Abstract
Polyphosphate kinase (PPK), encoded by the ppk gene, is the principal enzyme in many bacteria for the synthesis of inorganic polyphosphate (poly P) from ATP. A knockout mutant in the ppk gene of Pseudomonas aeruginosa PAO1 is impaired in flagellar swimming motility on semisolid agar plates. The mutant is deficient in type IV pili-mediated twitching motility and in a “swarming motility” previously unobserved in P. aeruginosa. In swarming cultures, the polar monotrichous bacteria have differentiated into elongated and polar multitrichous cells that navigate the surface of solid media. All of the motility defects in the ppk mutant could be complemented by a plasmid harboring the ppk gene. Because bacterial motility is often crucial for their survival in a natural environment and for systemic infection inside a host, the dependence for motility on PPK reveals important roles for poly P in diverse processes such as biofilm formation, symbiosis, and virulence.
Inorganic polyphosphate (poly P) is a linear chain of tens or many hundreds of phosphate residues linked by high-energy phosphoanhydride bonds. It is found in every cell in nature: bacterial, archaeal, fungal, protozoan, plant, and animal (1, 2). Poly P has numerous and varied biological functions depending on where it is (species, cell, or subcellular compartment) and when it is needed. Among these functions are substitution for ATP in kinase reactions; reservoir of phosphate; chelation of divalent metals; capsule of bacteria; and regulatory roles in growth, development, stress, and deprivation (1, 2). In our studies of Escherichia coli, the most significant function observed thus far is its regulatory role in adapting to nutritional stringencies and environmental stresses, and for survival in the stationary phase of growth (3). This role has been inferred from the behavior of mutant cells lacking polyphosphate kinase (PPK), the enzyme responsible for the synthesis of poly P from ATP (4).
Motility is arguably one of the most impressive features in microbial physiology. Movement in aqueous environments by swimming or along surfaces by using different modes of translocation has been classified into several distinct forms (5). Swimming on a surface takes place when the fluid film is sufficiently thick and the micromorphological pattern is unorganized. When the fluid layer on a surface is relatively thin, the swimming bacteria become elongated and hyperflagellated and move in a coordinated manner known as “swarming” (5–7). Twitching motility is another form of translocation on a solid surface in which the micromorphological pattern is less organized than in swarming (5, 8). Among these three modes of surface translocation, swimming and swarming depend on flagella, whereas twitching depends on type IV pili (5).
These various forms of surface motility enable bacteria to establish symbiotic and pathogenic associations with plants and animals (9–11). Potential benefits of motility include increased efficiency of nutrient acquisition, avoidance of toxic substances, ability to translocate to preferred hosts and access to optimal colonization sites within them, and dispersal in the environment during the course of transmission. The cost of motility is significant considering the metabolic burden of synthesizing and assembling various flagellar and pili components and the energetic expense of fueling flagellar and presumably pili motors (12, 13). Thus, the synthesis and control of movement through the chemotactic signal transduction system of the motility apparatus are subject to strict control mechanisms that are usually redundant and multilayered (12, 13).
Pseudomonas aeruginosa is a Gram-negative, rod-shaped, motile bacterium that contains a single flagellum and several type IV pili, all usually located at the same pole. It is a highly versatile organism that survives in a wide variety of environments, and causes diseases in insects, plants, and animals. In humans, it is an opportunistic pathogen causing a variety of infections in immunocompromised hosts such as patients with cystic fibrosis, burns, cancer, and those requiring extensive stays in intensive care units (14). The essential roles of flagella and type IV pili and of motility and chemotaxis at various stages throughout the infectious cycle in pulmonary and burn infections have been described in detail (15–20).
We reported that poly P and PPK are required for flagella-mediated swimming motility of bacterial pathogens, including P. aeruginosa, Klebsiella pneumoniae, Vibrio cholerae, Salmonella typhimurium, and Salmonella dublin on semisolid agar plates (21). In this report, we show that the ppk mutant of P. aeruginosa is also deficient in type IV pili-mediated twitching and a newly discovered swarming motility. We also discuss the plausible mechanisms of poly P and PPK action on motility and how these findings impinge on the pathogenicity of P. aeruginosa.
Materials and Methods
Bacterial Strains and Plasmids.
The P. aeruginosa strains used in this study are as follows: wild type PAO1, PAOM5 [Δppk∷tetracycline resistance (TcR)], PAO-D (ΔfliD∷gentamycin resistance), PAO-NP (ΔpilA∷TcR), MS159 (ΔfliC∷gentamycin resistance), and PA14-fliF [ΔfliF∷Tn5 (TcR)]. The PAO-D, PAO-NP, and MS159 strains are gifts from S. K. Aurora and R. Ramphal (University of Florida, Gainesville, FL), and PA14-fliF from G. A. O'Toole and R. Kolter (Harvard University, Boston). Plasmid pHEPAK11 contains the P. aeruginosa ppk gene under tacP control and was derived from the E. coli-P. aeruginosa shuttle vector pMMB66HE (22).
