Abstract
Filamentous inclusions made of α-synuclein constitute the defining neuropathological characteristic of Parkinson's disease, dementia with Lewy bodies, and multiple system atrophy. Rare familial cases of Parkinson's disease are associated with mutations A53T and A30P in α-synuclein. We report here the assembly properties and secondary structure characteristics of recombinant α-synuclein. Carboxy-terminally truncated human α-synuclein (1–87) and (1–120) showed the fastest rates of assembly, followed by human A53T α-synuclein, and rat and zebra finch α-synuclein. Wild-type human α-synuclein and the A30P mutant showed slower rates of assembly. Upon shaking, filaments formed within 48 h at 37°C. The related proteins β- and γ-synuclein only assembled after several weeks of incubation. Synthetic human α-synuclein filaments were decorated by an antibody directed against the carboxy-terminal 10 amino acids of α-synuclein, as were filaments extracted from dementia with Lewy bodies and multiple system atrophy brains. Circular dichroism spectroscopy indicated that α-synuclein undergoes a conformational change from random coil to β-sheet structure during assembly. X-ray diffraction and electron diffraction of the α-synuclein assemblies showed a cross-β conformation characteristic of amyloid.
Parkinson's disease (PD) is the most common neurodegenerative movement disorder. Neuropathologically, it is defined by nerve cell loss in the substantia nigra and other brain regions and the presence there of Lewy bodies and Lewy neurites (1, 2). Abundant Lewy bodies and Lewy neurites in cerebral cortex are also the defining neuropathological characteristics of dementia with Lewy bodies (DLB), a common late-life dementia that is clinically similar to Alzheimer's disease (3, 4). Ultrastructurally, Lewy bodies and Lewy neurites are composed of filamentous and granular material (5). The discovery of a missense mutation in the α-synuclein gene in rare cases of familial early-onset PD (6) has led us to the finding that α-synuclein is the major component of the abnormal filaments of Lewy bodies and Lewy neurites in idiopathic PD and DLB (7, 8). Moreover, the filamentous glial and neuronal inclusions of multiple system atrophy (MSA) have been found to be made of α-synuclein (9–12). Taken together, this work has shown that PD, DLB, and MSA are α-synucleinopathies.
α-Synuclein is a 140-aa protein of unknown function that is abundantly expressed in brain, where it is concentrated in presynaptic nerve terminals (13, 14). Two related proteins, β-synuclein (phosphoneuroprotein 14) and γ-synuclein (breast cancer-specific gene 1 or persyn), have been described (14–17). The amino-terminal half of each synuclein is taken up by imperfect repeats with the consensus sequence KTKEGV. In α-synuclein, these repeats are followed by a hydrophobic middle region and a negatively charged carboxy terminus. The familial PD mutations A30P and A53T are located in the repeat region of α-synuclein (6,18). Surprisingly, rodent α-synuclein, which is 95% identical to the human protein, carries a threonine at position 53, like mutant human α-synuclein (19). The same is true of zebra finch α-synuclein (synelfin), which is 89% identical to its human homolog (20).
By immunoelectron microscopy, isolated filaments from DLB and MSA brains were labeled by an antibody directed against residues 117–131 of α-synuclein (8, 11). Recombinant human α-synuclein (1–120) was shown to assemble readily into filaments under physiological conditions, with morphologies and staining characteristics similar to those extracted from diseased brain (21). Two subsequent studies reported filament assembly from full-length human α-synuclein after incubations ranging from 1 wk to 3 mo at 37°C (22, 23). The A53T mutation was shown to increase the rate of filament assembly (23). More recently, improved methods requiring shorter incubation times have confirmed that full-length α-synuclein assembles into filaments (24–26). The influence of the A30P mutation on the rate of filament assembly is unclear, with one study (25) reporting a stimulatory and another (27) an inhibitory effect. Recombinant wild-type and A53T α-synuclein have been found to bind to brain vesicles, whereas A30P α-synuclein was devoid of significant vesicle-binding activity (28). These observations suggest that the two α-synuclein mutations may differ in their primary effects.
Structural studies have revealed that amyloid fibrils formed from diverse proteins have similar characteristics. By electron microscopy, the fibers are long, straight, and unbranched with a diameter of ≈10 nm (29). X-ray diffraction gives a characteristic cross-β pattern that indicates that the fibrils consist of a hydrogen-bonded β-sheet structure in which the β-strands run perpendicular to the long fiber axis (30, 31). The fiber diffraction pattern of α-synuclein filaments has not been previously investigated.
Here, we describe the circular dichroism and fiber diffraction patterns of synthetic filaments formed from full-length wild-type and mutant human α-synuclein, from full-length rat and zebra finch α-synuclein, and from truncated human protein. Carboxy-terminally truncated α-synucleins showed the fastest rates of assembly, followed by rat and zebra finch α-synuclein and human A53T α-synuclein. Wild-type human α-synuclein and the A30P mutant exhibited slower rates of assembly. Filaments extracted from DLB and MSA brains and synthetic filaments formed from wild-type α-synuclein were decorated by an antibody that recognizes the last 10 amino acids of α-synuclein, indicating the presence of the entire carboxy terminus of α-synuclein in a large proportion of Lewy body and MSA filaments. Circular dichroism revealed a random coil to β-sheet transition of α-synuclein that was not observed for the related proteins β- and γ-synuclein. The x-ray and, more specifically, the electron diffraction patterns from the filaments gave strong evidence for cross-β structure.
