Abstract
Metal cation homeostasis is essential for plant nutrition and resistance to toxic heavy metals. Many plant metal transporters remain to be identified at the molecular level. In the present study, we have isolated AtNramp cDNAs from Arabidopsis and show that these genes complement the phenotype of a metal uptake deficient yeast strain, smf1. AtNramps show homology to the Nramp gene family in bacteria, yeast, plants, and animals. Expression of AtNramp cDNAs increases Cd2+ sensitivity and Cd2+ accumulation in yeast. Furthermore, AtNramp3 and AtNramp4 complement an iron uptake mutant in yeast. This suggests possible roles in iron transport in plants and reveals heterogeneity in the functional properties of Nramp transporters. In Arabidopsis, AtNramps are expressed in both roots and aerial parts under metal replete conditions. Interestingly, AtNramp3 and AtNramp4 are induced by iron starvation. Disruption of the AtNramp3 gene leads to slightly enhanced cadmium resistance of root growth. Furthermore, overexpression of AtNramp3 results in cadmium hypersensitivity of Arabidopsis root growth and increased accumulation of Fe, on Cd2+ treatment. Our results show that Nramp genes in plants encode metal transporters and that AtNramps transport both the metal nutrient Fe and the toxic metal cadmium.
Metal cations are essential for plant nutrition (1). Metals such as iron (Fe), manganese (Mn), and copper (Cu) are necessary cofactors for many enzymatic reactions. Some metals such as zinc (Zn) play important structural roles in proteins. Furthermore, metal cations recently have been shown to be involved in signaling in animals (2) and plants (3). However, plants also need to control against excessive accumulation of essential cations and toxic heavy metals, such as cadmium (Cd2+), lead, mercury, and arsenic.
Metal transporters are essential to maintain intracellular metal homeostasis (4). In plants, metal cation transporters play important roles in several steps of metal nutrition. These transport proteins mediate metal uptake in root cells and metal transfer between cells and organs. Metal transporters are involved in metal detoxification by mediating the transport of metal cations or metal chelates from the cytosol to the vacuolar compartment (5–8). A better knowledge of the mechanisms of metal transport in plants is needed for understanding and manipulating plant nutrition and plant resistance to toxic heavy metals.
Uptake of metal cations has been investigated in various species. These studies point to multiple pathways for the low- and high-affinity and constitutive and inducible influx of metal cations, such as Zn and Fe (9–11). It has been suggested that toxic metals such as cadmium enter plant cells by transporters for essential cations, such as Fe and calcium (12–15). For example, in pea, Fe deficiency, which is known to stimulate high-affinity Fe uptake also stimulates Cd2+ uptake (12). Recently, screening of plant cDNA libraries for genes able to restore the growth defects of yeast metal transport mutants has led to the identification of plant genes encoding transporters that allow the uptake of Cu (16), Fe, Mn, and Zn (17–19) and various cations (14) in yeast. The ion transport function of these cloned transporters has not yet been directly analyzed in roots.
Genes encoding members of the Nramp family of integral membrane proteins were identified through very diverse genetic screens (20). Nramp1 was cloned in mouse as a locus involved in intracellular bacterial pathogen sensitivity (21). Nramp homologous sequences have now been identified in bacteria, fungi, plants, and animals. It was discovered recently that some Nramp proteins function as metal transporters. SMF1, a yeast Nramp homologue, was shown to encode a manganese transporter (22). Then, DCT1/Nramp2, a mammalian homologue of the Nramp genes, was isolated in a functional screen for Fe transport systems and shown to encode a broad specificity metal transporter (23). DCT1/Nramp2 was also shown to be a genetic determinant for anemia, suggesting a role in endosomal recycling of Fe (24). Nramp genes have been sequenced in some plant species (25) but have not yet been characterized functionally, with the exception of the ethylene insensitivity gene EIN2, which functions in signal transduction with no metal transport function (26).
In the present study, we have isolated plant genes homologous to the Nramp family in Arabidopsis. Furthermore, we have combined heterologous expression studies in yeast and molecular physiological studies in plants to show that Arabidopsis Nramp genes encode broad specificity metal transporters that contribute to metal sensitivity in plants. In Saccharomyces cerevisiae, Nramp gene expression complements the phenotype of yeast mutants impaired in Mn and Fe transport and increases Cd2+ sensitivity and Cd2+ content in yeast cells. AtNramp3 mRNA levels are increased on Fe starvation. Characterization of a mutant carrying a T-DNA insertion in AtNramp3 and overexpression of this gene demonstrate the contribution of this gene to Fe transport and Cd2+ sensitivity in planta.
Materials and Methods
Cloning of AtNramp Genes.
AtNramp1 cDNA was cloned by screening the λYES library of Arabidopsis thaliana, ecotype Columbia (27), using an expressed sequence tag clone (dBEST accession no. Z30530) as the probe. AtNramp3 and AtNramp4 were amplified by PCR using Advantage Taq cDNA polymerase (CLONTECH) from first-strand cDNA prepared from Arabidopsis roots using the primers NR1-1S (5′-AATGCCACAACTCGAGAACAACG-3′) and NR1-0R (5′-GAAAGCTAAGAACTCATGATCCTAAGG-3′) for AtNramp3, and the primers NR2-3S (5′-GAAATATGTCGGAGACTGATAGAG-3′) and NR2-3R (5′-CCTTACTCACTCATCATCCCTC-3′) for AtNramp4. The truncated version of AtNramp3 was generated by PCR using NR1-1S and muNR1-R (5′-CGGGATCCTTACACCACAATATATCCTGAACCAGCATATATCTCATTGGAGAAGAACTCC-3′). The PCR fragments were cloned in pGEMT-easy (Promega) and excised either with NotI for cloning in pDR195 or with EcoRI for cloning in pMON530 (Monsanto).