Construction of the ppk Knockout Strain.
The ppk gene was isolated by PCR amplification by using the forward primer 5′-GCGAAGCTTTCCCTTACCGCCTTCAAACG-3′ (GCG clamp in italics, HindIII site in bold, followed by a 20-mer stretch of DNA starting at 246 bp upstream of the GTG translational start codon of the ppk gene) and the reverse primer 5′-GCGTCTAGAGCAGAGCCCAAGAGCCTTTC-3′ (GCG clamp in italics, XbaI site in bold, followed by a 20-mer stretch of DNA starting at 99 bp downstream of the TGA stop codon of the ppk gene). The primers were designed according to the published sequence of the ppk locus of the P. aeruginosa chromosome (23, 24). The 2.43-kb PCR product was cloned into the HindIII/XbaI site of pBluescript II SK (+/−). A 0.8-kb BamHI fragment from the interior of the ppk gene was exchanged with a tetracycline-resistance (TcR) cassette as a 1.3-kb BglII fragment. The pBluescript II SK (+/−) vector containing the ppk gene knockout cassette was used to transform P. aeruginosa PAO1 and TcR transformants were selected. DNA was isolated from individual transformants and PCR analysis was used to check the replacement of the wild-type gene by a double homologous recombination event in the ppk locus.
Motility Assays.
Swimming.
Media used for assay was tryptone broth [10 g/liter tryptone (Difco)/5 g/liter NaCl] that contained 0.3% (wt/vol) agarose (GIBCO/BRL). Swim plates were inoculated with bacteria from an overnight culture in LB agar (1.5%, wt/vol) plates at 37°C with a sterile toothpick. The plates were then wrapped with Saran Wrap to prevent dehydration and incubated at 30°C for 12–14 h.
Swarming.
Media used for assay consisted of 0.5% (wt/vol) Difco bacto-agar with 8 g/liter Difco nutrient broth, to which 5 g/liter glucose was added. Swarm plates were typically allowed to dry at room temperature overnight before being used. Swarming efficiency was improved when cells were inoculated onto swarm plates from swim agar (0.3%, wt/vol) plates incubated overnight at 30°C; inoculation from overnight LB agar (1.5%, wt/vol) plates also supported swarming. Other media and agar such as Eiken broth and Eiken agar (Eiken Chemical, Tokyo), tryptone and LB broths, and Difco granulated agar could also be used for the swarming response.
Twitching.
Assays were by two methods with Difco LB broth (10 g/liter tryptone/5 g/liter yeast extract/10 g/liter NaCl) solidified with 1% (wt/vol) Difco granulated agar. (i) Stab assay. Twitch plates were briefly dried and strains were stab inoculated with a sharp toothpick to the bottom of the Petri dish from an overnight-grown LB agar (1.5%, wt/vol) plate. After incubation at 37°C for 24 h, the zone of motility at the agar/Petri dish interface was measured. (ii) Slide-culture assay. Strains were point inoculated with a toothpick onto the surface of a slab of LB agar (1%, wt/vol) placed on a microscope slide. The inoculated LB agar was then covered with a glass cover slip, and the slide cultures were incubated for 4–5 h at 37°C in a Petri dish.
Microscopy.
Bacteria were examined for general cell morphology and motility by phase-contrast optics with a Zeiss Axiophot2 microscope with a X40 lens objective. Samples were prepared either from plate or liquid culture as wet mounts in PBS. Bacteria examined by electron microscopy to confirm cell morphology and the presence of flagella and pili were prepared as described below. Carbon-coated grids were gently placed on the surface of the colony on plates. After 2 min, the grids were carefully removed, rinsed twice with distilled water, and stained with 1% uranyl acetate. The negatively stained cells were visualized with a CM-12 transmission electron microscope (FEI, Hillsboro, OR).
Results
The remarkably high conservation of the PPK sequence among many bacteria including some of the major pathogens prompted the creation of a ppk knockout mutation in P. aeruginosa PAO1 to examine its phenotypic features including virulence (21, 25). In the ppk gene cloned by PCR, a portion was replaced with a TcR cassette. This knockout cassette was integrated into the ppk locus by homologous recombination and verified by genomic PCR analysis. At the biochemical level, inactivation of the ppk gene was confirmed by a loss of PPK activity and a decrease in poly P accumulation (data not shown). Because motility is intimately involved in survival, symbiosis, and virulence of bacteria (9–11), the effects of the mutation on various forms of motility were examined.
