Abstract
We investigated the potential of double-stranded RNA interference (RNAi) with gene activity in Arabidopsis thaliana. To construct transformation vectors that produce RNAs capable of duplex formation, gene-specific sequences in the sense and antisense orientations were linked and placed under the control of a strong viral promoter. When introduced into the genome of A. thaliana by Agrobacterium-mediated transformation, double-stranded RNA-expressing constructs corresponding to four genes, AGAMOUS (AG), CLAVATA3, APETALA1, and PERIANTHIA, caused specific and heritable genetic interference. The severity of phenotypes varied between transgenic lines. In situ hybridization revealed a correlation between a declining AG mRNA accumulation and increasingly severe phenotypes in AG (RNAi) mutants, suggesting that endogenous mRNA is the target of double-stranded RNA-mediated genetic interference. The ability to generate stably heritable RNAi and the resultant specific phenotypes allows us to selectively reduce gene function in A. thaliana.
In Arabidopsis thaliana, reverse genetic techniques for isolating mutants corresponding to known sequences, such as antisense suppression (1–7), cosuppression by overexpression of the target gene (3, 8, 9), targeted gene disruption (10), or the PCR approach of screening for T-DNA insertion libraries (11, 12) have been developed, but are often insufficient and have many unanticipated difficulties. The widespread identification of differentially expressed genes, homologous genes, and interacting proteins have created a need for potent and efficient methods for obtaining their loss-of-function or reduction-of-function mutants.
Double-stranded RNA (dsRNA)-mediated interference with expression of specific genes has been observed in a number of organisms including Caenorhabditis elegans (13–17), plants (18, 19), Drosophila (20, 21), Trypanosoma brucei (22), and a planarian (23). Although the mechanism of RNA interference (RNAi) is not well understood, it seems to provide an effective way to discover gene function in many organisms (24–26).
To investigate the potential of dsRNA interference with gene activity in A. thaliana, we introduced dsRNA-expressing constructs of selected genes with previously defined functions into plants. Gene constructs delivered into plants with Agrobacterium-mediated transformation are stably integrated into the genome of host cells; thus, RNA expression from these constructs in transgenic plants can be persistent and heritable.
In this study, one gene from each of four major categories of genes involved in flower development was chosen, to determine the ability of RNAi to allow functional assessment of genes with diverse developmental functions in flowers. They are the floral organ identity gene AGAMOUS (AG), the floral meristem-size gene CLAVATA3 (CLV3), the floral meristem identity gene APETALA1 (AP1), and the floral organ number gene PERIANTHIA (PAN) (27–30). The phenotypes produced by dsRNAs corresponding to these genes are similar to those of their previously identified reduction-of-function or loss-of-function mutants (31–36). The progeny from fertile RNAi mutants, such as CLV3 (RNAi) and AP1 (RNAi) plants, also showed phenotypes. In addition to high specificity and heritability, a phenotypic series (weak, intermediate, and strong) was obtained from dsRNA interference. Furthermore, in situ hybridization indicates that endogenous target mRNA is decreased in RNAi mutants. Most constructs that are designed to produce only antisense or only sense RNA do not induce interference. Thus, specific and inheritable dsRNA interference may offer a useful alternative to classical reverse genetic screening of mutants in A. thaliana.
Materials and Methods
Constructs.
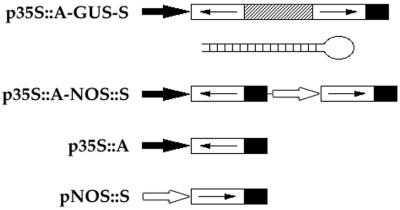
A summary of DNA constructs is shown in Fig. 1. In p35S∷A-GUS-S and p35S∷A, constructs were ligated to the BamHI and XbaI sites of pCGN1547 (37) into which an 842-bp fragment of the cauliflower mosaic virus 35S promoter and a 253-bp fragment of the 3′ end of nopaline synthase had previously been inserted in the Asp718/BamHI and XbaI/PstI sites, respectively (38). Constructs consisting of a 339-bp fragment of the nopaline synthase promoter, gene-specific sequences in the sense orientation and a 253-bp fragment of the 3′ end of nopaline synthase were ligated to the PstI and HindIII sites of pCGN1547 and p35S∷A to make pNOS∷S and p35S∷A-NOS∷S, respectively. In p35S∷A-GUS-S, the β-glucuronidase (GUS) fragment containing nucleotides 787–1,809 was used as a linker between gene-specific fragments in the antisense and sense orientations. AG, CLV3, AP1, and PAN cDNA coding sequences used in this study contain nucleotides 301–855 (27), 3–291 (28), 445–854 (29), and 27–396 (30), respectively.
Figure 1.
