Abstract
The phytopathogenic fungus Alternaria alternata f. sp. lycopersici (AAL) produces toxins that are essential for pathogenicity of the fungus on tomato (Lycopersicon esculentum). AAL toxins and fumonisins of the unrelated fungus Fusarium moniliforme are sphinganine-analog mycotoxins (SAMs), which cause inhibition of sphingolipid biosynthesis in vitro and are toxic for some plant species and mammalian cell lines. Sphingolipids can be determinants in the proliferation or death of cells. We investigated the tomato Alternaria stem canker (Asc) locus, which mediates resistance to SAM-induced apoptosis. Until now, mycotoxin resistance of plants has been associated with detoxification and altered affinity or absence of the toxin targets. Here we show that SAM resistance of tomato is determined by Asc-1, a gene homologous to the yeast longevity assurance gene LAG1 and that susceptibility is associated with a mutant Asc-1. Because both sphingolipid synthesis and LAG1 facilitate endocytosis of glycosylphosphatidylinositol-anchored proteins in yeast, we propose a role for Asc-1 in a salvage mechanism of sphingolipid-depleted plant cells.
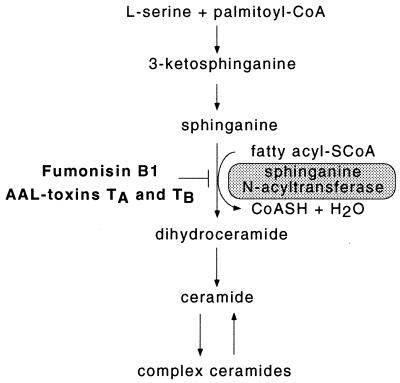
Mycotoxins are a continuous risk in the production and postharvest storage of crops, threatening animal and human health. They can be divided into the nonselective mycotoxins, phytotoxic to a broader range of organisms than the producing fungus infests and in the host-selective mycotoxins, primary determinants of phytopathogenicity (1). One class of host-selective mycotoxins is structurally related to sphinganine (2), a precursor in plant sphingolipid biosynthesis (Fig. 1) (3). Two nonrelated fungi are well known for the production of sphinganine-analog mycotoxins (SAMs): Fusarium moniliforme, a fungal pathogen of maize that produces fumonisin B1 (FB1) (4) and Alternaria alternata f. sp. lycopersici (AAL), a tomato pathogen that produces AAL toxins (5). SAMs competitively inhibit de novo sphingolipid (ceramide) biosynthesis in vitro (see Fig. 1), which leads to a variety of cellular responses, including the accumulation of sphingoid bases in animal cells (6) and plants (7). Besides their role as structural components of biomembranes, sphingolipids can be determinants in the proliferation or death of cells, depending on the type of cells studied (6). SAMs are toxic to all tissues of sensitive tomato cultivars at low concentrations (5, 8) and induce apoptosis in sensitive tomato lines (9). SAMs also induce apoptosis in some mammalian cell lines (2) and are toxic to animals and suspected to be conditionally carcinogenic in humans (4). These data indicate the necessity to understand the mechanism of action of SAMs, which has been performed by studying SAM resistance of tomato.
Figure 1.
De novo sphingolipid synthesis in plants. This scheme has been adapted from a recent report (3) in which enzyme activities and sphingolipid composition in plants were summarized. The conversion of sphinganine to ceramide is inhibited by FB1 and AAL toxins. Variations in plant ceramides exist by the presence of hydroxylated or (poly)unsaturated sphinganine as the sphingoid base moiety or by 2-hydroxylated fatty acyl groups. Predominant complex sphingolipids are glycosylceramides and inositolphosphorylceramides. No plant genes involved in this process have been characterized yet.
Until now, mycotoxin resistance of plants has been associated with detoxification and altered affinity or the absence of the toxin targets (1). Resistance or susceptibility to both AAL and SAMs in tomato is determined by the Alternaria stem canker (Asc) locus (5). It inherits in a single codominant fashion. Plants heterozygous for the Asc locus (Asc, asc) have an intermediate sensitivity to SAMs. AAL toxins are essential for the pathogenesis of AAL in tomato, because AAL toxin-deficient mutants cannot infest healthy leaves of the asc isoline (10). F. moniliforme, however, is not pathogenic on the asc isoline (8), indicating that FB1 produced by F. moniliforme is not sufficient for virulence on tomato. Our goal was to establish the molecular basis of the resistance and susceptibility of tomato to SAMs. Based on an established genetic analysis of the Asc locus (11), we have isolated the Asc gene by positional cloning. Sensitivity is associated with a mutation in the Asc gene that leads to a predicted aberrant ASC protein, a new plant member of the longevity assurance protein family.