PPK Is Required for Swimming Motility on Semisolid Agar.
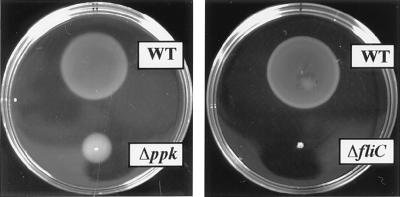
In liquid media, P. aeruginosa cells are motile rods with a single, polar flagellum. Characteristic of polarly flagellated bacteria, they reverse the flagellar rotation to “back-up” to swim in a new direction (26). In semisolid agar (0.2–0.4%) plates, cells swim through water-filled channels to create concentric chemotactic rings. As reported (21), the ppk mutant is impaired in swimming motility only in semisolid agarose plates despite possessing an apparently normal flagellum (Fig. 1). This failure in flagellar function results in a swimming defect that is less drastic than that seen in a nonmotile strain with a knockout mutation in a flagellin structural gene fliC (Fig. 1).
Figure 1.
Swimming motility of P. aeruginosa PAO1, its ppk mutant, and a fliC control strain derived from a P. aeruginosa PAK strain on a semisolid agarose plate. Cells were inoculated with a toothpick from an overnight LB agar plate onto a swim plate (tryptone broth plus 0.3% agarose) and photographed after a 14-h incubation at 30°C. Strains: WT, PAO1; Δppk, PAOM5; and ΔfliC, MS159.
Swarming Motility in P. aeruginosa and the Requirement for PPK.
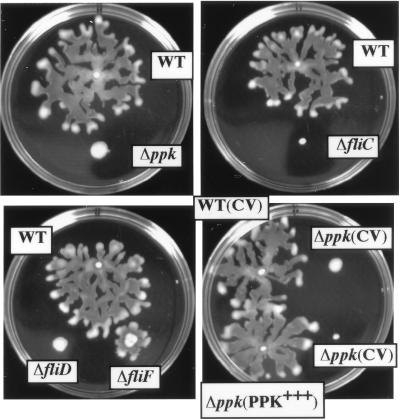
Surface translocation on the surface of agar 0.45% or more in concentration supports a swarming motility among some bacteria that is clearly distinguishable from swimming (5–7). We observe that P. aeruginosa also possess such a swarming motility. When inoculated on 0.5% agar at 30°C, wild-type (WT) PAO1 cells begin to grow as a colony with morphology similar to that seen at the usual agar concentration (1.5%). Irregular branching begins to appear at the periphery when the colony grows to 2–4 mm in diameter in 12–24 h depending on the medium and the agar (Fig. 2). Generally, one of the extensions elongates into a mucoid fluid tendril that moves directly away from the central colony. Other tendrils develop both from this initial stream or the central colony, eventually creating a characteristic dendritic pattern of branching fluid streams; the slime-like liquid gives the front a glistening appearance. Movement across the agar surface is rather rapid and the bacteria colonize the entire surface within several hours after the initiation event.
Figure 2.
Swarming motility in P. aeruginosa PAO1. The ppk mutant is completely deficient in swarming motility which could be restored to the WT level by providing a plasmid bearing the ppk gene. Cells were inoculated onto swarm plates (Difco nutrient broth plus 0.5% Difco bacto-agar supplemented with 0.5% glucose) from an overnight swim plate with a toothpick and photographed after a 24-h incubation at 30°C. Strains: WT, PAO1; Δppk, PAOM5; ΔfliC, MS159; ΔfliD, PAO-D; ΔfliF, PA14-fliF; WT (CV), PAO1 (pMMB66HE); Δppk (CV), PAOM5 (pMMB66HE); and Δppk (PPK+++), PAOM5 (pHEPAK11). CV represents the control vector pMMB66HE and PPK+++ the PPK-overproducing plasmid pHEPAK11.
The source of the agar and the composition of the medium greatly affect the swarming. Difco bacto-agar and Eiken agar consistently supported a strong swarming response, whereas Difco granulated agar never did. The optimal agar concentration to elicit swarming was 0.45–0.5%, whereas 0.6% was inhibitory. Eiken broth and Difco nutrient broth were better than LB and tryptone broth. In the latter two media, initiation of the swarming response was usually slower by 10–20 h and the morphology of the swarming colony front was concentric rather than dendritic (data not shown).