Gene constructs used to analyze dsRNA effects. In p35S∷A-GUS-S, gene-specific sequences (open boxes with arrows indicating the orientation) in the antisense (A) and sense (S) orientations were linked with a 1,022-bp fragment of the GUS gene (hatched box) and controlled by the 35S promoter (solid arrow). A schematic structure of the predicted dsRNA stem with a single-stranded loop generated by p35S∷A-GUS-S constructs is shown. In p35S∷A-NOS∷S, gene-specific sequences in the antisense and sense orientations were controlled by the 35S promoter and the nopaline synthase promoter, respectively (open arrow). p35S∷A contains gene-specific sequences in the antisense orientation under control of the 35S promoter. pNOS∷S contains gene-specific sequences in the sense orientation driven by the nopaline synthase promoter. Solid box, the 3′ end of nopaline synthase.
Agrobacterium-Mediated Transformation.
Agrobacterium strain ASE carrying DNA constructs in pCGN1547 was used to transform Arabidopsis plants (T0) by vacuum infiltration (39). Transformed Arabidopsis lines (T1) were selected on Murashige/Skoog (Sigma) plates containing kanamycin (50 μg/ml). Kanamycin-resistant seedlings were then transferred to soil. Phenotypic analysis of T1 and T2 plants is summarized in Table 1 and Table 2, respectively.
Table 1.
Effects of sense, antisense, and dsRNAs on transgenic plants
| Gene | Transformed background | Transformed construct | RNAi mutants/ total | % |
|---|---|---|---|---|
| AG | Ws* | p35S∷A-GUS-S | 235 /236 | 99.6 |
| pNOS∷A-GUS-S | 2 /32 | 6 | ||
| p35S∷A-NOS∷S | 3 /124 | 2 | ||
| p35S∷A | 0 /111 | 0 | ||
| pNOS∷S | 0 /95 | 0 | ||
| CLV3 | Ws | p35S∷A-GUS-S | 121 /137 | 88 |
| p35S∷A-NOS∷S | 2 /176 | 1 | ||
| p35S∷A | 0 /273 | 0 | ||
| pNOS∷S | ND† | ND | ||
| AP1 | L-er‡ | p35S∷A-GUS-S | 249 /260 | 96 |
| p35S∷A | 8 /140 | 6 | ||
| pNOS∷S | 0 /62 | 0 | ||
| PAN | crc-1 | p35s∷A-GUS-S | 110 /126 | 87 |
| p35S∷A-NOS∷S | 18 /66 | 27 | ||
| p35S∷A | 42 /76 | 55 | ||
| pNOS∷S | 2 /6 | 33 |
*Ws, Wassilewskija.
†ND, not determined.
‡L-er, Landsberg erecta.
Table 2.
Inheritance of genetic interference in CLV3 (RNAi) and AP1 (RNAi) mutants
| T1 plants | T2
plants
|
Copy no.† in T1 plants | ||
|---|---|---|---|---|
| Mutants | WT* | |||
| CLV3 (RNAi) | Plant 1 | 14 | 8 | ND‡ |
| AP1 (RNAi) | Plant 1 (W§) | 25 (W) | 8 | 1 |
| Plant 2 (W) | 22 (W) | 8 | 1 | |
| Plant 3 (I¶) | 21 (I) | 6 | 1 | |
| Plant 4 (I/S∥) | 19 (I/S) | 7 | 1 | |
| Plant 5 (S**) | 17 (S) | 5 | 1 | |
| Plant 6 (S) | 20 (S) | 4 | 1 | |
*WT, wild type.
†Number of transgene copies estimated from segregation ratios.
‡ND, not determined.
§W, weak.
¶I, intermediate.
∥I/S, intermediate/strong.
**S, strong.
In Situ Hybridization.
The AG cDNA clone pCIT565 containing nucleotides 9–977 (27) was used to synthesize antisense and sense probes. 35S-labeled RNA probes were synthesized with Riboprobe in vitro Transcription Systems (Promega). The template was linearized with HindIII and transcribed by T7 RNA polymerase (antisense probe), or linearized with XhoI and transcribed by SP6 RNA polymerase (sense probe). Tissue was fixed in 1× PBS containing 4% paraformaldehyde/0.1% Triton X-100/0.1% Tween 20 at 4°C overnight. Fixed tissue was dehydrated with ethanol, cleared with xylene, embedded in paraffin (Paraplast Plus, Oxford Labware, St. Louis), and sectioned at 8 μm. In situ hybridization was performed as described by Drews et al. (40), with modifications by Sakai et al. (41). Exposure time was 8–10 days.
Western Blot Analysis.
