Abstract
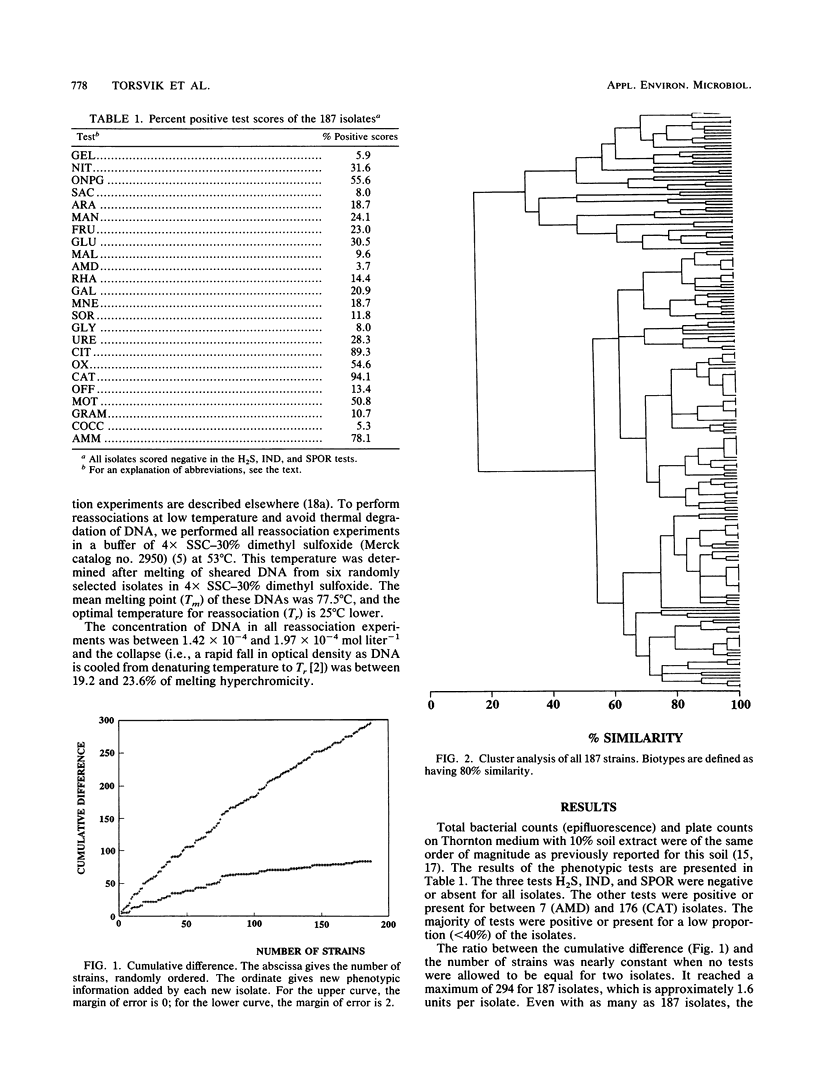
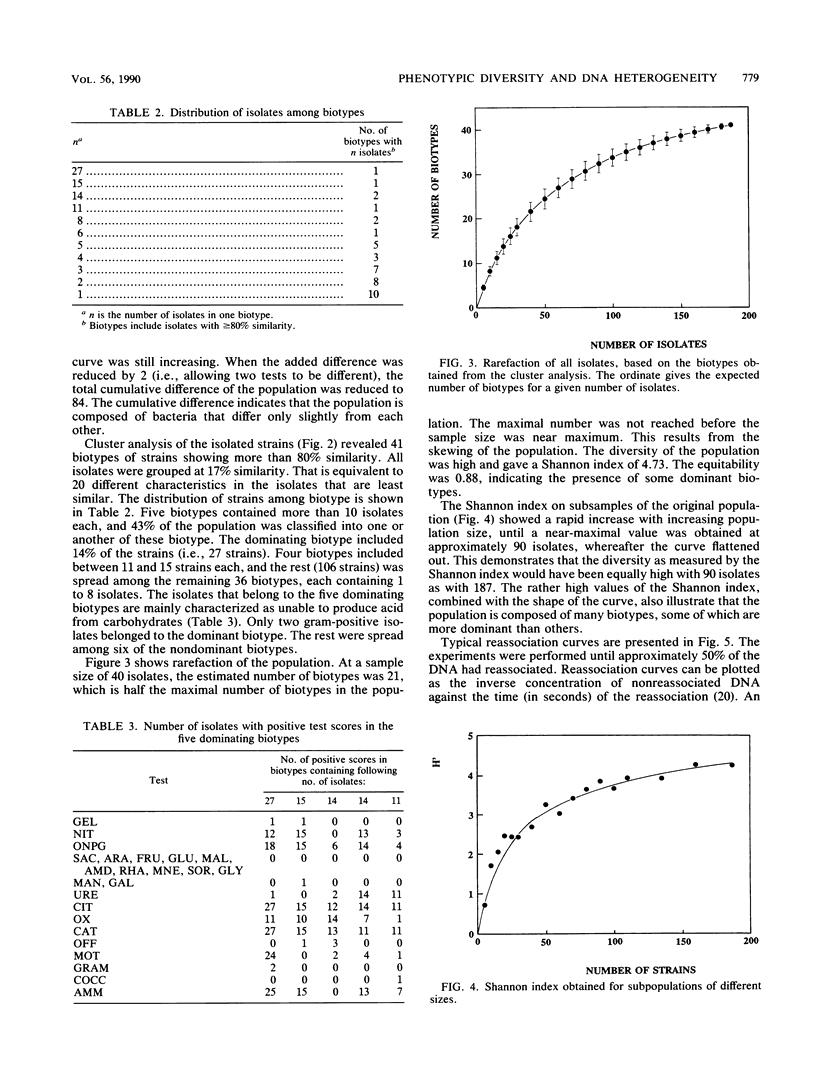
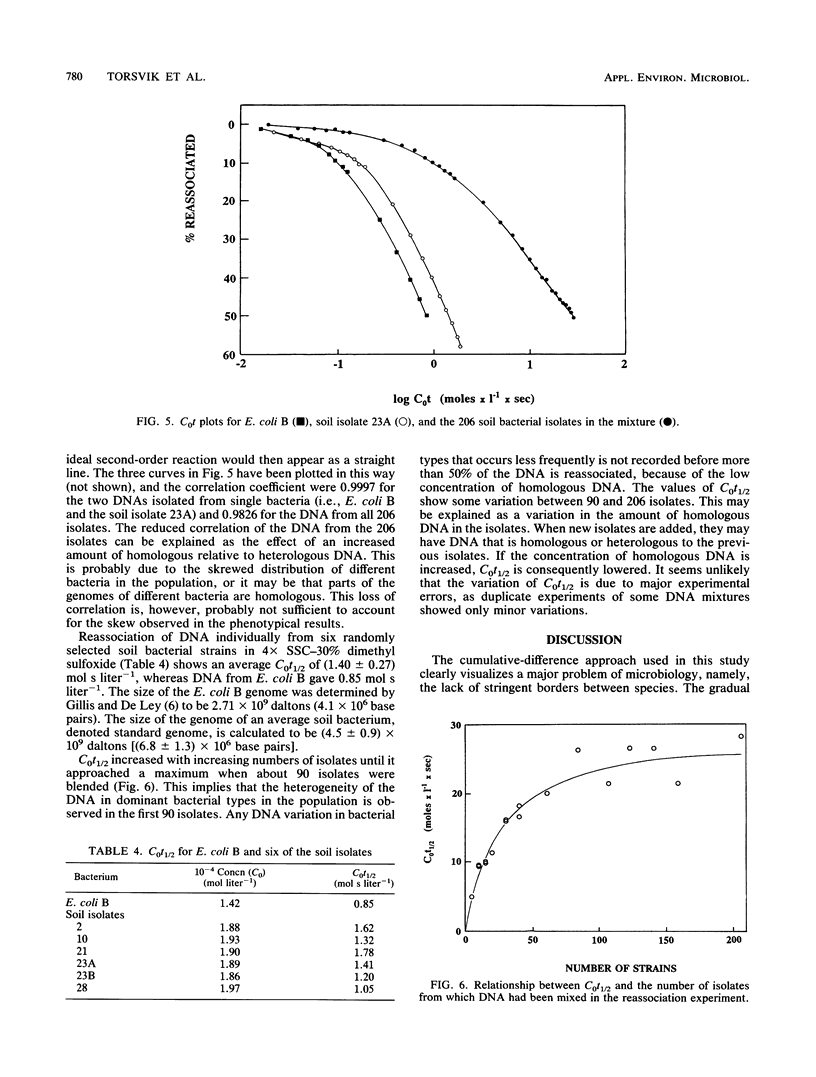
The phenotypic diversity of about 200 bacterial strains isolated from soil was compared with the genotypic diversity of the same population. The strains were phenotypically characterized by the API 20B test system. The results of these tests were subjected to cluster analysis, which revealed 41 biotypes at 80% similarity. The five dominating biotypes contained 43% of the strains. The phenotypic diversity as determined by the Shannon index, equitability, rarefaction, and cumulative differences was high, but indicated some dominant biotypes. The genetic diversity was measured by reassociation of mixtures of denatured DNA isolated from the bacterial strains (C0t plots). The observed genetic diversity was high. Reassociation of DNA from all bacterial strains together revealed that the population contained heterologous DNA equivalent to 20 totally different bacterial genomes (i.e., genomes that have no homology). This study showed that reassociation of DNA isolated from a collection of bacteria gave a good estimate of the diversity of the collection and that there was good agreement with different phenotypic diversity measures. The Shannon index in particular has features in common with the genetic diversity measure presented here.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Britten R. J., Graham D. E., Neufeld B. R. Analysis of repeating DNA sequences by reassociation. Methods Enzymol. 1974;29:363–418. doi: 10.1016/0076-6879(74)29033-5. [DOI] [PubMed] [Google Scholar]
