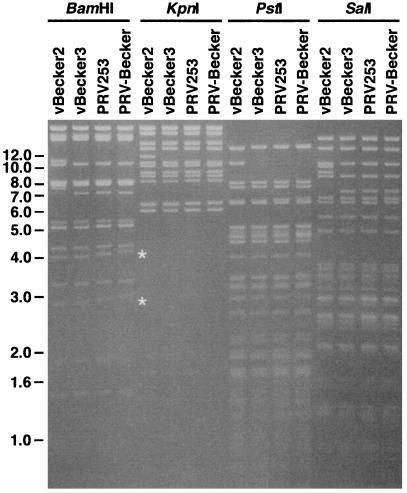
Plaque-forming virus harvested from PK15 cells transfected with pBecker3 did not express the β-gal marker associated with the BAC vector backbone (<0.1% of the virus resulting from transfection harbored the BAC vector sequence). Therefore, transfection of pBecker3 resulted in the production of virus lacking the BAC vector, vBecker3, without the need for further plaque purification or serial passage of the virus in culture. This was confirmed by restriction digest analysis with restriction enzymes that frequently cut PRV DNA (Fig. 6). Unlike vBecker2, which contains the pBluelox sequence, the only foreign DNA in vBecker3 is a single 34-bp loxP site; vBecker3 is identical to both PRV253 and PRV259.
Figure 6.
Gel electrophoresis of BamHI-, KpnI-, PstI-, and SalI-digested vBecker2, vBecker3, PRV253, and PRV-Becker viral nucleocapsid DNA. Restriction fragments unique to vBecker2 are all accounted for by the pBluelox insertion (data not shown). The restriction fragment length polymorphisms observed at ≈3.0 kb and ≈4.0 kb in the BamHI samples (marked by the white asterisks) map to the viral inverted repeats immediately flanking either side of the unique-short region, which contain small direct repeats that fluctuate in copy number during viral replication in mammalian cells (24, 25). Size standards are indicated in kb.
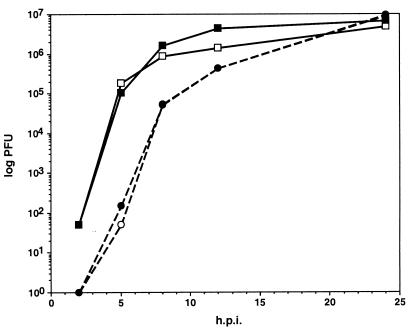
Single-step growth curves of vBecker3 were indistinguishable from the parental PRV-Becker wild-type isolate (Fig. 7). Furthermore, the neuroinvasiveness and neurovirulence of vBecker3 in a rat eye model of infection was identical to that previously described for PRV-Becker (data not shown) (21, 22).
Figure 7.
Single-step growth curves of wild-type PRV-Becker and vBecker3 in PK15 cells. Virus was harvested from both the media and cells at 2, 5, 8, 12, and 24 h postwash (squares, cells; circles, supernatants; open symbols, PRV-Becker; solid symbols, vBecker3).
Discussion
Full-length infectious clones of herpesviruses promise to be powerful tools to researchers studying the pathogenesis of this virus family. However, before this report, only one infectious clone had been made that results in a wild-type infection upon delivery of the plasmid DNA into mammalian cells (9). The BAC vector was excised from the viral genome by homologous recombination between a duplicated 527-bp viral sequence flanking the BAC vector in the viral genome. Complete excision of the BAC vector from the viral population required five serial passages of the virus in cell culture, and was suggested to be driven by a genome packaging size restriction intrinsic to the virus capsid volume. The 527-bp sequences will not mediate excision in recombination deficient E. coli. However, further modification or mutagenesis of the clone will be complicated by loss of the herpes DNA in E. coli that is recombination proficient.
We have described an alternative method for producing phenotypically wild-type herpesviruses by a self-excising BAC plasmid. Because self-excision of the BAC vector from the viral genome is by site-specific recombination mediated by the cre/lox system, there is no concern of undesired self-excision when using homologous recombination in E. coli. In addition, the stability of the BAC clone is further ensured by a synthetic intron inserted in the cre ORF, which specifically blocks Cre expression in E. coli.
As Cre recombinase is expressed whenever a virus with an intact BAC vector infects a mammalian cell, excision of the BAC vector from an entire viral population is expected to be rapid and complete. Indeed, Cre-promoted excision of the BAC vector from vBecker3 goes to completion after only one round of viral growth in cultured cells. Therefore, serial passage or plaque purification of the recombinant virus is not required. In addition, nonspecific deletions that could result from viral capsid packaging limitations of oversized genomes should not occur because the self-excision is rapid and independent of viral function. Importantly, although vBecker3 contains a single loxP site, we see no effect of this 34-bp insertion on neighboring viral gene expression, viral growth, viral spread, or viral pathogenesis in any of our assays.