Bud clusters (stages 1–12, including the inflorescence meristem) from one inflorescence were frozen and ground in liquid nitrogen, thawed in 30 μl of lysis buffer (50 mM Tris, pH 7.5/1 mM EDTA/100 mM NaCl/1% Nonidet P-40/0.1% SDS/0.1% Triton X-100/0.7% 2-mercaptoethanol/1 mM PMSF). The extract was mixed with 15 μl of 3× sample buffer (187 mM Tris, pH 6.8/6% SDS/30% glycerol/3% 2-mercaptoethanol/0.06% bromophenol blue), boiled for 5 min, and centrifuged (16,000 × g for 10 min at room temperature). Twenty microliters of the supernatant was separated on an SDS/12.5% polyacrylamide gel. The protein was transferred to nitrocellulose membrane (Schleicher & Schuell), probed with an AG-specific polyclonal antibody (42) and horseradish peroxidase-labeled secondary antibody (Amersham International), and detected with the enhanced chemiluminescence system (ECL; Amersham International).
Results
To make constructs that produce dsRNA, gene-specific sequences in the antisense and sense configurations were either linked with the partial GUS gene and placed under control of the constitutive 35S promoter from cauliflower mosaic virus (p35S∷A-GUS-S), or controlled by the 35S promoter and the constitutive nopaline synthase promoter, respectively (p35S∷A-NOS∷S). A single RNA transcribed from the fusion gene in p35S∷A-GUS-S can potentially form a dsRNA stem with a single-stranded loop structure (Fig. 1). Genetic interference effects of sense, antisense, and dsRNAs corresponding to AG, CLV3, AP1, and PAN are outlined in Table 1. For each of these genes, p35S∷A-GUS-S constructs caused potent and specific genetic interference. However, p35S∷A-NOS∷S, p35S∷A and pNOS∷S constructs had either no, or weak, genetic interference effects. We will refer to transgenic plants carrying functional p35S∷A-GUS-S constructs by listing the gene name followed by RNAi. For unknown reasons, the sense construct corresponding to CLV3 caused toxicity in Agrobacterium and the sense construct of PAN resulted in very low transformation efficiency of crabs claw-1 (crc-1) plants. Therefore, interference effects of the CLV3 sense construct were not determined and only six crc-1 transgenic plants containing the PAN sense construct were analyzed.
AG dsRNA-Mediated Genetic Interference.
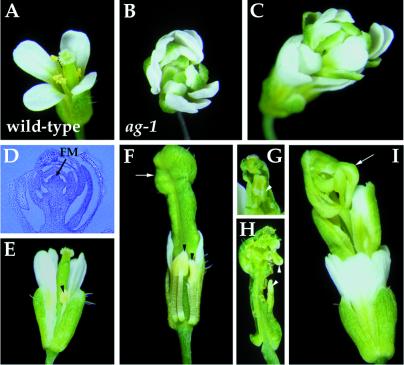
AG was chosen for initial characterization of RNAi in developing flowers. Arabidopsis flowers consist of four concentric whorls of organs. Most wild-type flowers have four sepals, four petals, six stamens, and two fused carpels, from the outermost first whorl to the innermost fourth whorl (Fig. 2A). Mutations in the AG gene cause homeotic alterations of the third and fourth whorls of organs in flowers (31). In severe ag loss-of-function mutants (Fig. 2B), the third whorl primordia develop into petals indistinguishable from those of the second whorl, and the fourth whorl develops into another ag flower, resulting in a repetitive pattern of sepals, petals, and petals (32).
Figure 2.
Flowers of wild-type, ag-1 and AG (RNAi) plants. (A) Wild-type flowers have four sepals, four petals, six stamens, and two carpels. (B) ag-1 flowers consist of an indeterminate number of whorls of sepals and petals in the pattern (sepals, petals, petals)n, with no staminoid or carpeloid tissue. (C–I) AG (RNAi) flowers with different severity of phenotypes. (C) Strong mutant flowers phenocopied ag-1. (D) Longitudinal section of a strong mutant flower showing a large number of sepals and petals produced by an indeterminate floral meristem (FM). (E) Weak mutant flower. The stamens fail to elongate and the anthers are slightly petaloid (arrowhead), with no pollen. (F) Intermediate mutant flower with some sepals and petals removed. Anthers are partially transformed into petaloid tissue (arrowheads). The gynoecium is bulged at the top (arrow, F), with inner organs such as carpels (arrowhead, G) and/or petals (arrowheads, H). (I) Intermediate/strong mutant flowers have the repeated pattern of sepals, petals, petals formed in outer whorls and an incomplete flower in the center (arrow). AG (RNAi) plants are in the Wassilewskija background; therefore, internode elongation between successive internal flowers are seen in intermediate/strong (I) and strong mutant flowers (C).