Yeast Strains and Growth Conditions.
The strains used in this study were INVSc1 (Invitrogen), DEY1453 (17), and smf1 (MATα his3 ade2 leu2 trp1 ura3smf1∷URA3ura3∷TRP1). This smf1 strain was derived from the smf1 from Supek et al. (22) by transformation with linearized pRH530, a plasmid containing ura3∷TRP1, and selection for cells that were trp+ and ura−. Yeast cells were grown on yeast extract/peptone/dextrose before transformation and synthetic dextrose-ura after transformation. Yeast cells were transformed according to standard procedures (Invitrogen). The fet3fet4 (DEY1453) strain was grown in media supplemented with 0.2 mM FeCl3. Complementation of the smf1 phenotype was tested by using media buffered at pH 6 with 50 mM Mes and supplemented or not with 20 mM EGTA.
Metal Accumulation in Yeast.
Yeast cells transformed with empty or AtNramp vector were grown for 24 h starting from an OD of 0.01 on synthetic dextrose-ura supplemented with 3 μM Cd2+ and 109Cd2+ at a specific activity of about 4 × 105 cpm/nmol. The cells were harvested on filters and washed twice with 0.1 M CaCl2 before the cell-associated radioactivity was quantified. Short-term uptake experiments were performed as described for Cd2+ (14) and for Fe (17).
Insertion Mutant Isolation.
For identification of a mutant carrying a T-DNA insertion in AtNramp3, superpools of genomic DNA representing 1,000 independent Arabidopsis transformants were screened by PCR using Ex-Taq polymerase (Takara Shuzo, Kyoto). Four pairs of primers were used, each including an AtNramp3 gene-specific primer (NR1-1S or NR1-0R) and a T-DNA-specific primer (5′-GATGCACTCGAAATCAGCCAATTTTAGAC-3′ for the left border of the T-DNA and 5′-TCCTTCAATCGTTGCGGTTCTGTCAGTTC-3′ for the right border of the T-DNA). The PCR product from one superpool (NR1-1S/Left border), which gave a positive signal after southern hybridization with an AtNramp3 cDNA probe, was reamplified by nested PCR using NR1-2S (5′-CAACGAGCCACTTCTAATCAACG-3′) and a nested primer specific for the left border of the T-DNA (5′-CCGCAATGTGTTATTAAGTTGTCTAAGCG-3′). Sequencing of the nested PCR product revealed that the T-DNA insertion in AtNramp3 was located between Glu-467 and Val-468. The DNA from the 10 pools of 100 plants from which the positive superpool was constituted were screened by PCR. Then, subpools of 20 plants were screened, leading to the identification of a single subpool containing the AtNramp3 disruption mutant. Finally, PCR screening of DNA samples from individual plants lead to the identification of two homozygous and one heterozygous plant carrying the insertion in the AtNramp3 gene. Their genotypes were confirmed by genomic Southern blot after digestion with EcoRI that yields a 7-kb fragment in the wild type and a 4-kb fragment in the mutant.
Gene Expression.
Arabidopsis seedlings were grown for 10 days vertically on plates containing Murashige and Skoog medium with 1% sucrose and 1% agar A (Sigma). Total RNA was extracted from about 60 seedlings by using the Trizol reagent (Life Technologies, Grand Island, NY). For Northern blots 15 μg total RNA was separated on denaturing 1.2% formaldehyde agarose gel, transferred to a Hybond-N nylon membrane (Amersham Pharmacia), and hybridized with probes made from full-length AtNramp cDNAs. For RT-PCR analysis, after digestion of the genomic DNA, the first cDNA strand from AtNramp3 was synthesized from 10 μg of total RNA extracted from wild-type, control T-DNA plants, or AtNramp3 mutant. A complementary oligonucleotide located 5′ of the insertion on AtNramp3 cDNA (NR1-RT: 5′-GAGAAGAACTCCAACAAAAGATAACC-3′) was used for reverse transcription. Subsequently the reverse transcription product was amplified using NR1-1S and NR1-RT primers.
Cd2+ Sensitivity of Root Growth.
Arabidopsis seedlings were grown vertically on plates containing a minimal medium without microelements (2.5 mM H3PO4/5 mM KNO3/2 mM MgSO4/1 mM Ca(NO3)2/1 mM Mes adjusted with KOH to pH 5.5/1% sucrose/1% agar A). This medium was supplemented with various concentrations of Cd2+. The plants to be compared were grown on the same plate. After 3 days of stratification at 4°C, the seedlings were grown for 10 days before measurement of the root length. A Student's t test was performed to evaluate the significance of the differences observed.
Metal Ion Content Measurements.
Two hundred seeds were germinated in a flask containing 50 ml of the minimal medium used for the root growth assays (without agar) and grown for 10 days on a shaker at 100 rpm. The plants were harvested by vacuum filtration and washed with 10 ml CaCl2 (0.1 M) and with 10 ml EDTA (10 mM). The dry weight (D.W.) of the samples was measured after drying overnight at 60°C. Subsequently, the samples were digested in 0.2–0.4 ml of 70% HNO3 (trace metal grade; Fisher) for 3 days, and complete digestion was achieved by heating the samples to 100°C for 30 min. After dilution, the metal content of the samples was determined by inductively coupled plasma optical emission spectroscopy. The results are given in ppm (mg metal/kg D.W.)
Results
Identification and Cloning of Arabidopsis Nramp Genes.