The ppk mutant is deficient in swarming motility with all media and agars tested (Fig. 2). The irregular branching that signals the initiation of swarming was never observed. Complementation of the mutant with plasmid pHEPAK11 containing the ppk gene restored normal swarming (Fig. 2). Because swarming motility is flagella dependent in other bacteria, we examined mutants lacking components of the flagellar apparatus. Mutations in the filament structural protein flagellin gene fliC and cap protein gene fliD abolished the swarming response whereas with a mutation in the basal-body MS (membrane/supramembrane) ring protein gene fliF, some swarming was retained (Fig. 2). Mutation in the type IV pili structural subunit pilin gene, pilA, did not affect swarming (data not shown). Thus, swarming motility in P. aeruginosa is a flagellum-dependent process.
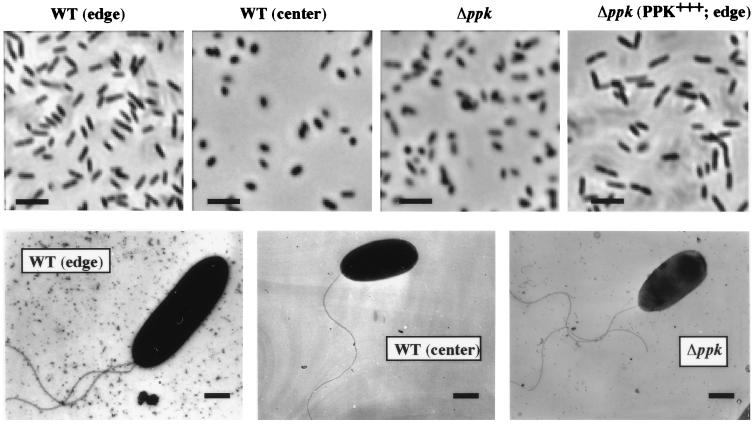
Differential cell elongation accompanied by hyperflagellation has been described as nearly universal for swarmer cells (6, 7, 27). These swarmer cells cooperatively migrate over surfaces en masse at the colony edge or as rafts of cells that temporarily leave the colony behind. In P. aeruginosa, as in other bacteria, surface migration at the colony periphery was generally en masse with rafts of cells that migrated away from the colony forefront preceded by a shiny mucoid layer which presumably consists of the exopolysaccharide, alginate, and/or the biosurfactant, rhamnolipid. Cells isolated from the periphery of a swarming colony appeared elongated compared with cells isolated either from the center of the colony or from the ppk mutant colony on the swarm plate (Fig. 3 Upper). Cells from the periphery of the swarming colony of the ppk mutant harboring the pHEPAK11 plasmid were elongated as were the cells from the periphery of the WT colony. Electron microscopy confirmed the morphology observed by phase-contrast microscopy (Fig. 3 Lower). WT edge cells are about twice as elongated as the WT center cells and the ppk mutant cells. The elongated WT edge cells often possessed two or more polar flagella (50% in a random count of 28 cells) as opposed to a single polar flagellum in WT center and ppk mutant cells (Fig. 3 Lower). Under these conditions, type IV pili could not be detected in the swarming cells by electron microscopy (data not shown).
Figure 3.
Morphology of P. aeruginosa swarm cells by (Upper) phase-contrast and (Lower) electron microscopy. (Upper) Images of cells suspended in PBS from swarm colonies (X50 magnification). (Lower) Electron micrographs of bacteria taken directly from swarm plates (×10,400.) Strains: WT (edge), edge cells of PAO1 colony; WT (center), center cells of PAO1 colony; Δppk, cells from PAOM5 colony; and Δppk (PPK+++; edge), edge cells of PAOM5 colony containing pHEPAK11 plasmid. [Bars = 5 μm (Upper) and 0.5 μm (Lower).]
PPK Is Required for Twitching Motility.
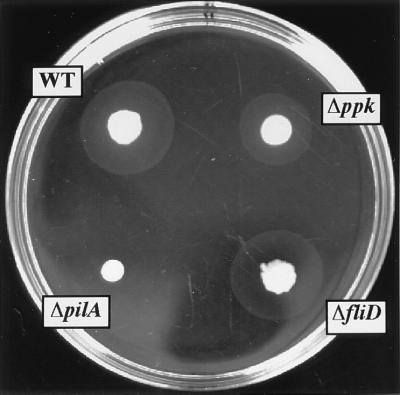
P. aeruginosa exhibit twitching motility, a form of translocation on a solid surface dependent on type IV pili (5, 8). The ppk mutant is deficient not only in flagella-mediated swimming and swarming (see above) but also in twitching motility. When cells are stabbed through an agar layer to the bottom of the Petri dish, colony expansion at the interstitial surface between the agar and the plastic occurs by twitching motility (5, 8). The interstitial twitch zone formed by colony expansion is reduced in the case of the ppk mutant compared with WT cells even though the surface colonies on top of the agar are similar in size and shape (Fig. 4). As expected, the pilA mutant showed no twitching zone unlike the fliD mutant that did in this macroscopic assay (Fig. 4). As with swimming (21) and swarming (Fig. 2) motility defects, the ppk mutant can be restored to essentially WT levels of twitching motility by complementation with a plasmid bearing the ppk gene (Table 1).