The phenotypes produced by AG dsRNA are frequent and specific (Fig. 2 C–I). All but one of 236 transgenic plants showed ag mutant phenotypes. These AG (RNAi) mutants can be arranged into a phenotypic series based on the severity of the homeotic transformation in the third whorl and the extent of floral indeterminacy in the fourth whorl. Weak and intermediate AG (RNAi) mutant flowers showed partial homeotic transformation in the third whorl organs and slight floral indeterminacy (Fig. 2 E–H). However, intermediate/strong and strong AG (RNAi) mutant flowers showed complete transformation of the third whorl organs from stamens to petals and severe floral indeterminacy (Fig. 2 C, D, and I). Particularly, flowers from strong AG (RNAi) plants (Fig. 2C) are indistinguishable from those of strong ag mutant alleles such as ag-1 (Fig. 2B). Weak, intermediate, intermediate/strong, and strong AG (RNAi) mutants represent 16, 32, 43, and 9%, respectively, of the transgenic plant populations. In contrast, pNOS∷A-GUS-S, in which the nopaline synthase promoter was used to drive the fusion gene, and p35S∷A-NOS∷S constructs for AG caused very weak genetic interference in 2 out of 32 and 3 out of 124 transgenic plants, respectively (data not shown). Flowers from transgenic plants containing the AG antisense (n = 111) or sense (n = 95) construct are indistinguishable from those of wild-type plants (data not shown).
dsRNA Interferes with mRNA Accumulation.
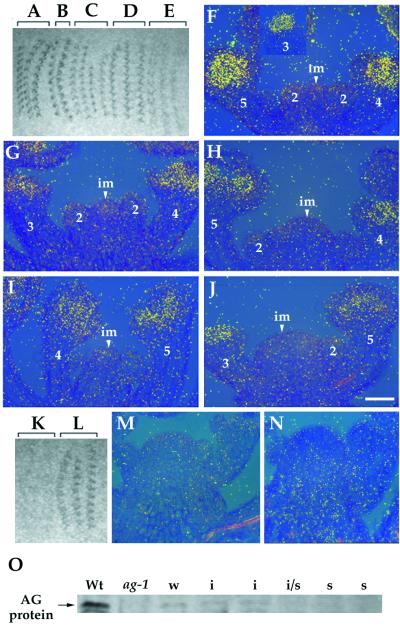
In situ hybridization was performed to determine the target of dsRNA interference. The earliest expression of AG in wild-type flowers is in stage three, in those cells that will give rise to the third- and fourth-whorl organ primordia. Later, AG expression is restricted to the stamen and the carpel primordia (Fig. 3F) (27). The autoradiogram of the tissue hybridized with an AG anti-mRNA probe showed that hybridization signals declined with increasingly severe phenotypes in AG (RNAi) mutants (Fig. 3 A–E), consistent with the observation that AG mRNA accumulation is decreased in AG (RNAi) mutants (Fig. 3 G–J). These results suggest that endogenous mRNA is a target of dsRNA-mediated genetic interference. When used as a standard control for in situ hybridization, an AG sense probe hybridized with the tissue from AG (RNAi) mutants but not with that from wild type (Fig. 3 K–N), suggesting that AG antisense RNA is produced in AG (RNAi) mutants. Reverse transcription–PCR analysis with GUS- and AG-specific primers also showed that expression levels of both strands of AG RNA from the fusion gene in p35S∷A-GUS-S increase with increasingly severe phenotypes (data not shown).
Figure 3.
Effects of AG dsRNA on levels of AG mRNA and AG protein. (A–E) An autoradiogram of the tissue hybridized with an AG anti-mRNA probe. The tissue is from wild-type (A and F), weak (B and G), intermediate (C and H), intermediate/strong (D and I), and strong (E and J) AG (RNAi) mutant plants. (A–E) Hybridization signals declined gradually with increasingly severe phenotypes. (F–J) The bright-field/dark-field double exposures of longitudinal section through the inflorescence meristems with stage 2–5 flowers. The silver grains representing AG mRNA expression were made to appear yellow with the use of a yellow filter. The number indicated corresponds to the development stage of flowers (43). im, inflorescence meristem. (Bar = 50 μm.) (K and L) An autoradiogram of the tissue hybridized with an AG sense probe. The tissue is from wild-type (K and M) and intermediate AG (RNAi) mutant plants (L and N). (O) Western blot analysis of AG protein. The anti-AG antibody recognizes the carboxyl-terminal part of the AG protein from aa 220–285 which is absent in the AG-1 protein (27, 42); thus, ag-1 is a control of the specificity of the antibody. Whereas AG protein is weakly expressed in weak (w) and intermediate (i) AG (RNAi) mutants compared with wild type (Wt), it is not detected at levels above background in intermediate/strong (i/s) and strong (s) AG (RNAi) mutants.
Furthermore, Western blot analysis of total protein from wild-type and AG (RNAi) mutants with an AG-specific polyclonal antibody (42) demonstrated that the severity of phenotypes is correlated with a reduction of the AG protein level in AG (RNAi) mutants. AG protein is weakly expressed in weak and intermediate AG (RNAi) mutants. In contrast, it is not detected at levels above background in intermediate/strong and strong AG (RNAi) mutants (Fig. 3O).
Genetic Interference by CLV3 and AP1 dsRNAs.