Materials and Methods
Plant Materials.
For molecular analysis of the Asc and asc alleles, Clouse and Gilchrist (12) donated paired tomato lines isogenic and homozygous for the Asc locus [the Asc (resistant) and asc (sensitive) isolines]. For the production of YAC442A, the SAM-resistant tomato line VFNT Cherry has been used by Bonnema et al. (13). The SAM-sensitive asc, asc line Tester-3 (11) has been used in the functional complementation assay.
Isolation of the Asc-1 Gene.
All nucleic acid manipulations were performed according to Sambrook et al. (14) unless otherwise stated. A phage Lambda and a binary plasmid library were produced by partial Sau3A digestion of genomic DNA from the YAC442A yeast strain (11), size selection for DNA fragments larger than 10 kb and ligation into BamHI-digested phage Lambda GEM-11 arms (Promega) or pCGN1548 (15), respectively. EcoRI DNA fragments (Fig. 2A) from Lambda GEM-11 clones were converted to cleaved amplified polymorphic sequence markers (16) and the approximate positions of the recombinant breakpoints were determined. The clones pBAsc1 and pBAsc2 were isolated from the pCGN1548 library to encompass the Asc region within the nearest recombinant breakpoints. Because the pCGN1548 backbone is incompatible with Agrobacterium rhizogenes, the pBAsc1 and pBAsc2 clones were further subcloned into the binary vector pGreen029 (details about the pGreen/pSoup binary vector system are available at http://www.pGreen.ac.uk) as BamHI and Asp718 fragments, resulting in pBAsc101 and pBAsc206, respectively. To get a binary construct from which ORF1 is exclusively expressed, two SacI fragments (encompassing the 3′ region of ORF1; 1,766 bp) and an ApaI fragment (Asc-1 upstream region; 4,511 bp) were subsequently deleted from pBAsc206 to obtain pBAsc404 (Fig. 2A). Partial putative ORFs were predicted at http://dot.imgen.bcm.tmc.edu:9331/gene-finder/gf.html (17) and further analyzed with the blast algorithms at the National Center for Biotechnology Information website (18). Full-length clones from the Newman PRL2 cDNA library were donated by the Arabidopsis Biological Resource Center of Ohio State University (Columbus, OH) for 220P10T7 [Longevity Assurance Gene 1 (LAG1) At-1] and 92N21T7 (LAG1 At-2) from Arabidopsis thaliana ecotype Columbia. The tomato DNA inserts of pBAsc1 and pBAsc2 and the above cDNA clones were sequenced in both directions by dye terminator cycle sequencing (Applied Biosystems). The asc-1 gene was amplified with Asc-specific oligonucleotides BASC86 (EcoRI tail, forward) 5′-CGGGATCCCGATCAGTCTTTGTGGTCATCATC-3′ and BASC87 (BamHI tail, reverse) 5′-GGAATTCCTGCAATTCATTTGAAACTACAAC-3′, subsequently cloned in pGEM-Teasy (Promega), and multiple clones were sequenced in both directions to find differences with the Asc-1 gene avoiding PCR artifacts. Contiguous DNA sequences were constructed with the sequencher 3.0 software (Gene Codes, Ann Arbor, MI). Transmembrane (TM) segments were predicted with the tmhmm1.0 server at the Center for Biological Sequence Analysis, Technical University of Denmark (http://www.cbs.dtu.dk/services/TMHMM-1.0/).
Figure 2.
Molecular isolation and characterization of Asc-1. (A) Isolation of Asc-1. Genetic distance of chromosome markers are in centimorgans (cM); overlapping YACs are light gray boxes, in the detail of YAC442A (dark gray) are the EcoRI fragments positioned at zero recombinants. The corresponding chromosome segment in the asc, asc isoline (asc-1) contains deletions (blank gaps). Tomato DNA fragments used in the A. rhizogenes-mediated transformation are black lines. Only DNA constructs which give functional complementation in the hairy root bioassay (+) are depicted. ORF1 is Asc-1 (2.5 kb), which contains six exons (black boxes). The second exon of the asc-1 gene contains a 2-bp deletion (ΔAG). (B) Hairy root bioassay with two representative transformants of pGreen029 (empty binary vector) and pBAsc404 (containing Asc-1) on SAMs. (C) Genomic DNA blot of the Asc isolines and YAC442A hybridized with the coding region of the Asc-1 gene (dashed line in A). The deletions in the asc-1 promoter region are detected by a shift of a 4.2- (Asc) to a 3.9-kb (asc) HindIII fragment. (D) Competitive RT-PCR for visualizing relative mRNA levels of Asc-1 and asc-1 in leaves of the Asc isolines. Experiments were repeated three times from individual plants with similar results. The upper fragment represents the 457-bp fragment amplified from added competitor genomic Asc-1 DNA; the lower band represents the Asc-1-specific 289-bp cDNA fragment.
