Abstract
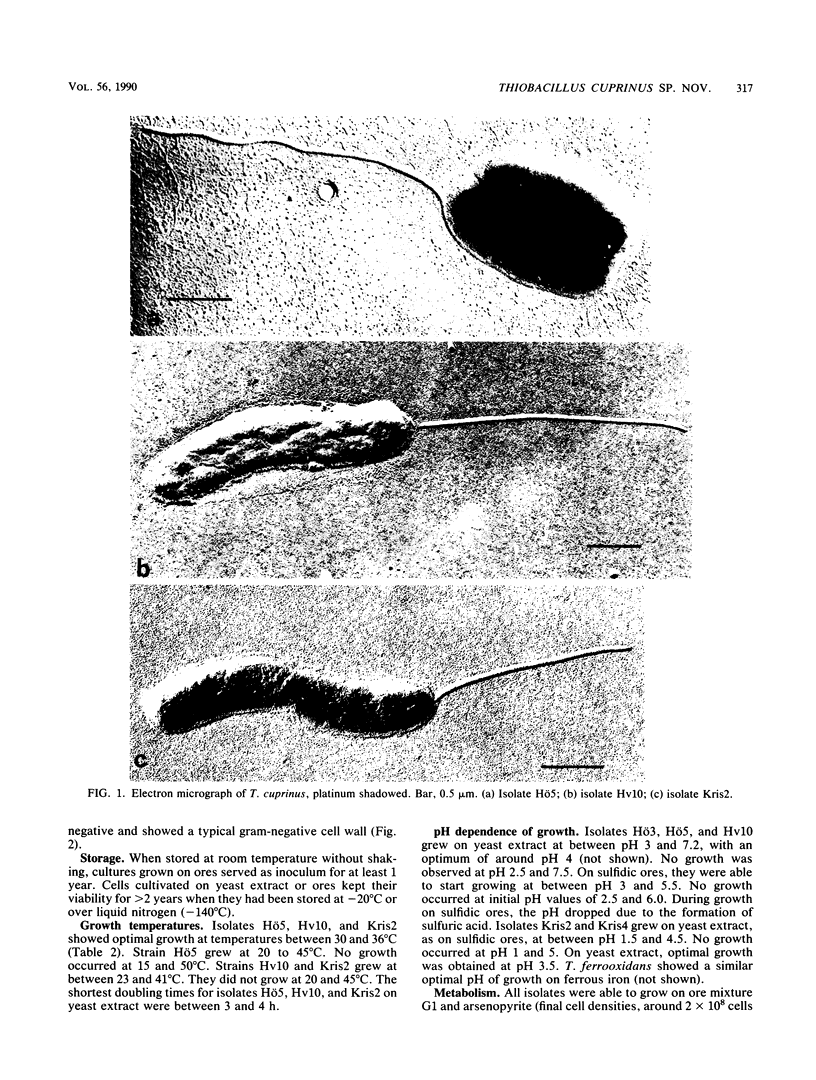
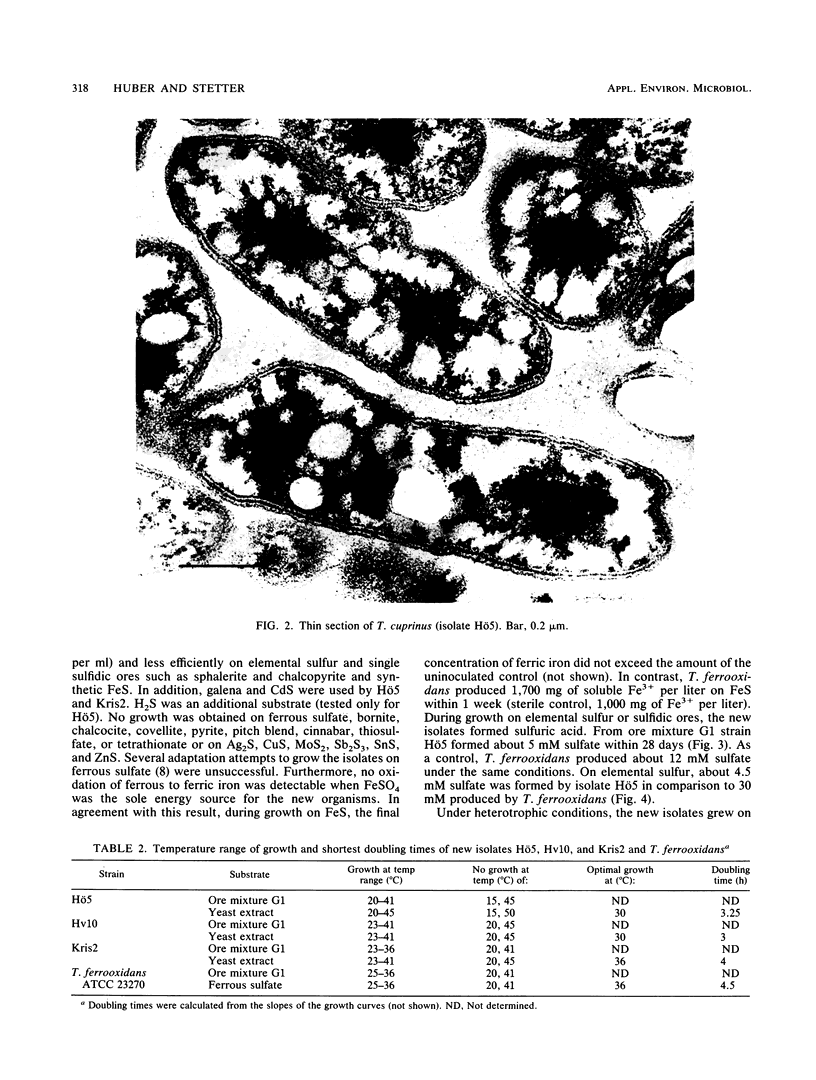
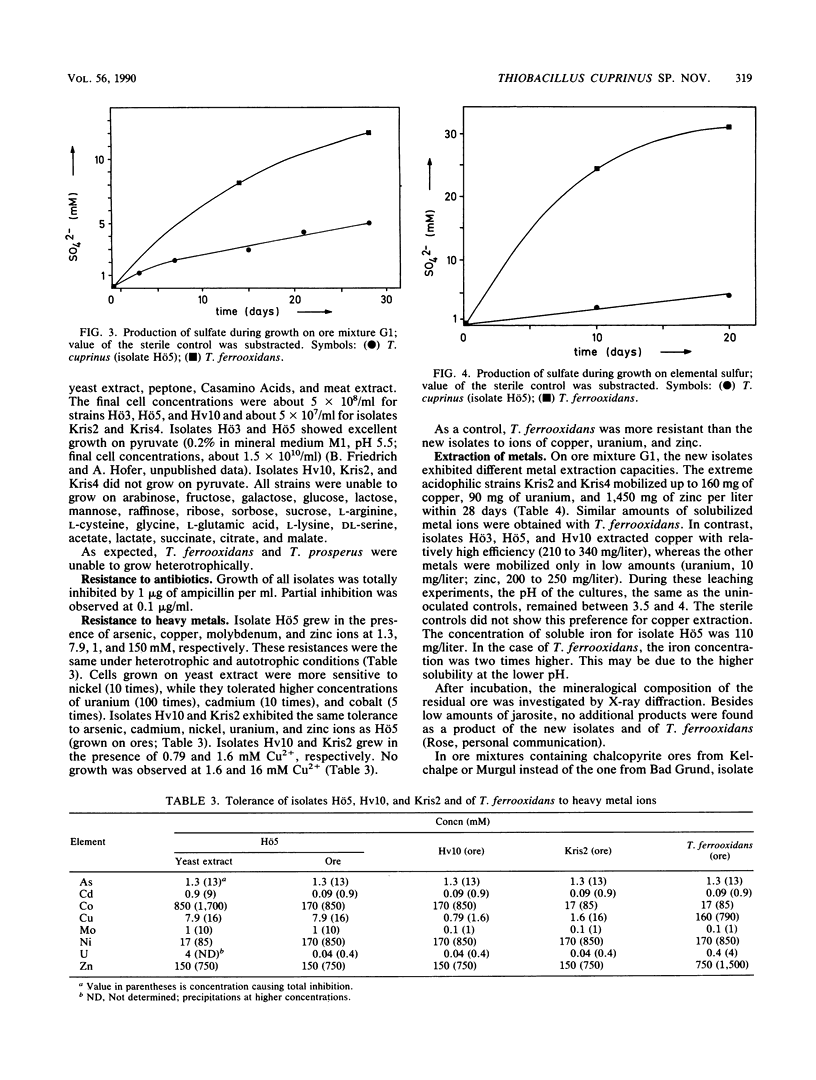
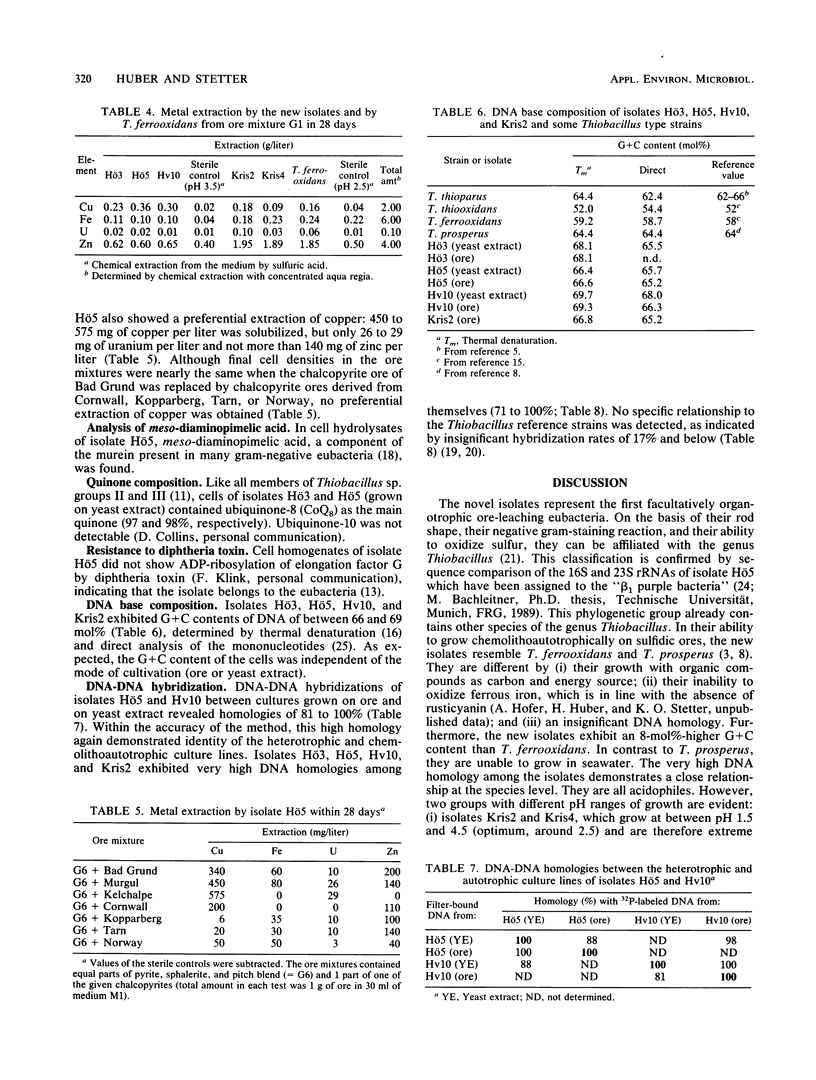
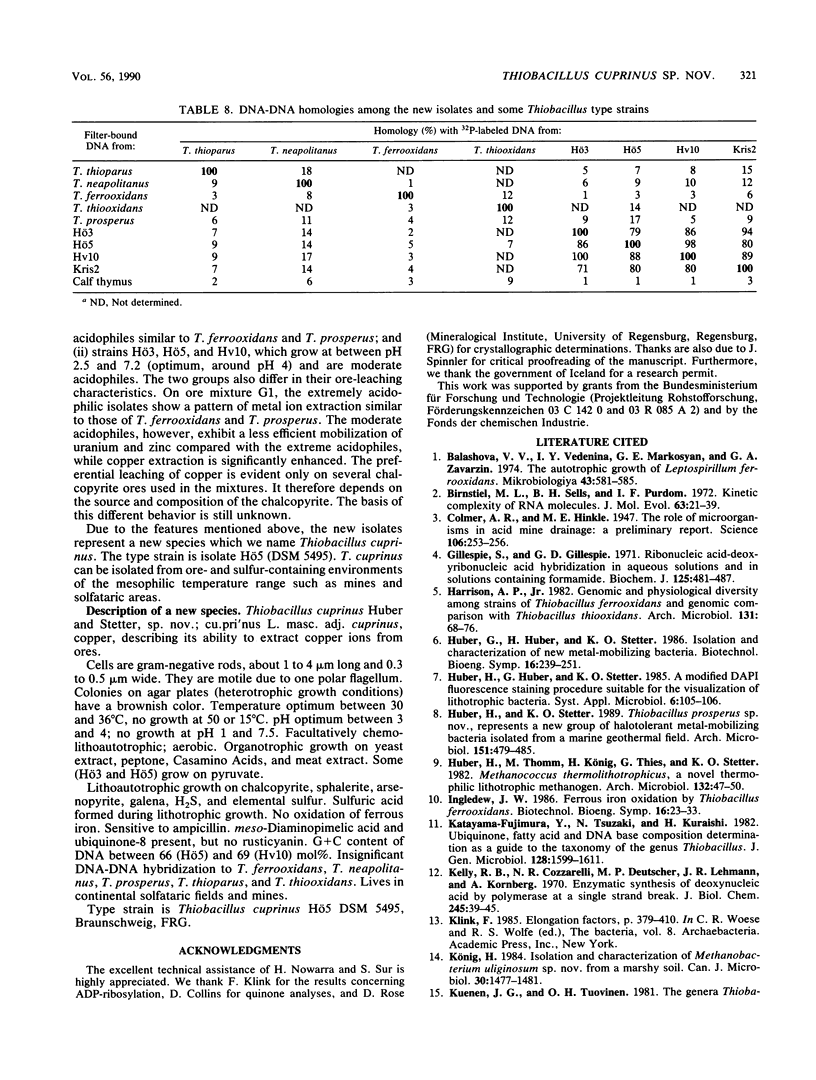
Five strains of mesophilic, facultatively organotrophic, ore-leaching eubacteria were isolated from solfatara fields in Iceland and a uranium mine in the Federal Republic of Germany. The new organisms are aerobic gram-negative rods. They can use sulfidic ores or elemental sulfur as sole energy source, indicating that they belong to the genus Thiobacillus. Alternatively, they grow on organic substrates such as yeast extract, peptone, and pyruvate. In contrast to the other leaching bacteria known so far, the new isolates are unable to oxidize ferrous iron. They consist of extreme and moderate acidophiles growing