Materials and Methods
Expression and Purification of Recombinant Synucleins.
Bacterial expression constructs encoding human, rat, and zebra finch α-synuclein, human A30P α-synuclein, A53T α-synuclein, α-synuclein (1–120), α-synuclein (1–130), β-synuclein, and γ-synuclein have been described (8, 14, 32). Human α-synuclein (1–87), α-synuclein (1–132), and α-synuclein (1–135) were produced by PCR amplification from the full-length construct. After DNA sequencing, they were subcloned as NdeI/HindIII fragments into the bacterial expression vector pRK172. Bacterial expression and purification of synuclein proteins, as well as procedures for immunoblotting, were as described (14). Purified proteins were kept at 4°C until use and their concentrations determined by quantitative amino acid analysis.
Assembly Experiments and Electron Microscopy.
For assembly, α-synuclein proteins were incubated for up to 48 h at 37°C in plastic or glass tubes placed in a shaking bacterial incubator (G24 environmental incubator shaker, New Brunswick Scientific, speed setting 450). β-Synuclein and γ-synuclein were incubated under the same conditions for up to 5 wk. In most experiments, synucleins were used at 400 μM in 150 μl 30 mM 3-[N-Morpholino]propanesulphonic acid (Mops), pH 7.2, containing 0.02% sodium azide. The samples were checked by SDS-PAGE immediately before microscopy to ensure that there had been no protein degradation. Aliquots of assembly mixtures were placed on carbon-coated 400-mesh grids and stained with 1% lithium phosphotungstate, and micrographs recorded at a nominal magnification of 40,000 on a Philips model EM208S microscope. Filaments were extracted from DLB and MSA brains, as described (8, 11). Procedures for immunoelectron microscopy were as described (21). The anti-synuclein antibody H3C (20) was used at 1:100. After reaction with the appropriate secondary gold-conjugated antibody (Sigma Fine Chemicals), the grids were stained with 1% lithium phosphotungstate.
Circular Dichroism Spectroscopy.
Synuclein proteins (400 μM in 30 mM Mops, 0.02% sodium azide) were incubated for 1–2 days at 37°C. They were diluted 1:10 and placed in a 0.1-cm quartz cuvette and spectra collected on a Jasco CD spectrophotometer (Easton, MD) at 20°C. Measurements were collected from the wavelength range 250–190 nm for each protein solution. Five scans were developed and averaged, with each sample being measured twice.
X-Ray Diffraction.
Fiber diffraction specimens in a partially dehydrated state were prepared on a stretch frame using starting protein solutions of approximately 10 mg/ml, as described previously (31). X-ray diffraction patterns were collected on MAR-Research (Hamburg, Germany) image plate x-ray detectors (345 or 300 mm diameter) using a Cu Kα Rigaku rotating anode x-ray source, with exposure times of 30–60 min and specimen to film distances of 148 or 175 mm. The images were displayed and examined by using mosflm (33).
Electron Diffraction.
Assembled filaments of wild-type human α-synuclein (≈2 mg starting material) were pelleted at 250,000 g for 1 h (Beckman 100.2 rotor). The pellet was resuspended overnight in 50 μl of distilled water. One microliter of sample was applied to a holey carbon film, blotted, and rapidly frozen in liquid ethane for cryo-microscopy. Alternatively, 0.5 μl of sample was mixed with 0.5 μl of 1% glucose and applied to a carbon coated grid, blotted, and allowed to air dry. Electron diffraction and imaging were performed on a Hitachi HF-2000 microscope, using a Gatan cold stage at 170 K under low dose conditions. Diffraction patterns were taken at 200 kV with a camera length of 1.2 m and a dose of 100 e/nm2. Patterns were collected from suitable regions ≈1 μm diameter, where filaments showed partial alignment. A subsequent image taken in defocused diffraction mode showed the geometrical relationship between the region diffracting and the diffraction pattern. Diffraction was radiation sensitive and had noticeably decayed after a dose of 300 e/nm2. Diffraction spacings were calibrated against evaporated thallous chloride (Agar Scientific). The small rotation (≈5°) between diffraction pattern and image was determined with molybdenum oxide crystals (Agar Scientific).
Results
Assembly of α-Synuclein into Filaments.