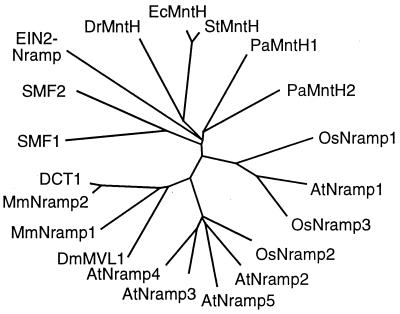
A search for sequences with homology to yeast and animal Nramp genes led to the identification of a family of Nramp homologous genes in Arabidopsis. AtNramp1 was identified as an expressed sequence tag sequence (212P17T7), and its cDNA was deposited later in the database first as AtNramp1 (AF165125), together with AtNramp2 (AF141204), and then as PMIT1 (AF181687). AtNramp3, AtNramp4 and, during the completion of this work, AtNramp5 and AtNramp6 were identified as genomic sequences on BACs T20D16, K8K14, F28A21, and T24D18, respectively. In addition, the sequence of EIN2 (AF 141202), a locus controlling ethylene sensitivity in Arabidopsis that shares homology with Nramp genes (26), was recently deposited in the database. The predicted EIN2 protein differs from AtNramp1 to AtNramp5 in that it includes a large hydrophilic C-terminal domain (26). Arabidopsis Nramp genes encode a family of highly hydrophobic membrane proteins. AtNramp2, AtNramp3, AtNramp4, and AtNramp5 are most closely related sharing between 67% and 75% conserved amino acid sequences, whereas AtNramp1 is more distantly related with only 33–37% amino acid conservation (Fig. 1). The Nramp domain within the EIN2 protein is even more divergent and shares only 17–20% identity at the amino acid level with AtNramp 1 to AtNramp 5 (Fig. 1). We cloned three AtNramp genes from Arabidopsis. A full-length AtNramp1 cDNA (AF 165125) was cloned by screening an Arabidopsis cDNA library using the expressed sequence tag. AtNramp3 (AF202539) and AtNramp4 (AF202540) were cloned by RT-PCR using the data from the genome sequencing project and sequenced.
Figure 1.
Phylogenetic tree of the Nramp gene family in animals, fungi, and plants. Nramp homologous genes: AtNramp1 (AF 165125), AtNramp2 (AF 141204), AtNramp3 (AF202539), AtNramp4 (AF202540), and AtNramp5 from Arabidopsis; OsNramp1 (L41217), OsNramp2 (L81152), and OsNramp3 (U60767) from rice (Oryza sativa). SMF1 (U15929) and SMF2 (U00062) from yeast (Saccharomyces cerevisiae). MmNramp1 (L13732) and MmNramp2 (L33415) from mouse (Mus musculus). DCT1 is the rat homologue of MmNramp2. DmMVL1 stands for Mavolio 1 (U23948) from Drosophila melanogaster. DrMntH (AE002012), EcMntH (AF161318), PaMntH1 (AF161319), PaMntH2 (AF161320), and StMntH (AF161317) from bacteria (Deinococcus radiodurans, Escherichia coli, Pseudomonas aeruginosa, and Salmonella typhimurium). The phylogenetic tree of Nramp sequences was drawn by using the treeview program after comparison of the deduced protein sequences with the clustalx program.
Arabidopsis Nramp Genes Complement Yeast Mutants Defective in Mn and Fe Transport.
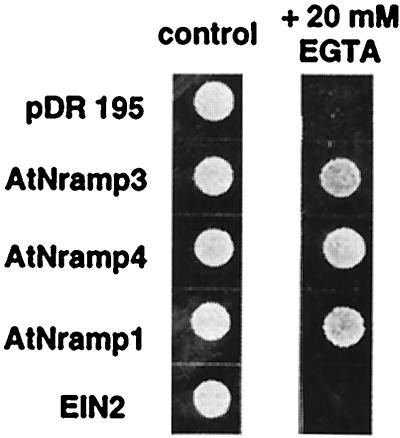
SMF1, a yeast Nramp homologue, has been shown to be a manganese transporter (22) with a broad metal specificity (28, 29). As a first step to investigate AtNramp function, we tested whether the plant AtNramp cDNAs are able to complement the phenotype of a S. cerevisiae strain disrupted in the SMF1 gene (smf1). smf1 yeast fail to grow on synthetic medium containing high concentrations of the divalent cation chelator EGTA. Fig. 2 shows that expression of AtNramp1, AtNramp3, and AtNramp4 under the control of a constitutive yeast promoter restored the growth of smf1 yeast on synthetic medium containing 20 mM EGTA, whereas a strain transformed with the empty vector pDR195 or EIN2 could not grow, as reported (26). Thus, AtNramp1, AtNramp3, and AtNramp4 complement the phenotype of smf1 yeast. These data demonstrate that AtNramp1, AtNramp3, and AtNramp4 cDNAs encode functional proteins and provide initial evidence that these proteins can mediate manganese transport.
Figure 2.
AtNramp genes complement a yeast mutant deficient in Mn uptake. Growth of smf1 yeast cells expressing AtNramp1, AtNramp3, or AtNramp4 on SD plates supplemented (Right) or not (Left) with 20 mM EGTA. smf1 yeast cells were transformed either with empty pDR195 or with pDR195 containing the cDNAs of AtNramp1, AtNramp3, AtNramp4 or of EIN2. Transformed strains were grown overnight in liquid synthetic dextrose-ura. The cultures were diluted to an OD of 0.1 and spotted on plates containing synthetic dextrose-ura (pH 6). The plates were incubated at 30°C for 3 days before photography.