Hairy Root Functional Complementation Assay.
The binary vector pGreen029 or its derivatives carrying tomato DNA inserts were transferred to A. rhizogenes LBA9402 together with the helper plasmid pSoup by electroporation and selected on yeast mannitol broth (YMB) (0.4 g/liter yeast extract/10 g/liter mannitol/0.1 g/liter NaCl/0.5 g/liter K2HPO4/0.2 g/liter MgSO4·7H2O) with 50 mg/liter kanamycin. Hypocotyl explants (3 cm) from 7-day-old Tester-3 tomato seedlings grown on root medium [half-strength Murashige and Skoog salts with vitamins/30 g/liter sucrose/8 g/liter plant agar (Duchefa, The Netherlands)] in vitro, were dipped in an A. rhizogenes suspension and cocultivated for 48 h on root medium. Transgenic hairy roots were selected and cleared from A. rhizogenes by subculturing on root medium supplemented with 50 mg/liter kanamycin/500 mg/liter carbenicillin/100 mg/liter vancomycin. The presence of the transgenes was verified by PCR and Southern blot analysis. A 95:5% mixture of AAL toxins TA/TB was isolated as described (19), and fumonisin B1 was obtained commercially (Sigma). Hairy root tips were tested for complementation to SAM resistance by determination of root growth (white root cap; 1 cm/day elongation) or death (brown root tip; less than 1 mm/day elongation) on root medium supplemented with 0.07 μM AAL toxins mixture or 0.7 μM fumonisin B1.
DNA and RNA Analysis.
Tomato Asc isoline plants were grown in pots under standard Dutch greenhouse conditions (20°C, 60% humidity, 15 kilolux light intensity maintained by automated artificial lights) and mature leaves were harvested 3 weeks after emergence of the seedlings. A DNA blot with XbaI- or HindIII-digested genomic DNA (7.5 μg) of these leaves was prepared with positively charged nylon membranes (Hybond N+, Amersham International) as described (20). The DNA blot was hybridized with a radiolabeled 1.5-kb PCR fragment prepared with the primers BASC86 and BASC87 encompassing the complete Asc-1 ORF, washed at low stringency (2× SSC at 60°C), and analyzed by a PhosphoImager (Molecular Dynamics). Total RNA was isolated from the above leaves in three independent experiments as described (20) and mRNA was subsequently isolated by the Oligotex procedure (Qiagen, Chatsworth, CA). Reverse transcription–PCR (RT-PCR) was performed by reverse transcription of 0.25 μg of mRNA with Expand by using the instructions of the manufacturer (Boehringer Mannheim). Sequences of ORF1 and ORF2 were determined by amplification of the cDNA with the primers BASC86 and BASC87 for ORF1, and with BASC76 (forward) 5′-AGAGGGAGCTGACATTTG-3′ and BASC77 (reverse) 5′-CCACAAATCTTGTCCTATCT-3′ for ORF2. In competitive RT–PCR amplifications of Asc (21), equal volumes of the cDNA preparation, a logarithmic dilution series of pBF103 [a pGEM3z (Promega)] plasmid containing the 5.0-kb EcoRI fragment with a complete genomic copy of Asc-1 and the Asc-specific oligonucleotides BASC88 5′-GCTGCGCAGATAATTCATCA-3′ (forward) and BASC89 5′-TCCGTTCCTTCTGTATATCC-3′ (reverse) were used.
Leaflet Bioassay.
Detached leaflets of greenhouse-grown Asc and asc isoline plants were incubated in triplicate in 9-cm Petri dishes on filter papers (Schleicher & Schüll #595, Dassel, Germany) saturated with 0.1% (vol/vol) Tween-80 (control), supplemented with 0.2 μM AAL toxins mixture, 100 μg/ml (wt/vol) ceramide type III or both. Ceramide type III (Sigma C2137) was dissolved at 50°C in 96% ethanol containing 0.1% Tween-80 before making dilutions. Tween-80 did not induce necrotic symptoms and was essential to keep the lipophilic ceramides in solution. Symptoms were observed 72 h after incubation in a climate cabin (22°C) in the light (3 kilolux).
Results
Functional Complementation of the asc Mutant.