We further assessed the effectiveness and specificity of dsRNA with the CLV3 and AP1 genes. Plants with mutations in the CLV3 gene have enlarged meristems and extra floral organs, especially carpels (Fig. 4 C, F, and I) (33). The majority of CLV3 (RNAi) mutants (89%, n = 121) have flowers with extra carpels (Fig. 4H); however, only 2% of those plants (n = 108) also have extra sepals, petals, and stamens (Fig. 4E). In addition, some CLV3 (RNAi) mutant plants (26%, n = 121) have enlarged shoot apical meristems with distortions in phyllotaxy (Fig. 4B) and bifurcation, flattening, and broadening of the stem (data not shown). In contrast, only 1% (n = 176) of plants transformed with p35S∷A-NOS∷S for CLV3 have the extra carpel phenotype. clv3 mutant phenotypes were not observed in transgenic plants containing the CLV3 antisense construct (n = 273).
Figure 4.
Phenotypes of wild-type, CLV3 (RNAi), and clv3–2 plants. Wild-type and CLV3 (RNAi) mutants are in the ecotype Wassilewskija, whereas clv3–2 is in the ecotype Landsberg erecta which has reduced internode elongation. The inflorescence meristems are enlarged in CLV3 (RNAi) (B) and clv3–2 (C) compared with wild type (A). (D) Wild-type flower. (E) CLV3 (RNAi) and (F) clv3–2 flowers have additional organs. (G–I) Cross section of gynoecia showing that the wild-type gynoecium (G) consists of two carpels, and gynoecia in CLV3 (RNAi) (H) and clv3–2 (I) have four carpels.
Mutations in the AP1 gene result in homeotic alterations of the outer two whorls and a partial or complete conversion of a floral meristem into an inflorescence meristem (34, 35). In weak ap1 mutant alleles, the first and second whorls consist of mosaic sepaloid organs and staminoid petals, respectively (Fig. 5B). In intermediate ap1 mutant alleles, flowers have leaf-like first whorl organs and leaf-like or staminoid second whorl organs (Fig. 5C). In strong ap1 mutant alleles, bract-like organs are produced in the first whorl and petals are usually absent in the second whorl (Fig. 5D). In addition, secondary flowers usually arise from the axils of the first whorl organs in flowers of intermediate and strong ap1 mutant alleles (Fig. 5 C and D). About 96% of transgenic plants (n = 260) containing the AP1 dsRNA-expressing construct, p35S∷A-GUS-S, produced flowers similar to those of ap1 mutant alleles (Fig. 5 E–H). Weak (Fig. 5E), intermediate (Fig. 5F), intermediate/strong (Fig. 5G), and strong (Fig. 5H) phenotypes were observed in 94, 1, 3, and 2%, respectively, of AP1 (RNAi) mutants (n = 249). In contrast, transgenic plants containing the AP1 construct in the antisense orientation (6%, n = 140) had very weak mutant phenotypes (data not shown). The AP1 sense construct did not cause mutant phenotypes in transgenic plants (n = 62; data not shown).
Figure 5.
Phenotypes of wild-type, ap1 and AP1 (RNAi) flowers. (A) Wild-type flower. (B) ap1–5 flower. (C) ap1–4 flower. (D) ap1–1 flower. (E–H) Flowers from weak (E), intermediate (F), intermediate/strong (G), and strong (H) AP1 (RNAi) plants. Arrowheads indicate leaf- or bract-like first whorl organs. The numbered arrows indicate the primary (1), secondary (2), and tertiary (3) flowers. The black arrows in C and G indicate leaf-like or staminoid second-whorl organs.
One CLV3 (RNAi) T1 plant and 6 AP1 (RNAi) T1 plants of variable severity were selfed to examine the inheritance of genetic interference (Table 2). The progeny from each selected plant showed similar severity of phenotypes to those of the selfed parents, and the severity of phenotypes is constant between mutant siblings of each lineage. In addition, the progeny of AP1 (RNAi) plants had 3:1 (mutant/wild type) segregation ratios, suggesting that each of the 6 AP1 (RNAi) T1 plants contained one copy of the transgene. This result indicates that dsRNA-expressing constructs, which are integrated into the plant genome, are stably inherited in a Mendelian fashion, and that the RNAi effect persists to, or recurs in, new generations of plants.
RNA-Mediated Interference with PAN.
Flowers of plants mutant for pan are characterized by an increase in the organ number in the first two whorls, and a decrease in the organ number in the third whorl. Mutant flowers usually have five sepals, five petals, five stamens, and two carpels (36). When introduced into wild-type plants, the dsRNA-expressing construct of PAN caused either no, or weak, extra organ phenotypes. Reverse transcription–PCR analysis showed that PAN mRNA was reduced by 30–90% in PAN (RNAi) plants compared with wild-type plants (data not shown), suggesting that a small portion of PAN activity is sufficient for its function in wild-type plants. This hypothesis is consistent with results from previous immunohistochemical analysis showing that the mutant pan-1 and pan-2 alleles, with high expressivity of the extra organ phenotype, are likely null alleles (30).