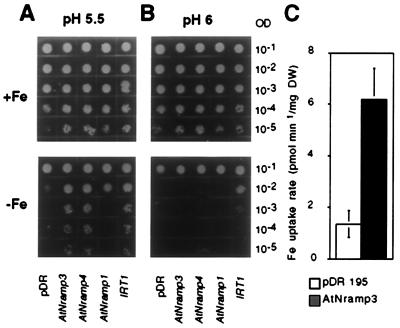
Subsequently, we tested whether the AtNramp genes are able to complement a yeast Fe transport mutant. The fet3fet4 yeast strain is defective in both low- and high-affinity Fe uptake systems (30), requires high Fe concentrations in its medium for growth, and shows reduced Fe uptake. Expression of AtNramp3 or AtNramp4 restored the ability of fet3fet4 to grow on a medium not supplemented with Fe (Fig. 3A). AtNramp3 or AtNramp4 complemented the phenotype of fet3fet4 as efficiently as IRT1, a well-characterized Fe uptake transporter gene from Arabidopsis (Fig. 3A; ref. 17), whereas AtNramp1 was less efficient. Interestingly, the ability of AtNramp3 or AtNramp4 to complement fet3fet4 was sharply pH dependent; whereas, at pH 5.5, AtNramp3 or AtNramp4 expression complemented the growth phenotype of fet3fet4 yeast as efficiently as IRT1 (Fig. 3A); at pH 6, only IRT1 was still able to complement Fe uptake (Fig. 3B). The pH dependence of the complementation correlates with the finding that the mammalian homologue Nramp2/DCT1 functions as a H+-metal symporter (23). When we compared FeII uptake rates in fet3fet4 yeast transformed with the empty vector or expressing AtNramp3, we found that AtNramp3 expression consistently increased the Fe uptake rate at pH 5.5 (Fig. 3C). On average, the uptake rate was 6.4 times higher in AtNramp3-expressing yeast cells. These results provide additional evidence that AtNramp3 and AtNramp4 are metal transporters and reveal functional differences between the different members of the Arabidopsis Nramp family: AtNramp3 and AtNramp4, which are closely related, complement fet3fet4, whereas AtNramp1, which is evolutionary more distant, is less efficient.
Figure 3.
AtNramp3 and AtNramp4 complement a Fe uptake deficient yeast mutant. Growth of fet3fet4 yeast cells expressing AtNramp1, AtNramp3, or AtNramp4 or IRT1 on synthetic dextrose-URA (pH 5.5) (A) or (pH 6) (B) supplemented 0.2 mM FeCl3 (Upper) or without added Fe (Lower). fet3fet4 yeast cells were transformed with the empty pDR195 vector, with pDR195 containing the cDNA of AtNramp1, AtNramp3, or AtNramp4, or with the vector pFL61 containing the IRT1 cDNA. Transformed strains were grown overnight in liquid synthetic dextrose-ura supplemented with 0.2 mM FeCl3. The cultures were diluted to ODs of 10−1 to 10−5 (as indicated) and spotted on synthetic dextrose-ura plates. The plates were incubated at 30°C for 3 days before photography. (C) FeII uptake rates in fet3fet4 yeast expressing AtNramp3 or transformed with the empty vector. FeII accumulation was measured after 10 min in 30 μM 55Fe at pH 5.5.
AtNramp Expression Enhances Cd2+ Sensitivity and Increases Cd2+ Content in Yeast.
Cadmium is a toxic heavy metal transported across plant membranes by physiological metal transporters (14, 15). Therefore, we tested the influence of AtNramp expression on Cd2+ toxicity in yeast. On plates containing synthetic medium supplemented with 10 μM Cd2+, the growth of wild-type yeast expressing AtNramp1, AtNramp3, and AtNramp4 was strongly impaired compared with yeast transformed with the empty vector (Fig. 4A). In liquid culture, 3 μM Cd2+ strongly reduced the growth of yeast expressing AtNramp1, AtNramp3, and AtNramp4 compared with the control. Measurement of the Cd2+ content after 24 h showed that Cd2+ inhibition of growth by AtNramps correlates with increased Cd2+ accumulation (Fig. 4B). However, when Cd2+ uptake was studied in short-term experiments (20 min), expression of AtNramp3 or AtNramp4 did not increase Cd2+ accumulation (data not shown), indicating a low rate of enhanced Cd2+ accumulation.
Figure 4.
AtNramp expression increases Cd2+ sensitivity and Cd2+ content in yeast. (A) Growth of INVSc1 yeast cells expressing AtNramp1, AtNramp3, or AtNramp4 on plates containing synthetic dextrose-ura buffered to pH 6 with 50 mM Mes without CdCl2 (Left) or supplemented with 10 μM CdCl2 (Right). The plates were incubated at 30°C for 3 days before photography. (B) Cd2+ content of INVSc1 yeast cells expressing AtNramp1, AtNramp3, or AtNramp4 grown for 24 h in liquid synthetic dextrose-ura supplemented with 3 μM Cd2+, as measured by the uptake of 109Cd2+. INVSc1 yeast cells were transformed either with the empty pDR195 vector, or with pDR195 containing the AtNramp1, AtNramp3, or AtNramp4 cDNA. Transformed strains were grown overnight in synthetic dextrose-ura. The cultures were then spotted on plates or inoculated in 10 ml synthetic dextrose-ura at an OD of 0.01. Error bars represent the SE from four independent experiments (P < 0.03, 0.06, and 0.07 for AtNramp1, AtNramp3, and AtNramp4, respectively).
AtNramp Gene Expression in Arabidopsis Seedlings.