Position-based molecular isolation of Asc-1 (Fig. 2A) was initiated from adjacent chromosome markers TG134 and TG442 and a plant population with recombinations around the Asc locus (11). This approach enabled the isolation of a YAC442A encompassing the Asc locus. In addition, the YAC442A molecular markers P6 and C18 have been produced, residing at one and three recombinations from Asc, respectively (11). Chromosome walking from P6 and C18 in a Lambda phage YAC442A library produced a 70-kb contiguous stretch of Lambda clones spanning the Asc locus. By positioning the nearest recombinant breakpoints adjacent to Asc, the genomic region to be used for functional complementation was confined to 17 kb (Fig. 2A). Functional complementation by YAC442A DNA fragments in a sensitive asc, asc background was accomplished by the induction of A. rhizogenes-induced transgenic hairy roots and subsequently testing root viability on SAMs. Before transgenic roots were tested, it was crucial to optimize the SAM concentrations to discriminate between hairy roots induced with a wild-type A. rhizogenes on explants of the Asc, asc and the asc, asc tomato genotype. In general, SAM concentrations above 1 μM lead to growth inhibition of Asc, asc hairy roots, whereas SAM concentrations below 20 nM had no effects on asc, asc hairy roots. For AAL toxins, a concentration of 70 nM and for FB1, a concentration of 700 nM was found to be optimal for discrimination. This is in agreement with the observation that FB1 is approximately 20 times less phytotoxic for detached tomato leaflets than AAL toxins (8). Two genomic DNA fragments (pBAsc102 and pBAsc206) were identified that, on transformation, resulted in the generation of SAM-resistant roots at the discriminating concentrations, indicating that SAM resistance is a dominant property (Fig. 2A). Roots transformed with the control vector pGreen029 could be distinguished from SAM-resistant roots by the inhibition of root elongation, necrosis of the root tip, and deformation of root hairs (Fig. 2B).
Identification and Characterization of the Asc Gene.
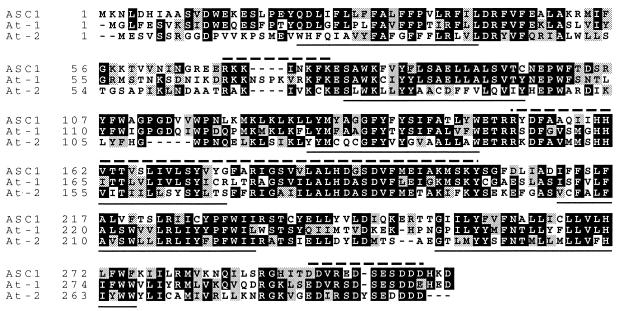
By sequencing of the pBAsc102 and pBAsc206 plasmid inserts which complemented the asc, asc mutant phenotype, two closely linked putative ORFs potentially encoding Asc have been detected in the 17-kb contiguous DNA sequence. Both ORFs are expressed in resistant tomato lines, because the corresponding mRNAs have been detected by RT–PCR analysis. DNA sequence determination of RT–PCR products of ORF2 did not reveal any differences between the Asc isolines (data not shown). To obtain a binary construct from which ORF1 is exclusively expressed, a deletion construct was made from pBAsc206 to obtain pBAsc404 (see Fig. 2A). Complementation assays of pBAsc404 confirmed that ORF1 driven by its endogenous promoter (Asc-1; Fig. 2B) mediates SAM resistance, demonstrating that ORF1 encodes the ASC1 protein. Analysis of DNA blots indicated that Asc-1 is a single-copy gene and one homolog is present in the asc isoline, further referred to as the asc-1 gene (Fig. 2C). Because the asc isoline has no obvious visible phenotype in the absence of SAM, this gene does not appear to be essential for plants under standard greenhouse conditions. Intron positions in Asc-1 were determined by comparison of the cDNA with the genomic DNA sequences, because several positions were not predicted by genefinder algorithms. The predicted ASC1 36-kDa protein contains six TM helices, a basic domain between TM1 and TM2, and a C-terminal acidic domain (Fig. 3). Expression of Asc-1 and asc-1 could not be detected on blots containing mRNA isolated from leaves, flowers, and immature fruits (data not shown). However, in a competitive RT-PCR amplification with Asc-specific oligonucleotides, the expression of Asc-1 and asc-1 was found to be equal (Fig. 2D). To further investigate the difference between SAM-resistant and -sensitive genotypes, PCR with Asc-1-specific oligonucleotides and subsequent sequencing revealed five deletions ranging from 4 to 133 bp (in total 300 bp) in the promoter region of the asc-1 gene. These deletions are visible as a single restriction fragment length polymorphism on Southern blots of HindIII-digested genomic DNA of the Asc isolines (Fig. 2C). Apparently, these deletions do not affect transcription of asc-1. The most significant mutation detected in the sensitive genotype is a 2-bp deletion in the second exon of the asc-1 gene that leads to a premature stop codon in exon 2 (ΔAG in Fig. 2A). Thus, sensitivity of asc, asc plants to SAM is likely associated with a truncated aberrant 10-kDa protein, which provides additional support that ORF1 mediates SAM insensitivity. The intermediate SAM sensitivity of heterozygous Asc, asc plants can be explained by a gene-dose effect, assuming that the steady-state level of Asc-1 mRNA determines the abundance of ASC1 protein.