The self-excising BAC plasmid can be used to make infectious clones of any herpesvirus that have first been modified to carry a loxP site in the viral genome. The only requirement is that the loxP site be positioned in the viral genome such that it does not interfere with viral gene expression. This deserves some consideration, as herpesvirus genomes are very compact with few regions that are not transcribed (23). We chose to place the loxP site in PRV-Becker in a relatively large intergenic space between the Us9 and Us2 genes. By positioning the loxP site immediately adjacent to the Us9 ORF, and upstream the Us9 polyadenylation sequence, we avoided interference with the downstream Us2 promoter. The loxP site is therefore likely to be transcribed along with the Us9 ORF. Whereas a loxP sequence could destabilize the Us9 transcript by introducing a small stem-loop structure in the 3′ end of the mRNA, we observed no decrease in Us9 expression during infection of PK15 cells. This strategy of loxP placement should be amenable for any herpesvirus genome, and the application of this method and the pBluelox and pCREin plasmids should simplify the construction of wild-type encoding herpesviruses.
Acknowledgments
We thank Audrey Liu for her excellent help with portions of the subcloning work, Amy Brideau for providing antibody 1A8 and the PRV161 virus, and Rebecca Tirabassi, Jane Flint, and Tom Silhavy for insightful discussions concerning the experimental approach. This work was supported by National Institute of Neurological Disorders and Stroke Grant 1RO133506 (to L.W.E.). G.A.S. is a Lilly Fellow of the Life Sciences Research Foundation.
Abbreviations
- BAC
bacterial artificial chromosome
- PRV
pseudorabies virus
- MCMV
mouse cytomegalovirus
- β-gal
β-galactosidase
- GFP
green fluorescent protein
- PK15
porcine kidney 15 cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.080502497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.080502497
References
- 1.O'Connor M, Peifer M, Bender W. Science. 1989;244:1307–1312. doi: 10.1126/science.2660262. [DOI] [PubMed] [Google Scholar]
- 2.Shizuya H, Birren B, Kim U-J, Mancino V, Slepak T, Tachiiri Y, Simon M. Proc Natl Acad Sci USA. 1992;89:8794–8797. doi: 10.1073/pnas.89.18.8794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Delecluse H J, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. Proc Natl Acad Sci USA. 1998;95:8245–8250. doi: 10.1073/pnas.95.14.8245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Horsburgh B C, Hubinette M M, Qiang D, MacDonald M L, Tufaro F. Gene Ther. 1999;6:922–930. doi: 10.1038/sj.gt.3300887. [DOI] [PubMed] [Google Scholar]
- 5.Saeki Y, Ichikawa T, Saeki A, Chiocca E A, Tobler K, Ackermann M, Breakefield X O, Fraefel C. Hum Gene Ther. 1998;9:2787–2794. doi: 10.1089/hum.1998.9.18-2787. [DOI] [PubMed] [Google Scholar]
- 6.Stavropoulos T A, Strathdee C A. J Virol. 1998;72:7137–7143. doi: 10.1128/jvi.72.9.7137-7143.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suter M, Lew A M, Grob P, Adema G J, Ackermann M, Shortman K, Fraefel C. Proc Natl Acad Sci USA. 1999;96:12697–12702. doi: 10.1073/pnas.96.22.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith G A, Enquist L W. J Virol. 1999;73:6405–6414. doi: 10.1128/jvi.73.8.6405-6414.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner M, Jonjic S, Koszinowski U H, Messerle M. J Virol. 1999;73:7056–7060. doi: 10.1128/jvi.73.8.7056-7060.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Card J P, Rinaman L, Schwaber J S, Miselis R R, Whealy M E, Robbins A K, Enquist L W. J Neurosci. 1990;10:1974–1994. doi: 10.1523/JNEUROSCI.10-06-01974.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tirabassi R S, Enquist L W. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams D R, Young D I, Young M. J Gen Microbiol. 1990;136:819–826. doi: 10.1099/00221287-136-5-819. [DOI] [PubMed] [Google Scholar]
- 14.O'Gorman S, Dagenais N A, Qian M, Marchuk Y. Proc Natl Acad Sci USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp A C, Enquist L W. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brideau A D, Card J P, Enquist L W. J Virol. 2000;74:834–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tirabassi R S, Enquist L W. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brideau A D, del Rio T, Wolffe E J, Enquist L W. J Virol. 1999;73:4372–4384. doi: 10.1128/jvi.73.5.4372-4384.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H-J, Enquist L W. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Messerle M, Crnkovic I, Hammerschmidt W, Ziegler H, Koszinowski U H. Proc Natl Acad Sci USA. 1997;94:14759–14763. doi: 10.1073/pnas.94.26.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 22.Card J P, Dubin J R, Whealy M E, Enquist L W. J Neurovirol. 1995;1:349–358. doi: 10.3109/13550289509111024. [DOI] [PubMed] [Google Scholar]
- 23.Roizman B, Sears E. In: Fundamental Virology. Fields B N, Knipe D M, Howley P M, editors. Philadelphia: Lippincott-Raven; 1996. pp. 1043–1107. [Google Scholar]
- 24.Zhang G, Leader D P. J Gen Virol. 1990;71:2433–2441. doi: 10.1099/0022-1317-71-10-2433. [DOI] [PubMed] [Google Scholar]
- 25.van Zijl M, van der Gulden H, de Wind N, Gielkens A, Berns A. J Gen Virol. 1990;71:1747–1755. doi: 10.1099/0022-1317-71-8-1747. [DOI] [PubMed] [Google Scholar]