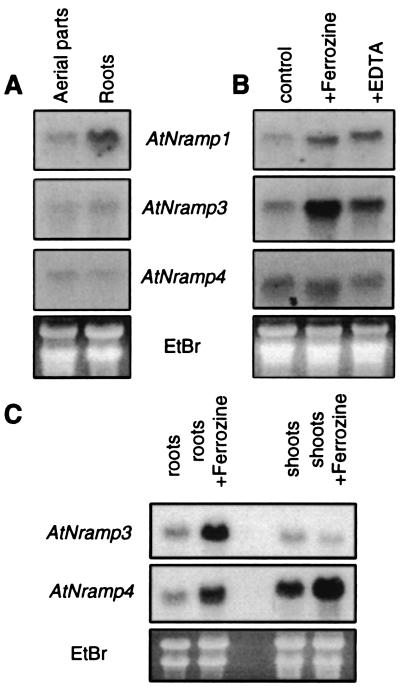
We examined the expression pattern of AtNramp genes. When Arabidopsis seedlings were grown on metal replete Murashige and Skoog medium, AtNramp1, AtNramp3, and AtNramp4 mRNAs could be detected by Northern blot in total RNA extracts from both roots and aerial parts of Arabidopsis seedlings (Fig. 5A). AtNramp1 was preferentially expressed in roots, whereas AtNramp3 and AtNramp4 were expressed at comparable levels in roots and aerial parts (Fig. 5A).
Figure 5.
Differential induction of AtNramps by Fe starvation. (A) Expression of AtNramp1, AtNramp3, or AtNramp4 in roots and aerial parts of Arabidopsis seedlings. RNA was extracted separately from roots and aerial parts (cotyledons, hypocotyls, and primary leaves). (B) Effect of Fe or metal starvation on AtNramp1, AtNramp3, or AtNramp4 expression in whole Arabidopsis seedlings. (C) On Fe starvation, AtNramp3 and AtNramp4 are up-regulated in roots. RNA was extracted from seedlings grown on (MS) or on MS supplemented with either 1 mM ferrozine or 1 mM EDTA. Arabidopsis were grown on MS medium for 10 days before RNA extraction. Then, 15 μg of total RNA was loaded in each lane and transferred on a nylon membrane. The membrane was probed with full-length DNA probes corresponding to AtNramp1, AtNramp3, or AtNramp4. Ethidium bromide staining of the gel is shown as a loading control (EtBr).
We further tested whether AtNramp gene expression could be enhanced by metal starvation. For this purpose, Arabidopsis seedlings were grown on media supplemented with 1 mM EDTA, a divalent cation chelator, or 1 mM ferrozine, a specific chelator of Fe2+. AtNramp1 and AtNramp3 expression were significantly increased when seedlings were grown on 1 mM EDTA, and this increase was even more obvious for AtNramp3 when they were grown on 1 mM Ferrozine (Fig. 5B, n = 4). Furthermore, we showed that the up-regulation of AtNramp3 mRNA by Fe starvation is root-specific (Fig. 5C), whereas AtNramp4 mRNA levels were increased in both roots and shoots by Fe starvation (Fig. 5 B and C, n = 3 of 4 experiments). We chose to focus initial molecular physiological studies in Arabidopsis on AtNramp3, which complements a yeast mutant deficient in Fe transport and is up-regulated by Fe starvation.
AtNramp3 Disruption Leads to Moderate Cd2+ Resistance in Arabidopsis Seedlings.
To investigate the physiological function of AtNramp3 in plants, we used PCR to screen a population of Agrobacterium-transformed Arabidopsis for individuals carrying a T-DNA insertion in this gene. A plant with a T-DNA insertion in the 3′ end of the AtNramp3 gene was identified (Fig. 6A). Both PCR and genomic Southern experiments confirmed the mutant genotype. After a backcross with wild-type Arabidopsis, segregation of the kanamycin resistance phenotype indicated that the original plant carried a T-DNA insertion at a single locus. RT-PCR experiments using primers 5′ of the insertion site allowed the detection of AtNramp3 mRNA in wild-type but not in mutant plants, suggesting that the truncated gene was either not transcribed or that its mRNA was unstable. The insertion results in the truncation of the last 42 C-terminal amino acids of the AtNramp3 protein and replacement by 10 amino acids encoded by the left border of the T-DNA before reaching a stop codon. The truncated cDNA corresponding to this mutant gene failed to complement the smf1 yeast strain phenotype (Fig. 6B).
Figure 6.
Characterization of a mutant carrying a T-DNA insertion in AtNramp3. (A) Localization of the T-DNA insertion site in the AtNramp3 gene and cDNA. Gray boxes represent introns, black box represents T-DNA sequence putatively translated in the mutant. (B) The T-DNA disrupted allele of AtNramp3 cannot complement smf1, a Mn uptake deficient yeast mutant. smf1 yeast cells were transformed with empty pDR195, pDR195 containing the cDNA of AtNramp3, or a truncated AtNramp3 cDNA corresponding to the predicted mRNA product of the disrupted allele of AtNramp3. (C) Root growth shows increased resistance to Cd2+ in AtNramp3-1 mutant. Root length of Arabidopsis seedlings from control with a T-DNA insertion outside AtNramp3 (open bars) or mutant with T-DNA insertion in AtNramp3 (filled bars) grown vertically for 10 days on plates containing minimal medium supplemented with various concentrations of Cd2+. The length of the roots grown on plates without Cd2+ were 52.0 ± 5.0 mm for the control and 56.2 ± 2.4 mm for the AtNramp3Δ mutant. The graph represents the results of one representative experiment of four. Values are means of 12–16 root lengths, and error bars represent SE.