Figure 4.
Macroscopic stab assay for twitching motility. Cells from an overnight LB agar (1.5%, wt/vol) plate were inoculated with a toothpick to the bottom of the LB agar (1%, wt/vol) plate and incubated for 24 h at 37°C. The diffuse interstitial zone is a measure of twitching motility. The denser and smaller zone represents surface colony growth. Strains: WT, PAO1; Δppk, PAOM5; ΔfliD, PAO-D; and ΔpilA, PAO-NP.
Table 1.
Type IV pili-mediated twitching motility of P. aeruginosa strains
| Strain | Relevant genotype | Twitch area, % WT ± SEM* |
|---|---|---|
| PAO1 | WT | 100 ± 4 |
| PAOM5 | Δppk | 63 ± 2 |
| PAO-D | ΔfliD | 93 ± 4 |
| PAO-NP | ΔpilA | 0 |
| PAO1 (pMMB66HE) | WT + vector | 100 ± 5 |
| PAOM5 (pMMB66HE) | Δppk + vector | 55 ± 2 |
| PAOM5 (pHEPAK11) | Δppk + PPK+++ | 97 ± 4 |
Twitch area was measured after a 24-h incubation at 37°C
on LB agar (1%, wt/vol) plates. The standard error of the mean (SEM)
equals σn−1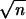 , where
n = 6.
, where
n = 6.
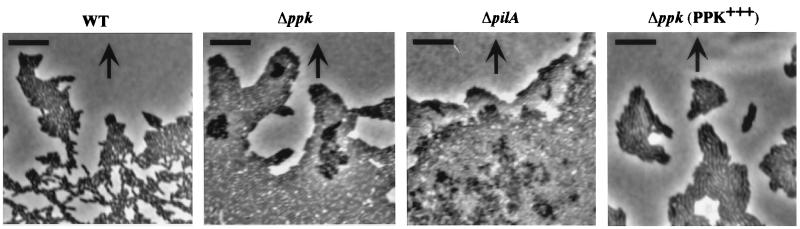
Microscopic analysis of twitching motility by a slide-culture assay revealed the leading edge of the WT twitching zone to be made up of large rafts of ≈10–50 cells moving away from the colony edge; the cells are monolayered, characteristic of twitching motility in this bacterium. Behind these rafts, a strikingly intricate lattice-work pattern is formed comprised of structures consisting of only one to five cells in width (Fig. 5). The ppk mutant cells were motile (Fig. 4), formed rafts (Fig. 5), and possessed apparently normal type IV pili as examined by electron microscopy (data not shown). However, the rafts of the ppk mutant contained very large numbers of cells, were often multilayered, and the highly structured fine lattice-like network behind the rafts of microcolonies was not observed (Fig. 5). (The pilA mutant that lacks the type IV pili exhibits neither raft nor lattice formation.) When the ppk mutant was complemented with the ppk gene, the twitching features of WT cells were restored.
Figure 5.
Light microscopy of zones of twitching motility by slide-culture assay. The strains were inoculated onto slide cultures and typical colony expansion zones were obtained at the interstitial surface between glass cover slip and medium after 3–5 h incubation at 37°C. Strains: WT, PAO1; Δppk, PAOM5; ΔpilA, PAO-NP; and Δppk (PPK+++), PAOM5-harboring plasmid pHEPAK11. The bar is 10 μm and the arrow indicates the radial direction of colony expansion.
Discussion
Of the many functions of poly P (1, 2, 28–30), our studies with E. coli suggest that regulatory controls are among the most significant (3, 4, 31). The E. coli mutant lacking PPK, and thus diminished in poly P, fails to activate the major regulatory gene rpoS that encodes the sigma factor that prepares cells to survive in the stationary phase (31). The mutant, deficient in adapting to nutritional stringencies and environmental stresses, fails to survive (3). In some bacterial pathogens, a defect in swimming motility is also manifested in ppk mutants lacking poly P (21). We have extended those observations to demonstrate that poly P and/or PPK control the swarming and twitching as well as the swimming motility of P. aeruginosa.