Figure 3.
Amino acid alignment of the L. esculentum ASC1 (Le) and A. thaliana LAG1 (At-1 and At-2) protein homologs. The ASC-1 and LAG1 At-1 homologs are 53% identical (black) and 73% similar (gray). The predicted TM spanning helices are underlined, the basic domain at amino acids 57–83 (in ASC1), the LAG1 motif at amino acid 151 (in ASC1), and the C-terminal acidic domain are indicated with dashed lines.
Asc Belongs to the Longevity Assurance Gene Family.
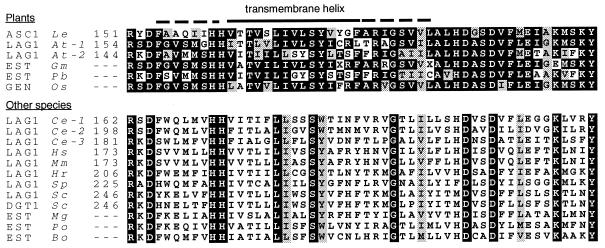
Asc-1 has no homology to any published plant disease resistance gene, which has been expected because resistance is based on insensitivity to SAMs, as opposed to the specific response to pathogen-encoded elicitor molecules in gene-for-gene host-pathogen interactions. Interestingly, Asc-1 is homologous to the Saccharomyces cerevisiae LAG1 that has been associated with life span in yeast (22). By accessing public DNA databases for expressed sequence tags and full-length cDNAs, LAG1 homologs were found to be transcribed in fish, mammals, insects, higher fungi, and monocotyledonous and dicotyledonous plants (Fig. 4). The cDNAs for two A. thaliana homologs (LAG1 At-1 and At-2) were donated by the Arabidopsis Biological Resource Center and sequenced. Although the overall homology between the known LAG1 proteins among distantly related species is limited (for instance, 26% identity and 46% similarity for the S. cerevisiae LAG1 with ASC1 over 141 amino acids), they share a similar six or seven TM helices profile and the LAG1 motif (23). In addition, a C-terminal acidic domain is present in most known LAG1 homologs. The LAG1 motif is conserved in evolution, suggesting a role for LAG1 proteins in a basic cellular function in eukaryotes (Fig. 4). The LAG1 homologs of the SAM-resistant A. thaliana and tomato show a similar intron/exon and protein domain structure (Fig. 3). Rice, soybean, and poplar tree were also found to transcribe Asc-1 homologs (Fig. 4). Monocotyledonous species, including maize and rice, have a high level of resistance (24) and only few dicotyledonous plant species are susceptible to SAMs (5), suggesting that Asc-1 homologs are functional in most plant species, particularly monocotyledonous species.
Figure 4.
Alignment of the 52-aa LAG1 motif of various organisms (GenBank accession nos. are mentioned between parentheses; identical and similar aa are boxed black or gray, respectively). Plant motifs have been aligned separately from the other species motifs. (Le) Lycopersicon esculentum (tomato, AF198177); (At) Arabidopsis thaliana [thale cress, AF198179 (At-1) and AF198180 (At-2)]; (Gm) Glycine max (soybean, AI965754); (Pb) Populus balsamifera ssp. trichocarpa (poplar tree, AI166337); (Os) Oryza sativa (rice, AQ365516); (Ce) Caenorhabditis elegans [U42438 (Ce-1); U40415 (Ce-2), and CAA21723.1 (Ce-3)]; (Hs) Homo sapiens (human, M62302); (Mm) Mus musculus (mouse, M62301); (Hr) Halocynthia roretzi (sea squirt, BAA81907); (Sp) Schizosaccharomyces pombe (fission yeast, U76608); (Sc) Saccharomyces cerevisiae [bakers' yeast, U08133 (LAG1); YKL008c (DGT1)]; (Mg) Magnaporthe grisea (rice blast fungus, AI069387); (Po) Paralichthys olivaceus (Japanese flounder, C82217); (Bo) Bombyx mori (domestic silk worm, AU002679). The numbers correspond with amino acid position in proteins predicted from completely sequenced LAG1 cDNAs. Expressed Sequence Tags (EST) are LAG1 motifs predicted from partial cDNAs; GEN is a rice genomic survey sequence for which a corresponding EST (AU064495) exists.