When grown in soil or in minimal or Murashige and Skoog medium, the mutant did not display any clearly visible phenotype. However, detailed analyses of root growth on plates containing 1–30 μM Cd2+ showed that root growth in the mutant was moderately but significantly more resistant to Cd2+ than the control (Fig. 6C). For example, the roots of the AtNramp3-1 mutant were 30.5 ± 13.0% longer than controls on 1 μM Cd2+ (n = 4 experiments, P < 0.004) and 57.0 ± 9.5% longer than controls on 10 μM Cd2+ (n = 5, P < 0.002; Fig. 6C). Thus, AtNramp3 contributes to Cd2+ toxicity in Arabidopsis. Measurements of the metal content in whole plants using inductively coupled plasma optical emission spectroscopy did not show any significant difference in Fe, Mn, and Zn content and Cd2+ accumulation between controls and mutants under the imposed conditions (data not shown). It is possible that the differences in metal accumulation between mutant and control were too small to be resolved at the whole plant level. For example, AtNramp3 disruption might alter metal levels in specific tissues, cell types, or cell compartments.
Overexpression of AtNramp3 Leads to Cd2+ Hypersensitivity and Fe Overaccumulation.
To further test a role of AtNramp3 in Arabidopsis, we generated plants ectopically overexpressing this gene. The AtNramp3 cDNA was placed under the strong constitutive CaMV35S promoter and introduced into Arabidopsis through Agrobacterium-mediated transformation. Two independent lines with elevated AtNramp3 mRNA levels (Fig. 7A) carrying single T-DNA insertions were selected and self-crossed to obtain stable homozygous overexpressing lines. Both lines displayed Cd2+ hypersensitivity as assayed by root growth measurements (Fig. 7 B and C). On Cd2+-free medium, the root length was the same in overexpressing lines and a control line transformed with the empty vector, pMON530 (0.8 ± 2.4%, n = 4, P > 0.3). However on media containing 1 and 10 μM Cd2+, the root length of the overexpressing plants was reduced by 24.4 ± 6.5% (n = 3, P < 0.007) and 31.0 ± 3.0% (n = 4, P < 0.004), respectively. The phenotype was maintained from the T3 to T4 generation as a genetically inheritable trait. This result confirms that AtNramp3 can modulate heavy metal toxicity in plants.
Figure 7.
Overexpression of AtNramp3 confers Cd2+ hypersensitivity in Arabidopsis. (A) Northern blot analysis of an overexpressing line. Conditions are as described in Fig. 5. (B) Root growth hypersensitivity to Cd2+. Arabidopsis seedlings were grown vertically for 10 days on plates containing minimal medium supplemented (Right) or not (Left) with 1 μM Cd2+. (C) Root length of Arabidopsis seedlings grown vertically for 10 days on plates containing minimal medium supplemented with 0, 1, or 10 μM Cd2+. The graph represents the results of one representative experiment of four with one of the two lines analyzed in detail. Values are mean of measurements on 12–16 roots, and error bars represent SE. (D) Iron over-accumulation in 35S-AtNramp3 plants on Cd2+ treatment. Arabidopsis seedlings were grown for 10 days in liquid minimal medium supplemented or not with 1 μM Cd2+. The Fe content was quantified by inductively coupled plasma optical emission spectroscopy. The mean ± SE of three independent experiments is represented. Control (open bars): homozygous, single insertion line transformed with empty pMON530. 35S-AtNramp3 (filled bars): homozygous, single insertion line transformed with pMON530 containing AtNramp3 cDNA driven by the 35S CaMV promoter.
To determine whether AtNramp3 affects Fe accumulation, we measured the iron content of control and AtNramp3 overexpressing seedlings grown in minimal medium supplemented or not with 1 μM Cd2+. AtNramp3 overexpressing plants accumulated 77 ± 15% (n = 3 experiments) more Fe than control plants transformed with empty vector, when exposed to Cd2+ (Fig. 7D). In contrast, no increase in Cd2+ content or reproducible change of the whole plant content in other metals such as Mn and Zn could be detected under these conditions. These findings reveal a role for AtNramp3 in Fe transport in Arabidopsis.
Discussion
We have cloned three members of the Arabidopsis Nramp gene family: AtNramp1, AtNramp3, and AtNramp4. Heterologous expression in yeast points to a role of AtNramp proteins in transition metal transport: AtNramp genes are able to complement yeast mutants altered in Mn and Fe uptake. Furthermore, expression of AtNramp genes in yeast leads to an increase in Cd2+ sensitivity and Cd2+ accumulation. Investigation of AtNramp expression as well as manipulation of these genes in planta also points to a role in metal homeostasis in plants: AtNramp3 and AtNramp4 are induced by Fe starvation; disruption of AtNramp3 in Arabidopsis reduces Cd2+ sensitivity of root growth, and, conversely, overexpression of AtNramp3 leads to an increase in Cd2+ sensitivity and a conditional increase in Fe accumulation.
The Nramp homologues SMF1 in yeast and DCT1/Nramp2 in mammals mediate the uptake of a broad range of metals (22, 23, 28, 29). However, no evidence for metal transport was found for other genes that belong to the Nramp family or contain Nramp-like domains. Neither Nramp1 from mice, which encodes the natural resistance macrophage associated protein and determines sensitivity to mycobacterium infection such as tuberculosis or leprosy (21), nor the recently isolated Nramp homologue EIN2, which is a determinant of ethylene sensitivity in plants (26), can complement a yeast mutant disrupted in the Nramp homologue SMF1 (26, 31). In contrast, the mouse Nramp2 and the three AtNramp genes that we have cloned are able to rescue smf1 yeast growth (31). Although not all Nramp proteins seem to function as metal transporters, these results provide initial data suggesting that the members of the AtNramp family in Arabidopsis function as metal transporters.