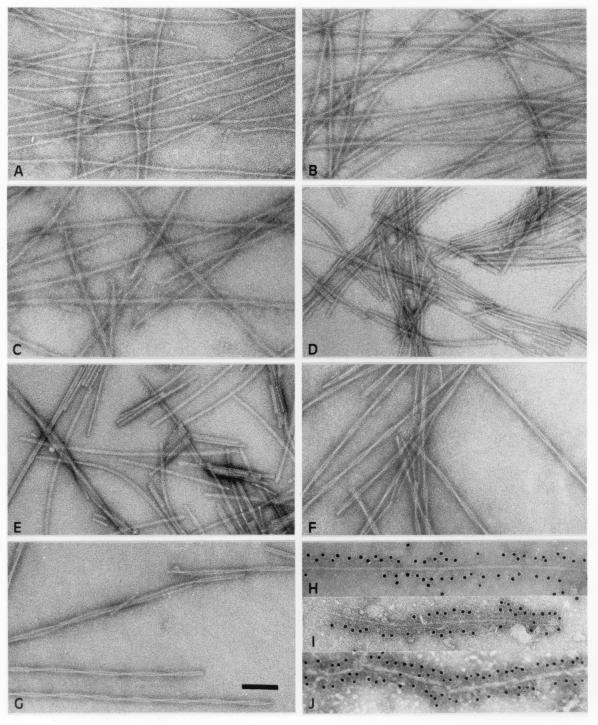
Incubation of full-length recombinant human α-synuclein, as well as of A30P α-synuclein and A53T α-synuclein led to bulk assembly into filaments, when the protein solution was subjected to vigorous shaking (Fig. 1A–C). The same was true of human α-synuclein (1–87) and α-synuclein (1–120), as well as of rat and zebra finch α-synuclein (Fig. 1D–G). Filaments formed within 48 h at 37°C. Human β-synuclein and γ-synuclein failed to form filaments under these conditions. Only after 5 wk of incubation did they assemble into small numbers of filaments. The β-synuclein filaments were of similar morphology to the α-synuclein filaments, whereas γ-synuclein assembled into short fragments and some twisted ribbons (data not shown).
Figure 1.
Electron micrographs of α-synuclein filaments. Filaments were assembled from recombinant wild-type human α-synuclein (A), human A30P α-synuclein (B), human A53T α-synuclein (C), human α-synuclein (1–120) (D), human α-synuclein (1–87) (E), rat α-synuclein (F), and zebra finch α-synuclein (G). Synthetic wild-type human α-synuclein filaments were decorated by antibody H3C (H), as were filaments extracted from DLB (I) and MSA (J) brains. (Scale bar, 100 nm).
Time course experiments were carried out to gain a semiquantitative estimate of the relative rates of filament formation, with their presence being monitored by electron microscopy. The assembly rates were as follows: human α-synuclein (1–87) > human α-synuclein (1–120) > human A53T α-synuclein = rat α-synuclein = zebra finch α-synuclein > human A30P α-synuclein = human α-synuclein. Recombinant α-synuclein proteins were used at 400 μM, and at least three independent protein preparations were used for each experiment. The protein concentrations required for assembly were determined by incubating varying concentrations of each recombinant α-synuclein for 48 h at 37°C and by monitoring the presence of filaments by electron microscopy. The minimal concentration for assembly of human α-synuclein was ≈200 μM, with similar values for the A30P and A53T mutants, as well as for rat and zebra finch α-synucleins. This contrasted with minimal concentrations of ≈50 μM for human α-synuclein (1–87) and α-synuclein (1–120).
For human α-synuclein, the filaments were typically 6–9 nm in width and several microns long (Fig. 1A). In some filaments, the overall width varied slightly along the length of the filament. In general, little substructure could be seen, apart from some irregular long pitch helical strands in places. These filaments were of similar appearance to those found in the α-synuclein-linked neurodegenerative diseases, particularly DLB. By immunoelectron microscopy, filaments formed from recombinant human α-synuclein were decorated by antibody H3C, as was a substantial proportion of the filaments extracted from DLB and MSA brains (Fig. 1H–J). H3C is a mAb that was produced against a synthetic peptide corresponding to residues 128–143 of zebra finch α-synuclein (20). We mapped the epitope of H3C on the human protein by using recombinant synuclein proteins and immunoblotting. H3C recognized full-length human α- and β-synuclein, but not γ-synuclein. It failed to recognize human α-synuclein (1–130), α-synuclein (1–132), and α-synuclein (1–135), demonstrating that its epitope resides in the carboxy-terminal 10 amino acids of human α-synuclein. These findings thus indicate that DLB and MSA filaments contain at least some α-synuclein molecules with the carboxy terminus and that this region is located on the surface of both native and synthetic filaments.
The morphologies of the filaments formed from A30P α-synuclein were similar to those formed from the wild-type protein (Fig. 1B). This contrasted with the filaments formed from A53T α-synuclein and from rat and zebra finch α-synucleins, for which the images showed a more marked variation in width between ≈5 and 14 nm with a crossover spacing of ≈110 nm (Fig. 1 C, F, and G). These noticeably twisted filaments resembled those seen in MSA. The filaments formed by the truncated human α-synucleins (1–87) and (1–120) were shorter and more irregular and had a tendency to aggregate laterally to make ribbon-like structures (Fig. 1 D and E).
Circular Dichroism Measurements.