Discussion
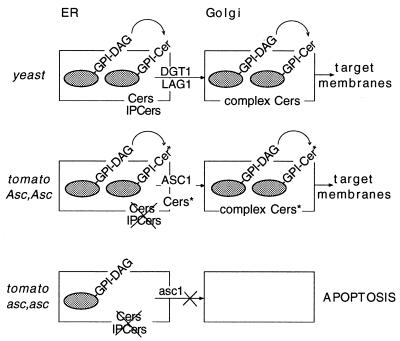
A knockout mutant of LAG1 in yeast shows no visual phenotype (22) because of the presence of the delayed endoplasmic reticulum (ER)-to-Golgi transport (DGT1) functional homolog. A double-knockout mutant (ΔLAG1/ΔDGT1) leads to slow growth and cell-wall defects (25). Expression of the Homo sapiens and Caenorhabditis elegans LAG1 homologs in a ΔLAG1/ΔDGT1 yeast strain showed that, despite low amino acid homology, they could functionally complement the mutant phenotype (23). LAG1 and DGT1 facilitate ER-to-Golgi transport of glycosylphosphatidylinositol (GPI)-anchored proteins (EGGAP transport), but are not involved in GPI-anchor attachment or general translocation of proteins across the ER membrane (25). In addition, EGGAP transport is dependent on de novo sphingolipid synthesis in yeast (26–29) and humans (30). GPI anchors provide an alternative to TM domains for the anchoring of proteins to membrane surfaces. Both sphingolipids and GPI-anchored proteins are present in lipid rafts, sphingolipid-cholesterol-enriched membrane subdomains postulated to be involved in membrane trafficking, endocytosis and polarized sorting of proteins (31). In plants, the arabinogalactan proteins have recently been discovered to be GPI anchored (32). Treatment of A. thaliana cell cultures with the arabinogalactan-binding Yariv agent leads to programmed cell death, implicating a role for GPI-anchored proteins in apoptosis of plant cells (33). Together, these data suggest that after SAM-induced inhibition of de novo sphingolipid (ceramide) biosynthesis, Asc-1 prevents apoptosis in resistant plants by the restoration of EGGAP transport (Fig. 5). This would be a unique response to a pathogen-encoded molecule in plants which is neither based on known types of toxin resistance (1) nor the specific recognition of an elicitor and initiation of defense reactions mediated by the majority of plant disease-resistance genes identified until now (34).
Figure 5.
Working hypothesis for SAM resistance in tomato plants. In yeast, ceramides (Cers) and inositolphosphorylceramides (IPCers) are synthesized on the ER membrane (rectangle). GPI anchors are synthesized in the ER and covalently linked to certain proteins (gray ovals) (32). In a subset of yeast GPI proteins, the diacylglycerol (DAG) membrane anchor moiety is remodeled to Cer (35). In resistant tomato plants, ASC1 could salvage ER-to-Golgi transport of GPI-anchored proteins (EGGAP transport) by the production of alternative ceramides (Cer*) in the absence of SAM-sensitive ceramide synthesis after SAM exposure. In sensitive plants, EGGAP transport could be halted after SAM exposure because of a lack of alternative ceramide production by the dysfunctional ASC1 protein, leading to apoptosis.
It remains enigmatic how ASC1 mediates resistance to SAM-induced effects on the concentration and composition of sphingolipids at the biochemical level. Viable yeast suppressor mutants that lack sphingolipids, but display a normal EGGAP transport could provide clues to this question (26, 28). For instance, a suppressor mutation in the sphingolipid compensation gene (SLC1–1) is involved in the production of nonceramide lipids which enables yeast to grow without making sphingolipids (28). In analogy, Asc-1 may act as a salvage mechanism to restore EGGAP transport by encoding a similar enzyme, although SLC1 has no significant homology with ASC1. Sphingolipid precursors accumulate in both AAL toxin-treated Asc and asc isoline plants (7), but it has not been tested if complex ceramides are depleted. It is thus not clear which of the two events is tolerated in SAM-resistant tomato plants. Interestingly, ceramides rescue detached sensitive tomato leaves from SAM-induced apoptosis (Fig. 6), suggesting that only ceramide depletion is responsible for SAM toxicity in the asc isoline. Consequently, ceramides may not be depleted completely in the Asc isoline as a result of an enzymatic activity encoded by Asc-1. The presence of both SAM-insensitive and -sensitive sphinganine-N-acyltransferase activities has been reported for beans and squash (3). Alternative to the first hypothesis, ASC1 could thus be a SAM-insensitive sphinganine-N-acyltransferase, assuming the presence of a SAM-sensitive sphinganine-N-acyltransferase encoded by an Asc-1 homolog in both Asc isolines. Because ceramide precursors such as sphinganine (Fig. 1) accumulate in the Asc isoline after SAM exposure (7), the relative contribution of a possible SAM-insensitive sphinganine-N-acyltransferase to the overall de novo ceramide synthesis should be limited. The possible presence of an Asc-1 homolog is in contrast with the apparent single-copy nature of Asc-1 (Fig. 2D). However, in S. cerevisiae, C. elegans, and A. thaliana, at least two copies of LAG1 homologs exist (Fig. 3). The human and C. elegans LAG1 genes could not be isolated by using the yeast LAG1 gene as a probe, likely because of limited nucleic acid homology (23). By using lower-stringency hybridization conditions, however, a single Asc-1 homolog has been discovered in both Asc isolines (not shown). In contrast to Asc-1 (Fig. 1C), no restriction fragment polymorphisms were detected for this homolog, which may be redundant with the function of Asc-1, analogous to the DGT1 gene in yeast (23, 25). We propose that ASC1 compensates for a depletion in ceramides after SAM exposure and thereby protects the cell from arrest of EGGAP transport. In conclusion, the LAG1-like Asc-1 gene rescues tomato hairy roots from SAM-induced cell death which opens the way to study the role of sphingolipid-dependent transport of GPI-anchored proteins in the apoptosis of plant cells.