Possible Roles of AtNramp3 and AtNramp4 as Iron Transporters in Plants.
Although it has been proposed that Nramps may function as broad specificity transition metal transporters (23, 28), the mammalian homologue Nramp2/DCT1 has been more specifically implicated in Fe homeostasis (24). AtNramp3 and AtNramp4 complement the phenotype of fet3fet4, a yeast mutant deficient in Fe uptake, as effectively as the well-characterized Arabidopsis Fe uptake transporter IRT1 (17). Furthermore, AtNramp3 mediates FeII uptake in yeast. Our data lead to a model in which two families of Fe transporters, IRTs and AtNramps, may contribute to Fe homeostasis in planta. Iron transport by AtNramp3 and AtNramp4 was stimulated by extracellular acidification, suggesting that the contribution of AtNramp and IRT transporters to physiological Fe transport may vary depending on pH.
Physiological Functions of AtNramps in Plants.
Expression studies show that AtNramp genes are expressed in both roots and aerial parts of Arabidopsis seedlings under metal replete conditions. In rice, OsNramp3 is expressed primarily in roots, and OsNramp2 in leaves, whereas OsNramp3 is present in both (25). The expression of plant Nramps is distinct with respect to the Fe uptake transporter gene of the ZIP family, IRT1, which is expressed specifically in roots in response to metal starvation (17). The expression of AtNramp suggests a contribution of Nramp transporters to constitutive metal transport in plants, whereas IRT1 has been proposed to function as an inducible metal uptake system in roots (17).
Interestingly, AtNramp1 and AtNramp3 mRNA levels are increased by metal starvation, and AtNramp3 and AtNramp4 are strongly up-regulated by iron starvation. Therefore, the increased accumulation of Mn and Cd observed on Fe starvation in pea and Arabidopsis (12, 19) might be because of the induction of both the multispecific transport system IRT1 (12, 17, 19) and Nramp transporters.
The induction of AtNramp3 on Fe starvation suggests a possible role of AtNramp3 in inducible Fe uptake. The Atnramp3-1 insertion mutant did not display a clearly visible phenotype on low Fe minimal medium or rich medium supplemented with the Fe chelating molecule Ferrozine. This is possibly because of functional redundancy with other inducible Fe transport systems, such as IRT1. However, the role of AtNramp3 in Fe transport in planta is supported by the observation that AtNramp3 overexpressing plants contained elevated levels of Fe, on Cd2+ treatment. It is possible that this phenotype could be revealed in the presence of Cd2+ because Cd2+ blocks other Fe transporters, such as IRT1 (17, 19). Together, these data indicate a contribution of AtNramp3 to Fe transport in plants.
Disruption of the AtNramp3 gene leads to a moderate increase in Cd2+ resistance. Considering that AtNramp3 has at least four closely related homologues in the Arabidopsis genome, it is remarkable that knocking out a single Nramp gene leads to an observable phenotype. In agreement with the phenotype of the mutant with a disrupted AtNramp3, overexpression of this gene confers increased Cd2+ sensitivity in Arabidopsis. In conclusion, the phenotype of plants disrupted or overexpressing AtNramp3 point to a role of AtNramps in physiological Cd2+ transport and Cd2+ sensitivity in plants.
In conclusion, we have functionally characterized three members of the Nramp gene family in Arabidopsis. Our data provide evidence that AtNramp genes encode multispecific metal transport systems in plants that can transport Fe, Mn, and Cd2+. Analyses of the expression of AtNramp genes in Arabidopsis suggest that all these Nramp genes play roles in constitutive metal transport. Furthermore, the complementation of fet3fet4 yeast mutant by AtNramp3 together with the induction of AtNramp3 gene expression by Fe starvation and the increased accumulation of Fe in AtNramp3 overexpressing seedlings on Cd2+ treatment suggest that AtNramp3 plays a role in inducible Fe transport in plants.
Acknowledgments
We thank Nathan Nelson (Tel Aviv University) for providing the smf1 mutant, Mary Lou Guerinot (Dartmouth College) for providing the fet3fet4 mutant and the IRT1 cDNA, the Arabidopsis Biological Resource Center for supplying seeds of T-DNA mutagenized lines, Randy Hampton (University of California, San Diego) for the plasmid pRH530, June Kwak and Eugene Kim for help with molecular biological techniques and yeast manipulation, and David Lee for reading of the manuscript. This research was supported by U.S. Department of Energy Grant DE-FG07-96ER20253 (to J.I.S.) and U.S. Department of Agriculture Grant 98-353-04-6684 (to J.I.S.), S.T. was supported by a fellowship from the Human Frontier Science Program Organization.
Note Added in Proof:
Another paper describing the molecular characterization of AtNramp1 and AtNramp2 is soon to be published (32).