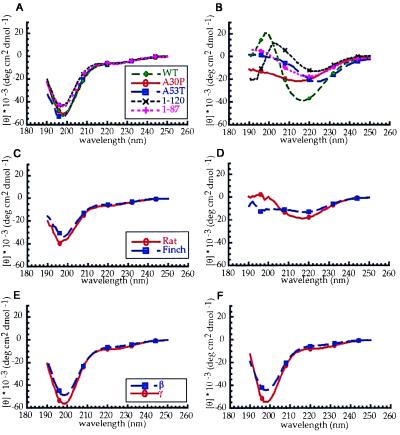
Measurements of the circular dichroism spectra of human, rat and zebra finch α-synucleins, and of the human mutants A30P and A53T and of the truncated human proteins (1–87) and (1–120) were consistent with a random coil structure, with a minimum at 195 nm (Fig. 2 A and C). After incubation for 1–2 days at 37°C with shaking, all of the spectra changed to give a strong indication of β-sheet structure. Wild-type α-synuclein showed a typical β-sheet spectrum with a minimum at 217 nm (Fig. 2B). A slight shift in minimum was observed for human A53T α-synuclein and human α-synuclein (1–87) and (1–120), which may have resulted from the extended β-sheet within the filaments (Fig. 2B). Zebra finch and human A30P α-synuclein showed a loss of signal at lower wavelengths, consistent with protein precipitation (Fig. 2D). The related proteins β- and γ-synuclein gave no indication of a shift from random coil structure after a similar incubation (Fig. 2 E and F).
Figure 2.
Circular dichroism spectra of synuclein proteins before (A, C, and E) and after (B, D, and F) incubation for 1–2 days at 37°C while shaking. (A and B) Human wild-type α-synuclein, A30P α-synuclein, and A53T α-synuclein, α-synuclein (1–87) and (1–120); (C and D) Rat and zebra finch α-synuclein; (E and F) β- and γ-synuclein. Before incubation all spectra indicated a random coil structure. After incubation, the α-synuclein spectra indicated a transition to β-sheet structure, whereas β- and γ-synuclein remained in the random coil conformation.
X-Ray Diffraction.
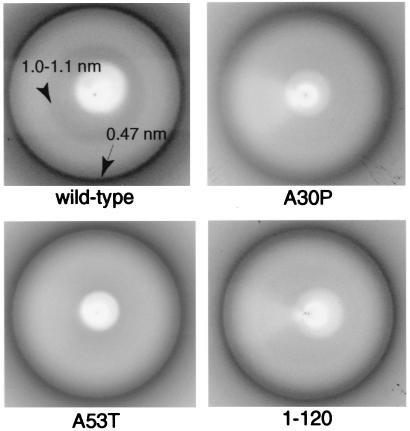
The x-ray diffraction patterns from dried pellets of filaments assembled from human wild-type α-synuclein, and from the human mutants A30P and A53T, as well as from the partial human sequence (1–120) all showed a strong ring at a spacing of ≈0.47 nm (Fig. 3). The pattern from the wild-type α-synuclein filaments also showed weak diffraction in the 1.0- to 1.1-nm region. Similar patterns were obtained from filaments of the rat and zebra finch proteins and of human α-synuclein (1–87) (data not shown).
Figure 3.
X-ray diffraction patterns from human α-synuclein filaments. Wild-type α-synuclein, mutant A30P α-synuclein, mutant A53T α-synuclein, and α-synuclein (1–120) are shown. Each pattern shows a reflection at 0.47 nm. A more diffuse, weaker reflection at 1.0–1.1 nm is seen in the wild-type sample.
Electron Diffraction.
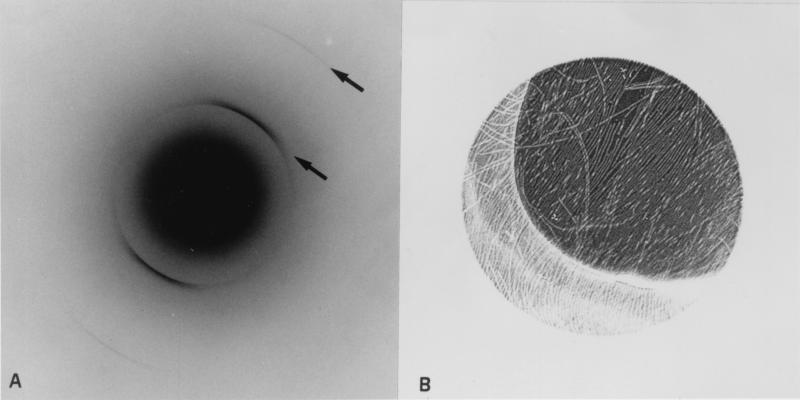
The advantages of electron diffraction over x-ray diffraction are that the diffracting volume can be much smaller and that the region from which the diffraction has been collected also can be imaged, allowing the geometrical relationship between specimen and diffraction pattern to be established. In addition, by working with a rapidly frozen specimen in vitreous ice, the material can be kept in a natively hydrated state. The electron diffraction pattern from a partially oriented raft of filaments of wild-type human α-synuclein showed a strong arc of diffraction at ≈0.47 nm, with a weaker second order spacing at 0.23 nm (Fig. 4A). In specimens dried in glucose, the third order spacing at 0.115 nm was also sometimes visible. These arcs clearly occurred in the same direction as the axes of the filaments (Fig. 4B) and therefore arose from features in the structure running perpendicular to the filament axis. The 0.47-nm arc sometimes appeared split, particularly at the outer tips of the arc. Using a range of camera lengths, no diffraction could be detected in the region of 0.9–1.1 nm, either meridionally or equatorially. Identical diffraction patterns were obtained from the twisted form of A53T α-synuclein filaments (data not shown).