optimally at pH 3 and 4, respectively. The extreme acidophiles showed leaching characteristics similar to those shown by Thiobacillus ferrooxidans, while the moderate acidophiles exhibited a pronounced preference for copper leaching on some chalcopyrite ores. The G+C content of the DNA is between 66 and 69 mol%, depending on the isolate. In DNA-DNA hybridization experiments, the new strains showed homologies among each other of >70%, indicating that they belong to the same species. No significant DNA homology to Thiobacillus reference strains was detectable. Therefore, the new isolates represent a new species of Thiobacillus, which we name Thiobacillus cuprinus. Isolate Hö5 is designated as the type strain (DSM 5495).
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balashova V. V., Vedenina I. Ia, Markosian G. E., Zavarzin G. A. Leptospirillum ferrooxidans i osobennosti ee avtotrofnogo rosta. Mikrobiologiia. 1974 Jul-Aug;43(4):581–585. [PubMed] [Google Scholar]
- Birnstiel M. L., Sells B. H., Purdom I. F. Kinetic complexity of RNA molecules. J Mol Biol. 1972 Jan 14;63(1):21–39. doi: 10.1016/0022-2836(72)90519-0. [DOI] [PubMed] [Google Scholar]
- Colmer A. R., Hinkle M. E. The Role of Microorganisms in Acid Mine Drainage: A Preliminary Report. Science. 1947 Sep 19;106(2751):253–256. doi: 10.1126/science.106.2751.253. [DOI] [PubMed] [Google Scholar]
- Gillespie S., Gillespie D. Ribonucleic acid-deoxyribonucleic acid hybridization in aqueous solutions and in solutions containing formamide. Biochem J. 1971 Nov;125(2):481–487. doi: 10.1042/bj1250481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly R. B., Cozzarelli N. R., Deutscher M. P., Lehman I. R., Kornberg A. Enzymatic synthesis of deoxyribonucleic acid. XXXII. Replication of duplex deoxyribonucleic acid by polymerase at a single strand break. J Biol Chem. 1970 Jan 10;245(1):39–45. [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- Schleifer K. H., Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972 Dec;36(4):407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleifer K. H., Stackebrandt E. Molecular systematics of prokaryotes. Annu Rev Microbiol. 1983;37:143–187. doi: 10.1146/annurev.mi.37.100183.001043. [DOI] [PubMed] [Google Scholar]
- Steigerwalt A. G., Fanning G. R., Fife-Asbury M. A., Brenner D. J. DNA relatedness among species of Enterobacter and Serratia. Can J Microbiol. 1976 Feb;22(2):121–137. doi: 10.1139/m76-018. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]