Whereas flowers homozygous for strong mutant alleles show high penetrance of the extra organ phenotype, only the first few flowers from homozygotes of weak mutant alleles show the phenotype. However, both strong and weak pan alleles cause high penetrance of unfused carpel phenotypes in a crc-1 genetic background (Fig. 6 A, B, D, and E) (30, 44), suggesting that crc mutants provide a more sensitive background than wild type in which to observe phenotypic effects of PAN reduction-of-function mutations. Therefore, RNA-expressing constructs corresponding to PAN were introduced into a crc-1 homozygous background to further assess the potential of RNA-mediated interference with PAN. Similar to pan alleles, PAN dsRNA-expressing constructs, p35S∷A-GUS-S (87%, n = 126) and p35S∷A-NOS∷S (27%, n = 66), caused extra organ number and unfused carpels in crc-1 (Fig. 6 C and F). Antisense (55%, n = 76) and sense (33%, n = 6) sequences corresponding to PAN have similar RNAi effects as well (data not shown), suggesting that low levels of dsRNAs might be produced in such a case and weak interference with PAN activity is sufficient to confer an unfused carpel phenotype in crc-1.
Figure 6.
Effects of PAN dsRNA on crc-1 transgenic plants. (A and D) crc-1. (B and E) crc-1 pan-3 and (C and F) crc-1; PAN (RNAi) flowers have extra organs and unfused gynoecia.
Discussion
This study shows that dsRNA-mediated genetic interference can operate in A. thaliana to efficiently induce sequence-specific inhibition of gene function. Although the technique of RNA microinjection has been widely used in C. elegans (13–15, 44), Drosophila (20, 21), and planarians (23), methods for RNA injection into zygotes of A. thaliana are not available. However, Agrobacterium-mediated transformation provides a convenient and efficient method to introduce dsRNA-expressing constructs into the plant genome. Therefore, RNAi in transgenic plants is persistent and inherited instead of being transient and unstable as in RNA-injected animals (13–15, 20, 21, 23, 45) and transiently transfected cells (22). In addition, inducible and tissue-specific promoters might be used to obtain regulated RNAi.
In this study, two kinds of dsRNA-expressing constructs, p35S∷A-GUS-S and p35S∷A-NOS∷S, were used to investigate RNAi effects. p35S∷A-NOS∷S is less potent at inducing genetic interference than p35S∷A-GUS-S, perhaps because of unequal expression levels of sense and antisense RNAs by two promoters of different strength. The nopaline synthase promoter is much weaker than the 35S promoter, suggested by the observation that pNOS∷A-GUS-S has weaker genetic interference than p35S∷A-GUS-S. These results suggest that equal and high levels of both strands of each RNA in each cell are essential for inducing potent RNAi. If this is true, use of two strong promoters of similar strength should improve RNAi effects of dsRNA-expressing constructs in which sense and antisense RNAs are produced separately; however, use of two identical promoters in a construct should be avoided to prevent cosuppression (46).
dsRNAs corresponding to four genes selected in this study caused potent and specific genetic interference, suggesting that dsRNA-mediated gene silencing can occur in the tissues where these genes normally function. In addition, a phenotypic series can be obtained from RNAi mutants. The fact that the severity of phenotypes varied between T1 individuals is possibly because of variable transgene copy number and/or positional effects of particular DNA insertion events. However, our results suggest that severity of phenotypes in AP1 (RNAi) T1 plants is not related to the transgene copy number.
CLV3 dsRNA seems predominantly to block gene function in a subset of the cells where it is normally expressed. CLV3 is expressed in the inflorescence and the floral meristems (28). Mutations in the CLV3 gene cause enlarged meristems and extra floral organs (33). About 89% of CLV3 (RNAi) plants have flowers with extra carpels but only 26% of CLV3 (RNAi) plants have enlarged inflorescence meristems. This result suggests a strong suppression of the CLV3 gene function in the center of the floral meristem but less suppression of its function in the inflorescence meristem. It is probably because of differential activity of the 35S promoter which drives expression of dsRNA in these tissues. It is also possible that some tissues could partially resist RNAi (25), or that some phenotypes (such as enlarged inflorescence meristems) are less sensitive to the level of gene activity.
When used as controls for RNAi experiments, the sense and antisense constructs of PAN also had the ability to induce genetic interference in a crc-1 homozygous background; so did the AP1 antisense construct in wild-type plants. It has been suggested that low levels of dsRNA might be produced from transgenes that are designed to produce only antisense or only sense RNA, via the readthrough transcription from transgenes arranged as an inverted repeat, or transcription from a transgene whose 3′ end is adjacent to an endogenous promoter (19, 24, 25, 47). Alternatively, it seems possible that cellular RNA-dependent RNA polymerase could be involved in the conversion of single-stranded RNA to dsRNA in a cell-specific manner, suggested by the cloning and in vitro catalytic analysis of an RNA-dependent RNA polymerase from tomato (25, 48).