Two forms of surface motility (swimming by a polar monotrichous flagellum and twitching by polar multitrichous pili) are well-documented in P. aeruginosa (26, 32). Swarming, another form of flagella-dependent surface motility, has been described in Proteus mirabilis, Vibrio parahaemolyticus, and Serratia mar-cescens (6, 27, 33), and more recently in E. coli, S. typhimurium, Serratia liquefaciens, Yersinia enterocolitica, and Pseudomonas syringae (34–37). We have now discovered that under appropriate conditions a similar swarming response occurs in P. aeruginosa, which, as expected, is lacking in the ppk mutant (Fig. 2).
The common characteristics of swarming cells are: (i) elongation accompanied by multiflagellation; (ii) movement in a coordinated fashion, either en masse at the colony edge or as rafts of migrating cells temporarily leaving the colony behind; and (iii) production of extracellular slime consisting of polysaccharide and/or biosurfactant (5–7, 33–37). Such multiflagellated strains of P. aeruginosa had been observed earlier but their capacity to swarm was not investigated (38). We now find that P. aeruginosa swarm cells elongate and multiflagellate polarly, produce the extracellular polysaccharide, alginate, and/or the biosurfactant, rhamnolipid, and move coordinately. Swarming differs from twitching in that swarm cells do not synthesize type IV pili, are unaffected in a pilA mutation, and depend on their flagella.
Swarming bacteria differ in that some are strong swarmers like P. mirabilis and V. parahaemolyticus. These flagellate profusely (up to 50-fold) during swarming, and show medium-independent and agar-independent swarming even on a 2% agar surface (6, 27). Other bacteria, such as S. marcescens, S. liquefaciens, E. coli, S. typhimurium, and Y. enterocolitica, are weak swarmers. They flagellate moderately (two- to threefold) during swarming, and show medium-dependent and agar-dependent swarming on a limit of 0.8% agar surface (7, 35, 36, 39). P. aeruginosa falls into the group of weak swarmers, but unlike all other swarmers, is polarly flagellated and not peritrichous.
Besides flagella-dependent swimming and swarming motility, the ppk mutant is also defective in type IV pili-mediated twitching motility despite possessing apparently normal pili. This behavior of the ppk mutant is similar to that of the pilH mutant that expresses surface pili but is impaired in twitching motility (40). The coordinate regulation of the synthesis of alginate and accumulation of poly P in P. aeruginosa by the AlgR2 pleiotropic regulatory protein (41) recalls the control by another alginate regulatory protein AlgR1 of twitching motility in addition to alginate synthesis (42). The stationary phase sigma factor RpoS is required for swimming and twitching motility as well as alginate production in P. aeruginosa (43). Because poly P and/or PPK are required for the induction of rpoS gene expression in E. coli (31), and assuming the same might be true in P. aeruginosa, the swimming, swarming, and twitching motility defects in the ppk mutant may be explained on the same basis.
Because the ppk mutants possess apparently normal flagella and pili under conditions favorable for swimming and twitching, it would appear that the defects are at the functional rather than structural levels. Yet in swarming, the mutant cells do fail to make an additional flagellum that may be caused by a biosynthetic defect.
Defects in various forms of motility because of abnormal function of flagella and/or pili could arise from defects in chemotaxis (chemosensing through the chemoreceptors) and/or the cytoplasmic signal transduction system (13). CheY, the response regulator of the two-component chemotactic signal transduction system, is the central control site of signal transduction in bacterial chemotaxis. The relative level of phospho-CheY regulates the direction of flagellar rotation, either clockwise or counterclockwise, by interacting with the switch complex at the base of the flagellar motor (13).
For the reversal of flagellar rotation in swimming, chemotaxis as well as the chemotaxis signal transduction system are essential (12, 13). In E. coli, the signal transduction system is essential for swarming motility but traditional chemotaxis is not (44). Recently, it has been proposed that the twitching motility of P. aeruginosa and the social gliding motility of Myxococcus xanthus are essentially the same type IV pili-based process (45). Chemotaxis and the chemotactic signal transduction system operating through the Frz proteins, which are homologous to enteric Che proteins, are required for the WT pattern of social gliding motility in M. xanthus (46). In a search for genes affecting twitching motility in P. aeruginosa, two cheY homologues, pilG and pilH, were found which do not affect swimming (39, 40). For reversing the flagellar rotation in swimming, a different cheY homologue is involved (47). P. aeruginosa possesses at least two distinct chemotaxis systems: one controls flagellar swimming and presumably swarming; the other supports type IV pili-based twitching.
How might poly P and/or PPK affect motility through chemotaxis and the signal transduction system? Poly P might substitute for ATP in CheY phosphorylation or phospho-PPK might directly transfer phosphate to some CheY-like proteins (phosphorylation by crosstalk), thus affecting the flagellum/pilus operations at a functional level (48). Poly P might also interfere with the cellular Ca2+ level to affect the activity of CheY-like protein(s) (48) or might act directly on the flagellar motor (49). In fact, poly P granules have been found at the base of the flagella in Helicobacter pylori which causes chronic gastritis and peptic ulcers in humans (50).