Figure 4.
Electron diffraction from hydrated wild-type human α-synuclein filaments. (A) Electron diffraction pattern with oriented arcs at 0.47 and 0.235 nm indicated by arrows. (B) Image of diffraction area showing partially oriented filaments with axes running roughly perpendicular to the arcs in A. The illuminated area is ≈1 μm in diameter.
Discussion
The present study describes the bulk assembly and secondary structure characteristics of α-synuclein upon shaking at 37°C for 48 h. Carboxy-terminally truncated proteins, such as human α-synuclein (1–87) and (1–120) assembled at a much faster rate than the full-length protein, with lower concentrations necessary for assembly. This indicates that α-synuclein assembles through its amino-terminal repeat region and that the presence of the carboxy-terminal region is partially inhibitory for assembly, confirming and extending our previous findings (21). Recombinant α-synuclein with the familial PD mutations A30P and A53T assembled into filaments, with the protein bearing the A53T mutation assembling at a faster rate than wild-type α-synuclein. Rat and zebra finch α-synuclein also assembled at a faster rate than wild-type human α-synuclein, probably because of the presence of a threonine residue at position 53. Thus, the other amino acid differences between rat or zebra finch and wild-type human α-synuclein were unable to compensate for the effects of threonine 53. Besides influencing the rate of assembly, the presence of a threonine residue at position 53 also changed the filament morphologies. Whereas wild-type human α-synuclein and the A30P mutant formed predominantly straight filaments similar to those found in DLB, human α-synuclein with the A53T mutation, as well as rat and zebra finch α-synuclein formed predominantly twisted filaments similar to those found in MSA. These findings underscore the previously noted (8, 11) polymorphic nature of the α-synuclein assemblies. The structural relationship between straight and twisted α-synuclein filaments may be similar to that between straight and paired helical τ filaments in Alzheimer's disease (34) and involve an alternative strand packing, perhaps based on interfaces formed by different sequence repeats.
By immunoelectron microscopy, filaments formed from wild-type α-synuclein were decorated by antibody H3C, which has been shown to recognize the carboxy-terminal 10 amino acids of human α-synuclein. H3C also decorated a large proportion of filaments extracted from DLB and MSA brains, indicating the presence of an intact carboxy terminus of α-synuclein which is exposed on the filament surface. We have previously shown that the amino-terminal region of α-synuclein is buried in synthetic filaments, as well as in filaments extracted from DLB and MSA brains (8, 11, 21). Strikingly, an antibody directed against residues 11–34 of α-synuclein labeled these filaments at one end (8, 11, 21), emphasizing the different availability of amino- and carboxy-terminal residues of α-synuclein in the filaments. By immunoblotting, filament preparations from DLB and MSA brains have been found to contain predominantly truncated α-synuclein (35, 36), suggesting that in the human diseases full-length α-synuclein becomes truncated after assembly. The same is true of microtubule-associated protein τ in the paired helical filaments of Alzheimer's disease (37, 38). However, unlike α-synuclein, the amino- and carboxy-termini of τ protein are both exposed on the filament surface (37).
Human β-synuclein and γ-synuclein failed to assemble into filaments during the 48-h incubation period. Only after incubation for several weeks did they begin to form small numbers of filaments. These results are in keeping with the finding that antibodies directed against β-synuclein and γ-synuclein do not stain the filamentous inclusions of PD, DLB, and MSA (7, 8) and that there is no genetic evidence linking β- and γ-synuclein to neurodegenerative disease (39–41).
The findings of the assembly of wild-type and mutant α-synuclein into straight and twisted filaments extend previous studies, which showed that full-length human α-synuclein assembles into filaments and that the A53T mutation increases the rate of assembly (22–27). Contrary to two recent studies (25, 27), we failed to observe a significant effect of the A30P mutation on the rate of filament assembly. Unlike A53T, this mutation has been shown to lead to a loss in the vesicle-binding activity of α-synuclein (28), suggesting that the primary effects of the two familial PD mutations may be different. The A30P mutation may lead to a partial loss of function that may in turn result in the accumulation of the mutant protein, followed by its assembly into filaments. Instead, the A53T mutation may have a direct stimulatory effect on filament assembly. It also may lead to a slower degradation by the ubiquitin-proteasome system, thus facilitating accumulation of the mutant protein (42).