Footnotes
References
- 1.Fox T C, Guerinot M L. Annu Rev Plant Physiol. 1998;49:669–696. doi: 10.1146/annurev.arplant.49.1.669. [DOI] [PubMed] [Google Scholar]
- 2.Orgad S, Nelson H, Segal D, Nelson N. J Exp Biol. 1998;201:115–120. doi: 10.1242/jeb.201.1.115. [DOI] [PubMed] [Google Scholar]
- 3.Hirayama T, Kieber J J, Hirayama N, Kogan M, Guzman P, Nourizadeh S, Alonso J M, Dailey W P, Dancis A, Ecker J R. Cell. 1999;97:383–393. doi: 10.1016/s0092-8674(00)80747-3. [DOI] [PubMed] [Google Scholar]
- 4.Nelson N. EMBO J. 1999;18:4361–4371. doi: 10.1093/emboj/18.16.4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rea P A, Li Z S, Lu Y P, Drozdowicz Y M, Martinoia E. Annu Rev Plant Physiol. 1998;49:727–760. doi: 10.1146/annurev.arplant.49.1.727. [DOI] [PubMed] [Google Scholar]
- 6.Salt D E, Rauser W E. Plant Physiol. 1995;107:1293–1301. doi: 10.1104/pp.107.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salt D E, Wagner G J. J Biol Chem. 1993;268:12297–12302. [PubMed] [Google Scholar]
- 8.Ortiz D F, Ruscitti T, McCue K F, Ow D W. J Biol Chem. 1995;270:4721–4728. doi: 10.1074/jbc.270.9.4721. [DOI] [PubMed] [Google Scholar]
- 9.Reid R J, Brookes J D, Tester M A, Smith F A. Planta. 1996;198:39–45. [Google Scholar]
- 10.Fox T C, Shaff J E, Grusak M A, Norvell W A, Chen Y, Chaney R L, Kochian L V. Plant Physiol. 1996;111:93–100. doi: 10.1104/pp.111.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hart J J, Norvell W A, Welch R M, Sullivan L A, Kochian L V. Plant Physiol. 1998;118:219–226. doi: 10.1104/pp.118.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen C K, Fox T C, Garvin D F, Kochian L V. Plant Physiol. 1998;116:1063–1072. doi: 10.1104/pp.116.3.1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart J J, Welch R M, Norvell W A, Sullivan L A, Kochian L V. Plant Physiol. 1998;116:1413–1420. doi: 10.1104/pp.116.4.1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clemens S, Antosiewicz D M, Ward J M, Schachtman D P, Schroeder J I. Proc Natl Acad Sci USA. 1998;95:12043–12048. doi: 10.1073/pnas.95.20.12043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J W W, Grunes D L, Kochian L V. Proc Natl Acad Sci USA. 1994;91:3473–3477. doi: 10.1073/pnas.91.8.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kampfenkel K, Kushnir S, Babiychuk E, Inze D, Van Montagu M. J Biol Chem. 1995;270:28479–28486. doi: 10.1074/jbc.270.47.28479. [DOI] [PubMed] [Google Scholar]
- 17.Eide D, Broderius M, Fett J, Guerinot M L. Proc Natl Acad Sci USA. 1996;93:5624–5628. doi: 10.1073/pnas.93.11.5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grotz N, Fox T, Connolly E, Park W, Guerinot M L, Eide D. Proc Natl Acad Sci USA. 1998;95:7220–7224. doi: 10.1073/pnas.95.12.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korshunova Y O, Eide D, Clark W G, Guerinot M L, Pakrasi H B. Plant Mol Biol. 1999;40:37–44. doi: 10.1023/a:1026438615520. [DOI] [PubMed] [Google Scholar]
- 20.Cellier M, Prive G, Belouchi A, Kwan T, Rodrigues V, Chia W, Gros P. Proc Natl Acad Sci USA. 1995;92:10089–10093. doi: 10.1073/pnas.92.22.10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vidal S M, Malo D, Vogan K, Skamene E, Gros P. Cell. 1993;73:469–485. doi: 10.1016/0092-8674(93)90135-d. [DOI] [PubMed] [Google Scholar]
- 22.Supek F, Supekova L, Nelson H, Nelson N. Proc Natl Acad Sci USA. 1996;93:5105–5110. doi: 10.1073/pnas.93.10.5105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunshin H, Mackenzie B, Berger U V, Gunshin Y, Romero M F, Boron W F, Nussberger S, Gollan J L, Hediger M A. Nature (London) 1997;388:482–488. doi: 10.1038/41343. [DOI] [PubMed] [Google Scholar]
- 24.Fleming M D, Romano M A, Su M A, Garrick L M, Garrick M D, Andrews N C. Proc Natl Acad Sci USA. 1998;95:1148–1153. doi: 10.1073/pnas.95.3.1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belouchi A, Kwan T, Gros P. Plant Mol Biol. 1997;33:1085–1092. doi: 10.1023/a:1005723304911. [DOI] [PubMed] [Google Scholar]
- 26.Alonso J M, Hirayama T, Roman G, Nourizadeh S, Ecker J R. Science. 1999;284:2148–2152. doi: 10.1126/science.284.5423.2148. [DOI] [PubMed] [Google Scholar]
- 27.Elledge S J, Mulligan J T, Ramer S W, Spottswood M, Davis R W. Proc Natl Acad Sci USA. 1991;88:1731–1735. doi: 10.1073/pnas.88.5.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X F, Supek F, Nelson N, Culotta V C. J Biol Chem. 1997;272:11763–11769. doi: 10.1074/jbc.272.18.11763. [DOI] [PubMed] [Google Scholar]
- 29.Chen X Z, Peng J B, Cohen A, Nelson H, Nelson N, Hediger M A. J Biol Chem. 1999;274:35089–35094. doi: 10.1074/jbc.274.49.35089. [DOI] [PubMed] [Google Scholar]
- 30.Dix D R, Bridgham J T, Broderius M A, Byersdorfer C A, Eide D J. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 31.Pinner E, Gruenheid S, Raymond M, Gros P. J Biol Chem. 1997;272:28933–28938. doi: 10.1074/jbc.272.46.28933. [DOI] [PubMed] [Google Scholar]
- 32.Curie, C., Alonso, J. M., Le Jean, M., Ecker, J. R. & Briat, J.-F. (2000) Biochem. J., in press. [PMC free article] [PubMed]