Consistent with the abnormal function of flagella caused by improper chemotaxis and the reduced virulence in S. typhimurium, Vibrio anguillarum, and Campylobacter jejuni (51–53), the ppk mutant of P. aeruginosa has been found to be avirulent in a burned-mouse pathogenesis model (A. N. Hamood, personal communication), even though the mutant possesses normal flagella and pili.
Acknowledgments
We thank S.-K. Chiu for help and advice with phase-contrast microscopy, N. Ghori for performing the electron microscopy, S. K. Aurora, R. Ramphal, G. A. O'Toole, and R. Kolter for gifts of bacterial strains, A. Kuroda for a gift of plasmids and reagents, A. N. Hamood for sharing unpublished results, and D. Kaiser, G. Schoolnik, and L. Bertsch for critical readings of the manuscript. These studies were supported by a grant from the National Institutes of Health.
Abbreviations
- poly P
inorganic polyphosphate
- PPK
polyphosphate kinase
- WT
wild type
- TcR
tetracycline resistance
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060030097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060030097
References
- 1.Kulaev I S. The Biochemistry of Inorganic Polyphosphates. New York: Wiley; 1979. [DOI] [PubMed] [Google Scholar]
- 2.Kornberg A, Rao N N, Ault-Riche D. Annu Rev Biochem. 1999;68:89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 3.Rao N N, Kornberg A. In: Progress in Molecular and Subcellular Biology. Schroder H C, Muller W E G, editors. Vol. 23. Heidelberg: Springer; 1999. pp. 183–195. [DOI] [PubMed] [Google Scholar]
- 4.Rao N N, Kornberg A. J Bacteriol. 1996;178:1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henrichsen J. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Allison C, Hughes C. Sci Prog. 1991;75:403–422. [PubMed] [Google Scholar]
- 7.Harshey R M. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 8.Henrichsen J. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 9.Ottemann K M, Miller J F. Mol Microbiol. 1997;24:1109–1117. doi: 10.1046/j.1365-2958.1997.4281787.x. [DOI] [PubMed] [Google Scholar]
- 10.Moens S, Vanderleyden J. Crit Rev Microbiol. 1996;22:67–100. doi: 10.3109/10408419609106456. [DOI] [PubMed] [Google Scholar]
- 11.Wall D, Kaiser D. Mol Microbiol. 1999;32:1–10. doi: 10.1046/j.1365-2958.1999.01339.x. [DOI] [PubMed] [Google Scholar]
- 12.Macnab R M. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 123–145. [Google Scholar]
- 13.Stock J B, Surette M G. In: Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. Neidhardt F C, Curtiss R III, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 1103–1129. [Google Scholar]
- 14.Deldon C V, Iglewski B H. Emerg Infect Dis. 1998;4:551–560. doi: 10.3201/eid0404.980405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drake D, Montie T C. J Gen Microbiol. 1988;134:43–52. doi: 10.1099/00221287-134-1-43. [DOI] [PubMed] [Google Scholar]
- 16.Sato H, Okinaga K, Saito H. Microbiol Immunol. 1988;32:131–139. doi: 10.1111/j.1348-0421.1988.tb01372.x. [DOI] [PubMed] [Google Scholar]
- 17.Tang H, Kays M, Prince A. Infect Immun. 1995;63:1278–1285. doi: 10.1128/iai.63.4.1278-1285.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang H B, DiMango E, Bryan R, Gambello M, Iglewski B H, Goldberg J B, Prince A. Infect Immun. 1996;64:37–43. doi: 10.1128/iai.64.1.37-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feldman M, Bryan R, Rajan S, Scheffler L, Brunnert S, Tang H, Prince A. Infect Immun. 1998;66:43–51. doi: 10.1128/iai.66.1.43-51.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Comolli J C, Hauser A R, Waite L, Whitchurch C B, Mattick J S, Engel J N. Infect Immun. 1999;67:3625–3630. doi: 10.1128/iai.67.7.3625-3630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rashid M H, Rao N N, Kornberg A. J Bacteriol. 2000;182:225–227. doi: 10.1128/jb.182.1.225-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furste J P, Pansegran W, Frank R, Blocker H, Scholz P, Bagdasarian M, Lanka E. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 23.Ishige K, Kameda A, Noguchi T, Shiba T. DNA Res. 1998;5:157–162. doi: 10.1093/dnares/5.3.157. [DOI] [PubMed] [Google Scholar]