The bulk assembly of full-length α-synuclein required vigorous shaking. It is possible that shaking leads to the shearing of α-synuclein assemblies, which then function as seeds, resulting in a marked acceleration of filament formation. Agitation-stimulated fibrillogenesis has been observed for other proteins (43, 44).
Circular dichroism showed that all α-synuclein sequences tested underwent a conformational change during fibril formation. Before incubation, they had a random coil structure, in confirmation of results obtained previously with human α-synuclein with or without the familial PD mutations (22, 23, 27, 45). Following shaking at 37°C for 48 h, the α-synuclein proteins showed a β-sheet structure, consistent with the presence of filamentous assemblies by electron microscopy. This is in contrast to a recent report which failed to find a significant random coil to β-sheet transition for wild-type human α-synuclein and the A30P mutant following 31 days of incubation at 37°C without agitation (27). A conformational change to β-sheet structure has previously been noted for the prion protein (46) and the Aβ peptide of Alzheimer's disease (47, 48). It is believed to be an important step in the pathogenesis of amyloid diseases.
X-ray diffraction of α-synuclein assemblies gave a cross-β pattern consisting of a 0.47-nm meridional reflection and in some samples a 1.0- to 1.1-nm equatorial reflection. The cross-β diffraction pattern was first described for insect silk (49) and is a well-established characteristic of amyloid fibrils (30, 31). The meridional reflection corresponds to the spacing between main chain β-strands and the broader equatorial reflection to the spacing between the β-sheets making up the fiber. Electron diffraction of hydrated filaments confirmed the cross-β diffraction pattern by showing the oriented 0.47-nm reflection and higher orders on the meridian. The observed splitting of the sharp reflection has been previously observed for transthyretin amyloid fibrils (50) and was taken to be due to twisting or slight imperfections in the ordering of the β-strands. In the best x-ray pattern, the scattering in the 1.0- to 1.1-nm region was diffuse and much weaker than the ring at 0.47 nm. The electron diffraction was recorded from small areas of thin layers of filaments to get good orientation, so that there was probably insufficient material in the beam for the diffuse scattering in the 1.0- to 1.1-nm region to be recorded. The fiber diffraction patterns clearly indicate that the α-synuclein filaments have a β-sheet conformation that is similar to the structure described for amyloid fibrils (31, 51).
Structural characterization of amyloid fibrils has been largely carried out on assemblies present in extracellular deposits (51). Much less is known about the intracellular filamentous deposits characteristic of the majority of late-onset neurodegenerative diseases. α-Synucleinopathies constitute one major class of neurodegenerative disease with intracellular deposits, the other two classes being diseases with intraneuronal filamentous τ protein deposits and diseases with filamentous deposits made of expanded glutamine repeats (52, 53). Whereas the secondary structure characteristics of τ filaments are not well-defined (54, 55), synthetic filaments formed from exon 1 of huntingtin with 51 glutamines gave a cross-β pattern by x-ray diffraction (53). The present study extends the structural characterization to α-synuclein assemblies and suggests that a cross-β conformation is shared by the extra- and intracellular filamentous deposits that define amyloid diseases.
Acknowledgments
We thank Drs. J. M. George and D. F. Clayton for kindly providing antibody H3C and the expression construct for zebra finch α-synuclein, C. Villa for help with photographic printing, and D. Owen for quantitative amino acid analysis.
Abbreviations
- PD
Parkinson's disease
- DLB
dementia with Lewy bodies
- MSA
multiple system atrophy
References
- 1.Lewy F. In: Handbuch der Neurologie. Lewandowsky M, Abelsdorff G, editors. Vol. 3. Berlin: Springer; 1912. pp. 920–933. [Google Scholar]