- Britten R. J., Kohne D. E. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968 Aug 9;161(3841):529–540. doi: 10.1126/science.161.3841.529. [DOI] [PubMed] [Google Scholar]
- Chassy B. M., Giuffrida A. Method for the lysis of Gram-positive, asporogenous bacteria with lysozyme. Appl Environ Microbiol. 1980 Jan;39(1):153–158. doi: 10.1128/aem.39.1.153-158.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escara J. F., Hutton J. R. Thermal stability and renaturation of DNA in dimethyl sulfoxide solutions: acceleration of the renaturation rate. Biopolymers. 1980 Jul;19(7):1315–1327. doi: 10.1002/bip.1980.360190708. [DOI] [PubMed] [Google Scholar]
- Gillis M., de Ley J. Determination of the molecular complexity of double-stranded phage genome DNA from initial renaturation rates. The effect of DNA base composition. J Mol Biol. 1975 Nov 5;98(3):447–464. doi: 10.1016/s0022-2836(75)80079-9. [DOI] [PubMed] [Google Scholar]
- Torsvik V., Goksøyr J., Daae F. L. High diversity in DNA of soil bacteria. Appl Environ Microbiol. 1990 Mar;56(3):782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmur J. G., Davidson N. Kinetics of renaturation of DNA. J Mol Biol. 1968 Feb 14;31(3):349–370. doi: 10.1016/0022-2836(68)90414-2. [DOI] [PubMed] [Google Scholar]


