Abstract
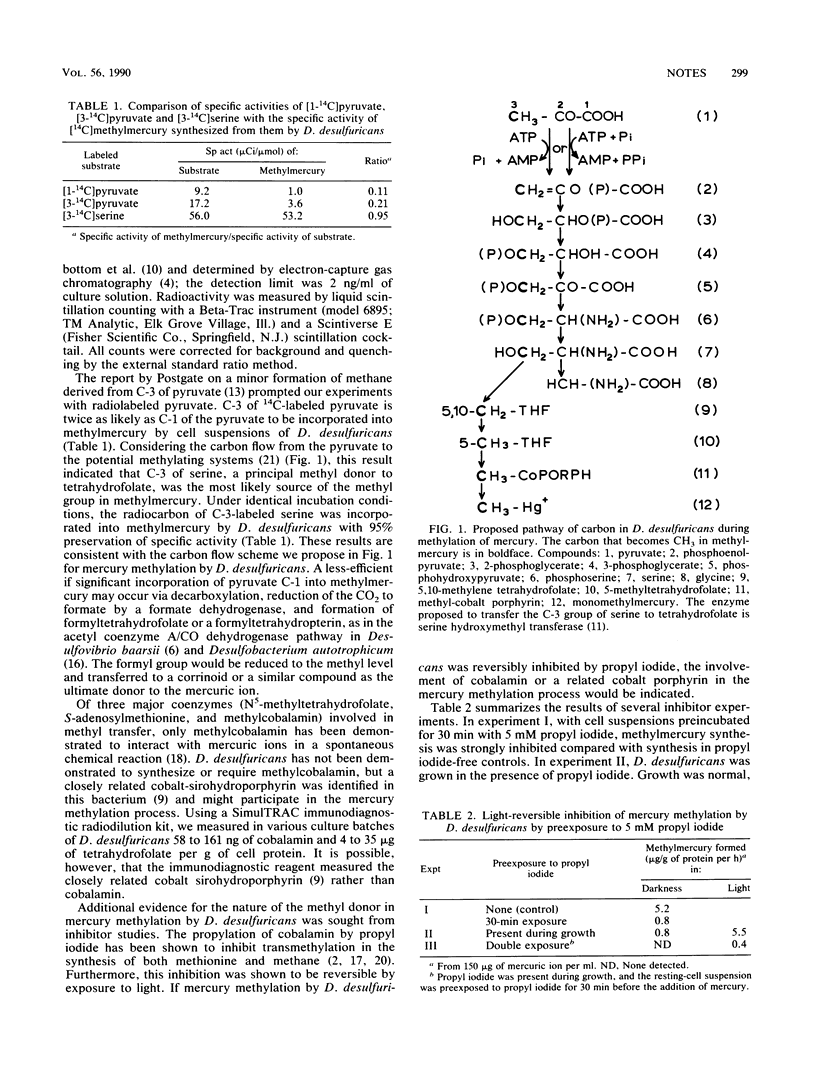
Radiocarbon incorporation from pyruvate and serine into monomethylmercury by Desulfovibrio desulfuricans was consistent with the proposal that the methyl group originates from C-3 of serine. Immunodiagnostic assays measured 4 to 35 μg of tetrahydrofolate and 58 to 161 ng of cobalamin or a closely related cobalt porphyrin per g of cell protein in D. desulfuricans. The light-reversible inhibition of mercury methylation by propyl iodide in D. desulfuricans indicates methyl transfer by a cobalt porphyrin.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BROT N., WEISSBACH H. ENZYMATIC SYNTHESIS OF METHIONINE. CHEMICAL ALKYLATION OF THE ENZYME-BOUND COBAMIDE. J Biol Chem. 1965 Jul;240:3064–3070. [PubMed] [Google Scholar]
- Berman M., Bartha R. Levels of chemical versus biological methylation of mercury in sediments. Bull Environ Contam Toxicol. 1986 Mar;36(3):401–404. doi: 10.1007/BF01623527. [DOI] [PubMed] [Google Scholar]
- Compeau G. C., Bartha R. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl Environ Microbiol. 1987 Feb;53(2):261–265. doi: 10.1128/aem.53.2.261-265.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compeau G. C., Bartha R. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl Environ Microbiol. 1985 Aug;50(2):498–502. doi: 10.1128/aem.50.2.498-502.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Landner L. Biochemical model for the biological methylation of mercury suggested from methylation studies in vivo with Neurospora crassa. Nature. 1971 Apr 16;230(5294):452–454. doi: 10.1038/230452a0. [DOI] [PubMed] [Google Scholar]
- Pan-Hou H. S., Imura N. Involvement of mercury methylation in microbial mercury detoxication. Arch Microbiol. 1982 Mar;131(2):176–177. doi: 10.1007/BF01054003. [DOI] [PubMed] [Google Scholar]
- Postgate J. R. Methane as a minor product of pyruvate metabolism by sulphate-reducing and other bacteria. J Gen Microbiol. 1969 Aug;57(3):293–302. doi: 10.1099/00221287-57-3-293. [DOI] [PubMed] [Google Scholar]
- Robinson J. B., Tuovinen O. H. Mechanisms of microbial resistance and detoxification of mercury and organomercury compounds: physiological, biochemical, and genetic analyses. Microbiol Rev. 1984 Jun;48(2):95–124. doi: 10.1128/mr.48.2.95-124.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEISSBACH H., REDFIELD B. G., DICKERMAN H., BROT N. STUDIES ON METHIONINE BIOSYNTHESIS. EFFECT OF ALKYLCOBAMIDE DERIVATIVES ON THE FORMATION OF HOLOENZYME. J Biol Chem. 1965 Feb;240:856–862. [PubMed] [Google Scholar]
- Wood J. M., Kennedy F. S., Rosen C. G. Synthesis of methyl-mercury compounds by extracts of a methanogenic bacterium. Nature. 1968 Oct 12;220(5163):173–174. doi: 10.1038/220173a0. [DOI] [PubMed] [Google Scholar]
- Wood J. M., Wolfe R. S. Propylation and purification of a B12 enzyme involved in methane formation. Biochemistry. 1966 Nov;5(11):3598–3603. doi: 10.1021/bi00875a031. [DOI] [PubMed] [Google Scholar]