- 2.Trétiakoff M C. Ph.D. thesis. University of Paris; 1919. [Google Scholar]
- 3.Okazaki H, Lipkin L E, Aronson S M. J Neuropathol Exp Neurol. 1961;21:442–449. doi: 10.1097/00005072-196104000-00007. [DOI] [PubMed] [Google Scholar]
- 4.Kosaka K. Acta Neuropathol. 1978;42:127–134. doi: 10.1007/BF00690978. [DOI] [PubMed] [Google Scholar]
- 5.Duffy P E, Tennyson V M. J Neuropathol Exp Neurol. 1965;24:398–414. [PubMed] [Google Scholar]
- 6.Polymeropoulos M H, Lavedan C, Leroy E, Ide S E, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, et al. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 7.Spillantini M G, Schmidt M L, Lee V M-Y, Trojanowski J Q, Jakes R, Goedert M. Nature (London) 1997;388:839–840. doi: 10.1038/42166. [DOI] [PubMed] [Google Scholar]
- 8.Spillantini M G, Crowther R A, Jakes R, Hasegawa M, Goedert M. Proc Natl Acad Sci USA. 1998;95:6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wakabayashi K, Yoshimoto M, Tsuji S, Takahashi H. Neurosci Lett. 1998;249:180–182. doi: 10.1016/s0304-3940(98)00407-8. [DOI] [PubMed] [Google Scholar]
- 10.Mezey E, Dehejia A, Harta G, Papp M I, Polymeropoulos M H, Brownstein M J. Nat Med. 1998;4:755–757. doi: 10.1038/nm0798-755. [DOI] [PubMed] [Google Scholar]
- 11.Spillantini M G, Crowther R A, Jakes R, Cairns N J, Lantos P L, Goedert M. Neurosci Lett. 1998;251:205–208. doi: 10.1016/s0304-3940(98)00504-7. [DOI] [PubMed] [Google Scholar]
- 12.Tu P-H, Galvin J E, Baba M, Giasson B, Tomita T, Leight S, Nakajo S, Iwatsubo T, Trojanowski J Q, Lee V M-Y. Ann Neurol. 1998;44:415–422. doi: 10.1002/ana.410440324. [DOI] [PubMed] [Google Scholar]
- 13.Uéda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero D A, Kondo J, Ihara Y, Saitoh T. Proc Natl Acad Sci USA. 1993;90:11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jakes R, Spillantini M G, Goedert M. FEBS Lett. 1994;345:27–32. doi: 10.1016/0014-5793(94)00395-5. [DOI] [PubMed] [Google Scholar]
- 15.Tobe T, Nakajo S, Tanaka A, Mitoya A, Omata K, Nakaya K, Tomita M, Nakamura Y. J Neurochem. 1992;59:1624–1629. doi: 10.1111/j.1471-4159.1992.tb10991.x. [DOI] [PubMed] [Google Scholar]
- 16.Ji H, Liu Y E, Jia T, Wang M, Liu J, Xiao G, Joseph B K, Rosen C, Shi Y E. Cancer Res. 1997;57:759–764. [PubMed] [Google Scholar]
- 17.Buchman V L, Hunter H J A, Pinon L G P, Thompson J, Privalova E M, Ninkina N N, Davies A M. J Neurosci. 1998;18:9335–9341. doi: 10.1523/JNEUROSCI.18-22-09335.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krüger R, Kuhn W, Müller T, Woitalla D, Graeber M, Kösel S, Przuntek H, Epplen J T, Schöls L, Riess O. Nat Genet. 1998;18:106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 19.Maroteaux L, Scheller R H. Mol Brain Res. 1991;11:335–343. doi: 10.1016/0169-328x(91)90043-w. [DOI] [PubMed] [Google Scholar]
- 20.George J M, Jin H, Woods W S, Clayton D F. Neuron. 1995;15:361–372. doi: 10.1016/0896-6273(95)90040-3. [DOI] [PubMed] [Google Scholar]
- 21.Crowther R A, Jakes R, Spillantini M G, Goedert M. FEBS Lett. 1998;436:309–312. doi: 10.1016/s0014-5793(98)01146-6. [DOI] [PubMed] [Google Scholar]
- 22.El-Agnaf O M A, Jakes R, Curran M D, Wallace A. FEBS Lett. 1998;440:67–70. doi: 10.1016/s0014-5793(98)01419-7. [DOI] [PubMed] [Google Scholar]
- 23.Conway K A, Harper J D, Lansbury P T. Nat Med. 1998;4:1318–1320. doi: 10.1038/3311. [DOI] [PubMed] [Google Scholar]
- 24.Giasson B I, Uryu K, Trojanowski J Q, Lee V M-Y. J Biol Chem. 1999;274:7619–7622. doi: 10.1074/jbc.274.12.7619. [DOI] [PubMed] [Google Scholar]
- 25.Narhi L, Wood S J, Steavenson S, Jiang Y, Wu G M, Anafi D, Kaufman S A, Martin F, Sitney K, Denis P, et al. J Biol Chem. 1999;274:9843–9846. doi: 10.1074/jbc.274.14.9843. [DOI] [PubMed] [Google Scholar]
- 26.Wood S J, Wypych J, Steavenson S, Louis J-C, Citron M, Biere A L. J Biol Chem. 1999;274:19509–19512. doi: 10.1074/jbc.274.28.19509. [DOI] [PubMed] [Google Scholar]
- 27.Conway K A, Lee S-J, Rochet J-C, Ding T T, Williamson R E, Lansbury P T. Proc Natl Acad Sci USA. 2000;97:571–576. doi: 10.1073/pnas.97.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jensen P H, Nielsen M H, Jakes R, Dotti C G, Goedert M. J Biol Chem. 1998;273:26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 29.Cohen A S, Calkins E. Nature (London) 1959;183:1202–1203. doi: 10.1038/1831202a0. [DOI] [PubMed] [Google Scholar]