- 24.Miyake T, Shiba T, Kameda A, Ihara Y, Munekata M, Ishige K, Noguchi T. DNA Res. 1999;6:103–108. doi: 10.1093/dnares/6.2.103. [DOI] [PubMed] [Google Scholar]
- 25.Tzeng C-M, Kornberg A. Mol Microbiol. 1998;29:381–382. doi: 10.1046/j.1365-2958.1998.00887.x. [DOI] [PubMed] [Google Scholar]
- 26.Taylor B L, Koshland D E. J Bacteriol. 1974;119:640–642. doi: 10.1128/jb.119.2.640-642.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarter L, Silverman M. Mol Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 28.Wood H G, Clark J E. Annu Rev Biochem. 1988;57:235–260. doi: 10.1146/annurev.bi.57.070188.001315. [DOI] [PubMed] [Google Scholar]
- 29.Kulaev I S, Vegabov V M. Adv Microb Physiol. 1983;24:83–171. doi: 10.1016/s0065-2911(08)60385-9. [DOI] [PubMed] [Google Scholar]
- 30.Kornberg A. J Bacteriol. 1995;177:491–496. doi: 10.1128/jb.177.3.491-496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiba T, Tsutsumi K, Yano H, Ihara Y, Kameda A, Tanaka K, Takahashi H, Masanobu M, Rao N N, Kornberg A. Proc Natl Acad Sci USA. 1997;94:11210–11215. doi: 10.1073/pnas.94.21.11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bradley D E. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 33.Alberti L, Harshey R M. J Bacteriol. 1990;172:4322–4328. doi: 10.1128/jb.172.8.4322-4328.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harshey R M, Matsuyama T. Proc Natl Acad Sci USA. 1994;91:8631–8635. doi: 10.1073/pnas.91.18.8631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eberl L, Christiansen G, Molin S, Givskov M. J Bacteriol. 1996;178:554–559. doi: 10.1128/jb.178.2.554-559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Young G M, Smith M J, Minnich S A, Miller V L. J Bacteriol. 1999;181:2823–2833. doi: 10.1128/jb.181.9.2823-2833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kinscherf T G, Willis D K. J Bacteriol. 1999;181:4133–4136. doi: 10.1128/jb.181.13.4133-4136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki T, Lino T. J Bacteriol. 1980;143:1471–1479. doi: 10.1128/jb.143.3.1471-1479.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eberl L, Molin S, Givskov M. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darzins A. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 41.Kim H-Y, Schlitman D, Shankar S, Xie Z, Chakrabarty A M, Kornberg A. Mol Microbiol. 1998;27:717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 42.Whitchurch C B, Alm R A, Mattick J S. Proc Natl Acad Sci USA. 1996;93:9839–9843. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Suh S J, Silo-Suh L, Woods D E, Hassett D J, West S E, Ohman D E. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/jb.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burkart M, Toguchi A, Harshey R M. Proc Natl Acad Sci USA. 1998;95:2568–2573. doi: 10.1073/pnas.95.5.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Semmler A B T, Whitchurch C B, Mattick J S. Microbiology. 1999;145:2863–2873. doi: 10.1099/00221287-145-10-2863. [DOI] [PubMed] [Google Scholar]
- 46.Ward M J, Zusman D R. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 47.Masduki A, Nakamura J, Ohga T, Umezaki R, Kato J, Ohtake H. J Bacteriol. 1995;177:948–952. doi: 10.1128/jb.177.4.948-952.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eisenbach M. Mol Microbiol. 1996;20:903–910. doi: 10.1111/j.1365-2958.1996.tb02531.x. [DOI] [PubMed] [Google Scholar]
- 49.Margolin Y, Barak R, Eisenbach M. J Bacteriol. 1994;176:5547–5549. doi: 10.1128/jb.176.17.5547-5549.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bode G, Mauch F, Ditschunet H, Malfertheiner P. J Gen Microbiol. 1993;139:3029–3033. doi: 10.1099/00221287-139-12-3029. [DOI] [PubMed] [Google Scholar]
- 51.Jones B D, Lee C A, Falkow S. Infect Immun. 1992;60:2475–2480. doi: 10.1128/iai.60.6.2475-2480.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.O'Toole R, Milton D L, Wolf-Watz H. Mol Microbiol. 1996;19:625–637. doi: 10.1046/j.1365-2958.1996.412927.x. [DOI] [PubMed] [Google Scholar]
- 53.Yao R, Burr D H, Guerry P. Mol Microbiol. 1997;23:1021–1031. doi: 10.1046/j.1365-2958.1997.2861650.x. [DOI] [PubMed] [Google Scholar]