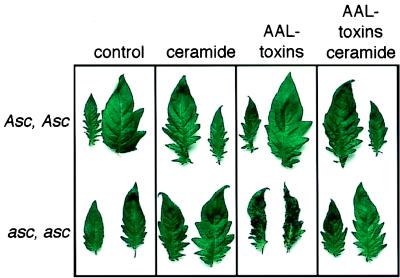
Figure 6.
Ceramide type III rescues AAL toxin-induced apoptosis. The control incubation or ceramide do not result in cell death in detached leaflets of both Asc isolines. At the concentrations used, AAL toxins only induce interveinal cell death and loss of turgor in the SAM-sensitive asc, asc isoline. The cell death is counteracted by the addition of exogenous ceramides during toxin treatment.
Acknowledgments
We thank Dr. D. Gilchrist for the L. esculentum Asc isolines, Drs. P. Vos and G. Bonnema for the tomato YAC libraries, Dr. P. Mullineaux for the pGreen binary vector system, Dr. P. Hooykaas for the A. rhizogenes strain, M. Kuipers for greenhouse assistance, Dr. D. Lynch for communicating unpublished data, and Ir. J. Folders and Dr. M. Joosten for critically reading the manuscript. L.A.M. received a grant from the European Community. B.F.B. and P.L.L. received grants from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek and the Associatie van Biotechnologische Onderzoeksscholen in Nederland.
Abbreviations
- AAL
Alternaria alternata f. sp. lycopersici
- SAMs
sphinganine-analog mycotoxins
- Asc
Alternaria stem canker
- YAC
yeast artificial chromosome
- RT-PCR
reverse transcription–PCR
- TM
transmembrane
- LAG1
longevity assurance gene 1
- ER
endoplasmic reticulum
- DGT
delayed ER-to-Golgi transport gene
- GPI
glycosylphosphatidylinositol
- EGGAP transport
ER-to-Golgi transport of GPI-anchored proteins
- FB1
fumonisin B1
Footnotes
References
- 1.Walton J D. Plant Cell. 1996;8:1723–1733. doi: 10.1105/tpc.8.10.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Jones C, Ciacci-Zanella J R, Holt T, Gilchrist D G, Dickman M B. Proc Natl Acad Sci USA. 1996;93:3461–3465. doi: 10.1073/pnas.93.8.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lynch D V. Methods Enzymol. 2000;311:130–149. doi: 10.1016/s0076-6879(00)11074-2. [DOI] [PubMed] [Google Scholar]
- 4.Dutton M F. Pharmacol Ther. 1996;70:137–161. doi: 10.1016/0163-7258(96)00006-x. [DOI] [PubMed] [Google Scholar]
- 5.Brandwagt B F, Mesbah L M, Laurent P J J, Takken F T, Kneppers T, Nijkamp H J J, Hille J. In: Molecular Genetics of Host-Specific Toxins in Plant Disease. Kohmoto K, Yoder O C, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 317–330. [Google Scholar]
- 6.Merrill A H, Jr, Schmelz E-M, Dillehay D L, Spiegel S, Shayman J A, Schroeder J J, Riley R T, Voss K A, Wang E. Toxicol Appl Pharmacol. 1997;142:208–225. doi: 10.1006/taap.1996.8029. [DOI] [PubMed] [Google Scholar]
- 7.Abbas H K, Tanaka T, Duke S O, Porter J K, Wray E M, Hodges L, Sessions A E, Wang E, Merrill A H, Riley R T. Plant Physiol. 1994;106:1085–1093. doi: 10.1104/pp.106.3.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gilchrist D G, Ward B, Moussato V, Mirocha C J. Mycopathologia. 1992;117:57–64. [Google Scholar]
- 9.Wang H, Li J, Bostock R M, Gilchrist D G. Plant Cell. 1996;8:375–391. doi: 10.1105/tpc.8.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akamatsu H, Itoh Y, Kodama M, Otani H, Kohmoto K. Phytopathology. 1997;87:967–972. doi: 10.1094/PHYTO.1997.87.9.967. [DOI] [PubMed] [Google Scholar]