- 30.Eanes E D, Glenner G G. J Histochem Cytochem. 1968;16:673–677. doi: 10.1177/16.11.673. [DOI] [PubMed] [Google Scholar]
- 31.Sunde M, Serpell L C, Bartlam M, Fraser P E, Pepys M B, Blake C C F. J Mol Biol. 1997;273:729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 32.Jakes R, Crowther R A, Lee V M-Y, Trojanowski J Q, Iwatsubo T, Goedert M. Neurosci Lett. 1999;269:13–16. doi: 10.1016/s0304-3940(99)00411-5. [DOI] [PubMed] [Google Scholar]
- 33.Leslie A G W. Joint CCP4 and ESF–EAMCB Newsletter on Protein Crystallography. 1992. , No. 26. [Google Scholar]
- 34.Crowther R A. Proc Natl Acad Sci USA. 1991;88:2288–2292. doi: 10.1073/pnas.88.6.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baba M, Nakajo S, Tu P-H, Tomita T, Nakaya K, Lee V M-Y, Trojanowski J Q, Iwatsubo T. Am J Pathol. 1998;152:879–884. [PMC free article] [PubMed] [Google Scholar]
- 36.Gai W P, Power J H T, Blumbergs P C, Culvenor J G, Jensen P H. J Neurochem. 1999;73:2093–2100. [PubMed] [Google Scholar]
- 37.Goedert M, Spillantini M G, Cairns N J, Crowther R A. Neuron. 1992;8:159–168. doi: 10.1016/0896-6273(92)90117-v. [DOI] [PubMed] [Google Scholar]
- 38.Novak M, Kabat J, Wischik C M. EMBO J. 1993;12:365–370. doi: 10.1002/j.1460-2075.1993.tb05665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lincoln S, Gwinn-Hardy K, Goudreau J, Chartier-Harlin M C, Baker M, Mouroux V, Richard F, Destée A, Becquet E, Amouyel P, et al. Neurosci Lett. 1999;259:65–66. doi: 10.1016/s0304-3940(98)00901-x. [DOI] [PubMed] [Google Scholar]
- 40.Lincoln S, Crook R, Chartier-Harlin M C, Gwinn-Hardy K, Baker M, Mouroux V, Richard F, Becquet E, Amouyel P, Destée A, et al. Neurosci Lett. 1999;269:107–109. doi: 10.1016/s0304-3940(99)00420-6. [DOI] [PubMed] [Google Scholar]
- 41.Flowers J M, Leigh P N, Davies A M, Ninkina N N, Buchman V L, Vaughan J, Wood N W, Powell J F. Neurosci Lett. 1999;274:21–24. doi: 10.1016/s0304-3940(99)00673-4. [DOI] [PubMed] [Google Scholar]
- 42.Bennett C M, Bishop J F, Leng Y, Chock P B, Chase T N, Mouradian M M. J Biol Chem. 1999;274:33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 43.Zagorski M G, Yang J, Shao H, Ma K, Zeng H, Hong A. Methods Enzymol. 1999;309:189–204. doi: 10.1016/s0076-6879(99)09015-1. [DOI] [PubMed] [Google Scholar]
- 44.Serio T R, Cashikar A G, Moslehi J J, Kowal A S, Lindquist S L. Methods Enzymol. 1999;309:649–673. doi: 10.1016/s0076-6879(99)09043-6. [DOI] [PubMed] [Google Scholar]
- 45.Weinreb P H, Zhen W, Poon A W, Conway K A, Lansbury P T. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 46.Pan K-M, Baldwin M A, Nguyen J T, Gaseet M, Serban A, Groth D, Mehlhorn I, Huang Z, Fletterick R J, Cohen F E, Prusiner S B. Proc Natl Acad Sci USA. 1993;90:10962–10966. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrow C J, Zagorski M G. Science. 1991;253:179–182. doi: 10.1126/science.1853202. [DOI] [PubMed] [Google Scholar]
- 48.Hilbich C, Kisters-Woike B, Reed J, Masters C L, Beyreuther K. J Mol Biol. 1991;218:149–163. doi: 10.1016/0022-2836(91)90881-6. [DOI] [PubMed] [Google Scholar]
- 49.Geddes A J, Parker K D, Atkins E D T, Beighton E A J. J Mol Biol. 1968;32:343–358. doi: 10.1016/0022-2836(68)90014-4. [DOI] [PubMed] [Google Scholar]
- 50.Blake C C F, Serpell L C. Structure. 1996;4:989–998. doi: 10.1016/s0969-2126(96)00104-9. [DOI] [PubMed] [Google Scholar]
- 51.Sunde M, Blake C C F. Quart Rev Biophys. 1998;31:1–39. doi: 10.1017/s0033583598003400. [DOI] [PubMed] [Google Scholar]
- 52.Goedert M, Spillantini M G, Davies S W. Curr Opin Neurobiol. 1998;8:619–632. doi: 10.1016/s0959-4388(98)80090-1. [DOI] [PubMed] [Google Scholar]
- 53.Perutz M F. Trends Biochem Sci. 1999;24:58–63. doi: 10.1016/s0968-0004(98)01350-4. [DOI] [PubMed] [Google Scholar]
- 54.Kirschner D A, Inouye H, Duffy L K, Sinclair A, Lind M, Selkoe D J. Proc Natl Acad Sci USA. 1987;84:6953–6957. doi: 10.1073/pnas.84.19.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schweers O, Schönbrunn-Hanebeck E, Marx A, Mandelkow E. J Biol Chem. 1994;269:24290–24297. [PubMed] [Google Scholar]