In situ hybridization revealed a correlation between decreasing levels of AG mRNA accumulation and increasing severity of phenotypes in AG (RNAi) plants, suggesting that the mechanism blocking mRNA accumulation could be responsible for dsRNA interference. This result is consistent with previous findings that endogenous mRNA is a target of dsRNA-mediated genetic interference (13, 14, 22, 23). In addition, a recent report of isolation of an RNaseD homolog from C. elegans mutants which are resistant to RNAi suggests that RNAi works by dsRNA-directed, enzymatic RNA degradation (49).
Whatever the mechanism by which RNAi acts to reduce specific mRNA levels, the experiments described here show that it is a useful method for determining the loss-of-function phenotypes of genes involved in development and meristem activity in A. thaliana.
Acknowledgments
We thank John Bowman and Yuval Eshed for critical reading of the manuscript; Xavier Ambroggio, Catherine Baker, Chieh Chang, Toshiro Ito, Jeff Long, Kazuaki Ohashi, Carolyn Ohno, G. Venugopala Reddy, Doris Wagner, Frank Wellmer, and Eva Ziegelhoffer for their comments on the manuscript; Amani Zewail for technical assistance; and Toshiro Ito for providing the anti-AG antibody. This work was supported by National Institutes of Health Grant GM45697 to E.M.M.
Abbreviations
- dsRNA
double-stranded RNA
- RNAi
RNA interference
- GUS
β-glucuronidase
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.060034297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.060034297
References
- 1.Baier M, Dietz K J. Plant Physiol. 1999;119:1407–1414. doi: 10.1104/pp.119.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coles J P, Phillips A L, Croker S J, Garcia-Lepe R, Lewis M J, Hedden P. Plant J. 1999;17:547–556. doi: 10.1046/j.1365-313x.1999.00410.x. [DOI] [PubMed] [Google Scholar]
- 3.Haldrup A, Naver H, Scheller H V. Plant Cell. 1999;17:689–698. doi: 10.1046/j.1365-313x.1999.00419.x. [DOI] [PubMed] [Google Scholar]
- 4.Huang N C, Liu K H, Lo H J, Tsay Y F. Plant Cell. 1999;11:1381–1392. doi: 10.1105/tpc.11.8.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nanjo T, Kobayashi M, Yoshiba Y, Sanada Y, Wada K, Tsukaya H, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K. Plant J. 1999;18:185–193. doi: 10.1046/j.1365-313x.1999.00438.x. [DOI] [PubMed] [Google Scholar]
- 6.Ni M, Tepperman J M, Quail P H. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- 7.Yoshizumi T, Nagata N, Shimada H, Matsui M. Plant Cell. 1999;11:1883–1896. doi: 10.1105/tpc.11.10.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bell E, Creelman R A, Mullet J E. Proc Natl Acad Sci USA. 1995;92:8675–8679. doi: 10.1073/pnas.92.19.8675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naver H, Haldrup A, Scheller H V. J Biol Chem. 1999;274:10784–10789. doi: 10.1074/jbc.274.16.10784. [DOI] [PubMed] [Google Scholar]
- 10.Kempin S A, Liljegren S J, Block L M, Rounsley S D, Yanofsky M F, Lam E. Nature (London) 1997;389:802–803. doi: 10.1038/39770. [DOI] [PubMed] [Google Scholar]
- 11.Mckinney E C, Ali N, Traut A, Feldmann K A, Belostotsky D A, McDowell J M, Meagher R B. Plant J. 1995;8:613–622. doi: 10.1046/j.1365-313x.1995.8040613.x. [DOI] [PubMed] [Google Scholar]
- 12.Winkler R G, Frank M R, Galbraith D W, Feyereisen R, Feldmann K A. Plant Physiol. 1998;118:743–750. doi: 10.1104/pp.118.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fire A, Xu S, Montgomery M K, Kostas S A, Driver S E, Mello C C. Nature (London) 1998;391:806–811. doi: 10.1038/35888. [DOI] [PubMed] [Google Scholar]
- 14.Montgomery M K, Xu S, Fire A. Proc Natl Acad Sci USA. 1998;95:15502–15507. doi: 10.1073/pnas.95.26.15502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Y, Mello C. Genes Dev. 1998;12:943–955. doi: 10.1101/gad.12.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabara H, Grishok A, Mello C C. Science. 1998;282:430–431. doi: 10.1126/science.282.5388.430. [DOI] [PubMed] [Google Scholar]
- 17.Timmons L, Fire A. Nature (London) 1998;395:854. doi: 10.1038/27579. (lett.). [DOI] [PubMed] [Google Scholar]