- 11.Mesbah L A, Kneppers T J, Takken F L, Laurent P, Hille J, Nijkamp H J. Mol Gen Genet. 1999;261:50–57. doi: 10.1007/s004380050940. [DOI] [PubMed] [Google Scholar]
- 12.Clouse S D, Gilchrist D G. Phytopathology. 1987;77:80–82. [Google Scholar]
- 13.Bonnema G, Hontelez J, Verkerk R, Zhang Y Q, Vandaelen R, van Kammen A, Zabel P. Plant J. 1996;9:125–133. doi: 10.1046/j.1365-313x.1996.09010125.x. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainville, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.McBride K E, Summerfelt K R. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 16.Konieczny A, Ausubel F M. Plant J. 1993;4:403–410. doi: 10.1046/j.1365-313x.1993.04020403.x. [DOI] [PubMed] [Google Scholar]
- 17.Smith R F, Wiese B A, Wojzynski M K, Davidson D B, Worley K C. PCR Methods Appl. 1996;6:454–462. [Google Scholar]
- 18.Altschul S F, Madden T L, Schaffer A A, Zhang J H, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clouse S D, Martensen A N, Gilchrist D G. J Chromatogr. 1985;350:255–263. doi: 10.1016/s0021-9673(01)93524-1. [DOI] [PubMed] [Google Scholar]
- 20.van der Biezen E A, Brandwagt B F, van Leeuwen W, Nijkamp H J J, Hille J. Mol Gen Genet. 1996;251:267–280. doi: 10.1007/BF02172517. [DOI] [PubMed] [Google Scholar]
- 21.Siebert P D, Larrick J W. Nature (London) 1992;359:557–558. doi: 10.1038/359557a0. [DOI] [PubMed] [Google Scholar]
- 22.D'Mello N P, Childress A M, Franklin D S, Kale S P, Pinswasdi C, Jazwinski S M. J Biol Chem. 1994;269:15451–15459. [PubMed] [Google Scholar]
- 23.Jiang J C, Kirchman P A, Zagulski M, Hunt J, Jazwinski S M. Genome Res. 1998;8:1259–1272. doi: 10.1101/gr.8.12.1259. [DOI] [PubMed] [Google Scholar]
- 24.Abbas H K, Tanaka T, Duke S O, Boyette C D. Weed Technol. 1995;9:125–130. [Google Scholar]
- 25.Barz W P, Walter P. Mol Biol Cell. 1999;10:1043–1059. doi: 10.1091/mbc.10.4.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath A, Sutterlin C, Manning-Krieg U, Movva N R, Riezman H. EMBO J. 1994;13:3687–3695. doi: 10.1002/j.1460-2075.1994.tb06678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Futerman A H. Trends Cell Biol. 1995;5:377–380. doi: 10.1016/s0962-8924(00)89078-9. [DOI] [PubMed] [Google Scholar]
- 28.Skrzypek M, Lester R L, Dickson R C. J Bacteriol. 1997;179:1513–1520. doi: 10.1128/jb.179.5.1513-1520.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sutterlin C, Doering T L, Schimmoller F, Schroder S, Riezman H. J Cell Sci. 1997;110:2703–2714. doi: 10.1242/jcs.110.21.2703. [DOI] [PubMed] [Google Scholar]
- 30.Ledesma M D, Simons K, Dotti C G. Proc Natl Acad Sci USA. 1998;95:3966–3971. doi: 10.1073/pnas.95.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons K, Ikonen E. Nature (London) 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 32.Schultz C, Gilson P, Oxley D, Youl J, Bacic A. Trends Plant Sci. 1998;3:426–431. [Google Scholar]
- 33.Gao M, Showalter A M. Plant J. 1999;19:321–331. doi: 10.1046/j.1365-313x.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- 34.Martin G B. Curr Opin Plant Biol. 1999;2:273–279. doi: 10.1016/S1369-5266(99)80049-1. [DOI] [PubMed] [Google Scholar]
- 35.Reggiori F, Conzelmann A. J Biol Chem. 1998;273:30550–30559. doi: 10.1074/jbc.273.46.30550. [DOI] [PubMed] [Google Scholar]