- 18.Voinnet O, Vain P, Angell S, Baulcombe D C. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 19.Waterhouse P M, Graham M W, Wang M-B. Proc Natl Acad Sci USA. 1998;95:13959–13964. doi: 10.1073/pnas.95.23.13959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennerdell J R, Carthew R W. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 21.Misquitta L, Paterson B M. Proc Natl Acad Sci USA. 1999;96:1451–1456. doi: 10.1073/pnas.96.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ngo H, Tschudi C, Guli K, Ullu E. Proc Natl Acad Sci USA. 1998;95:14687–14692. doi: 10.1073/pnas.95.25.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarado A S, Newmark P A. Proc Natl Acad Sci USA. 1999;96:5049–5054. doi: 10.1073/pnas.96.9.5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montgomery M K, Fire A. Trends Genet. 1998;14:255–258. doi: 10.1016/s0168-9525(98)01510-8. [DOI] [PubMed] [Google Scholar]
- 25.Fire A. Trends Genet. 1999;15:358–363. doi: 10.1016/s0168-9525(99)01818-1. [DOI] [PubMed] [Google Scholar]
- 26.Sharp P A. Genes Dev. 1999;13:139–141. [PubMed] [Google Scholar]
- 27.Yanofsky M F, Ma H, Bowman J L, Drews G N, Feldmann K A, Meyerowitz E M. Nature (London) 1990;346:35–39. doi: 10.1038/346035a0. [DOI] [PubMed] [Google Scholar]
- 28.Fletcher J C, Brand U, Running M P, Simon R, Meyerowitz E M. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 29.Mendel M A, Gustafson-Brown C, Savidge B, Yanofsky M F. Nature (London) 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 30.Chuang C-F, Running M P, Williams R W, Meyerowitz E M. Genes Dev. 1999;13:334–344. doi: 10.1101/gad.13.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bowman J L, Smyth D R, Meyerowitz E M. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman J L. In: Arabidopsis: An Atlas of Morphology and Development. Bowman J L, editor. New York: Springer; 1994. pp. 216–221. [Google Scholar]
- 33.Clark S E, Running M P, Meyerowitz E M. Development (Cambridge, UK) 1995;121:2057–2067. [Google Scholar]
- 34.Bowman J L, Alvarez J, Weigel D, Meyerowitz E M, Smyth D R. Development (Cambridge, UK) 1993;119:721–743. [Google Scholar]
- 35.Irish V F, Sussex I M. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Running M P, Meyerowitz E M. Development (Cambridge, UK) 1996;122:1261–1269. doi: 10.1242/dev.122.4.1261. [DOI] [PubMed] [Google Scholar]
- 37.McBride K E, Summerfelt K R. Plant Mol Biol. 1990;14:269–276. doi: 10.1007/BF00018567. [DOI] [PubMed] [Google Scholar]
- 38.Krizek B A, Meyerowitz E M. Proc Natl Acad Sci USA. 1996;93:4063–4070. doi: 10.1073/pnas.93.9.4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bechtold N, Ellis J, Pelletier G. C R Acad Sci. 1993;316:1194–1199. [Google Scholar]
- 40.Drews G N, Bowman J L, Meyerowitz E M. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- 41.Sakai H, Medrano L J, Meyerowitz E M. Nature (London) 1995;378:199–203. doi: 10.1038/378199a0. [DOI] [PubMed] [Google Scholar]
- 42.Ito T, Takahashi N, Shimura Y, Okada K. Plant Cell Physiol. 1997;38:248–258. doi: 10.1093/oxfordjournals.pcp.a029160. [DOI] [PubMed] [Google Scholar]
- 43.Smyth D R, Bowman J L, Meyerowitz E M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Alvarez J, Smyth D R. Development (Cambridge, UK) 1999;126:2377–2386. doi: 10.1242/dev.126.11.2377. [DOI] [PubMed] [Google Scholar]
- 45.Tabara H, Sarkissian M, Kelly W G, Fleenor J, Grishok A, Timmons L, Fire A, Mello C C. Cell. 1999;99:123–132. doi: 10.1016/s0092-8674(00)81644-x. [DOI] [PubMed] [Google Scholar]
- 46.Baulcombe D C, English J J. Curr Opin Biotechnol. 1996;7:173–180. [Google Scholar]
- 47.Kooter J M, Matzke M A, Meyer P. Trends Plant Sci. 1999;4:340–347. doi: 10.1016/s1360-1385(99)01467-3. [DOI] [PubMed] [Google Scholar]
- 48.Schiebel W, Pélissier T, Riedel L, Thalmeir S, Schiebel R, Kempe D, Lottspeich F, Sänger H L, Wassenegger M. Plant Cell. 1998;10:2087–2101. doi: 10.1105/tpc.10.12.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ketting R F, Haverkamp T H, van Luenen H G, Plasterk R H. Cell. 1999;99:133–141. doi: 10.1016/s0092-8674(00)81645-1. [DOI] [PubMed] [Google Scholar]