Abstract
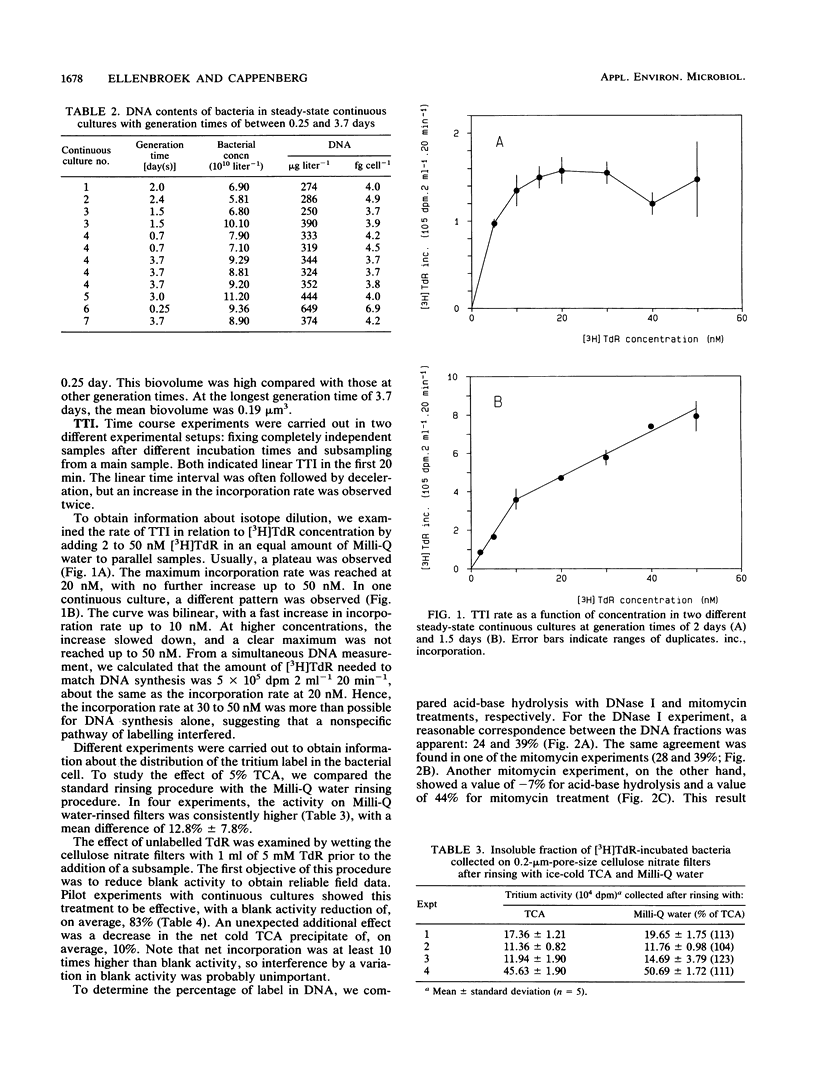
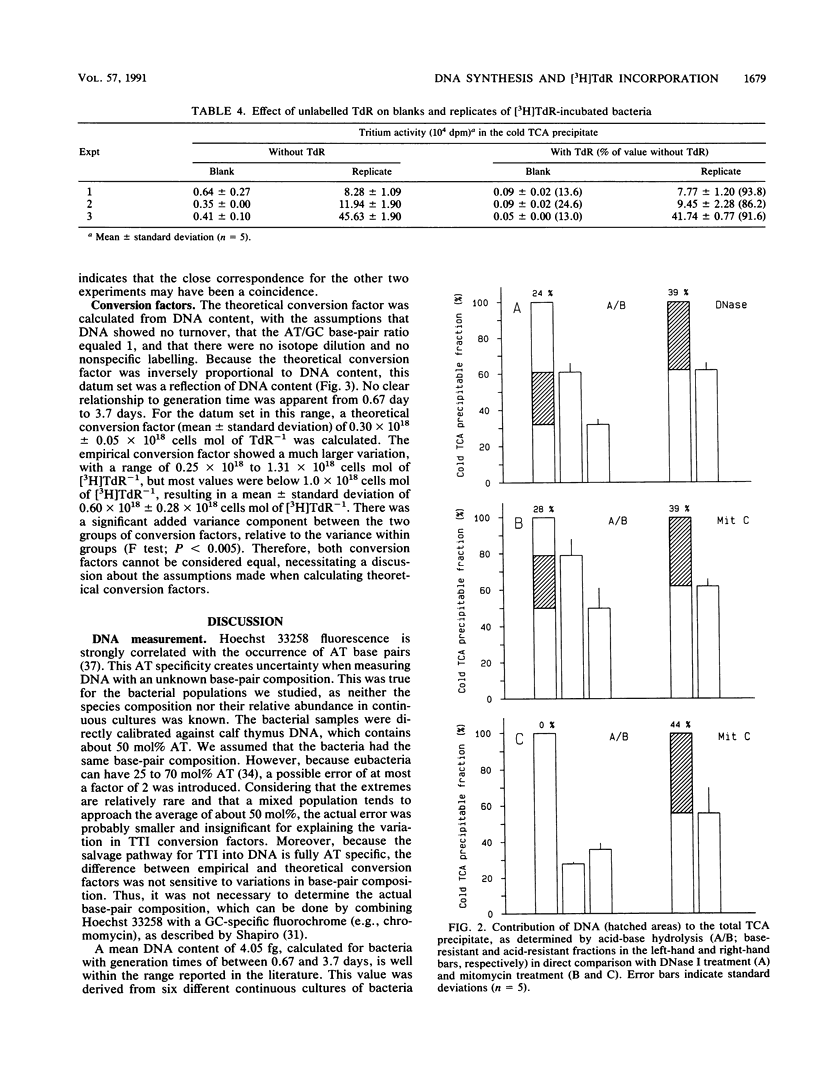
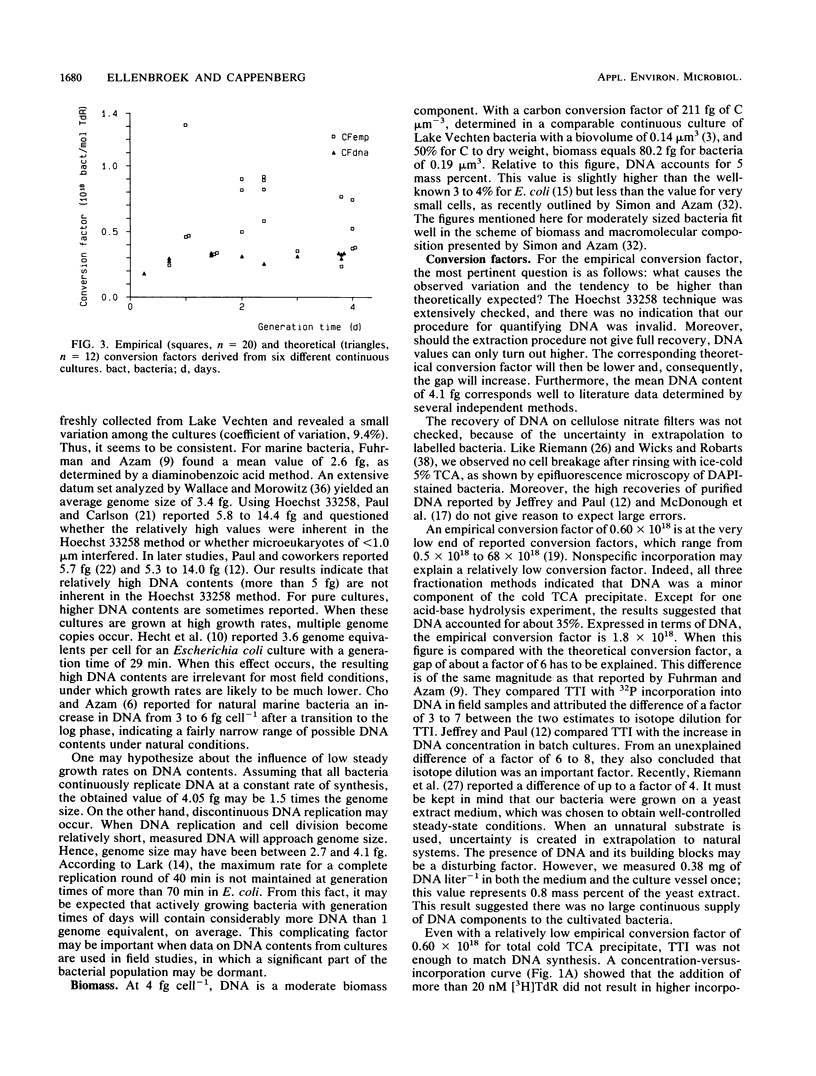
Continuous cultivation of heterotrophic freshwater bacteria was used to assess the relationship between DNA synthesis and tritiated thymidine incorporation. The bacteria were grown on a yeast extract medium with generation times of 0.25 to 3.7 days. In six different continuous cultures, each inoculated with a grazer-free mixed bacterial sample from Lake Vechten (The Netherlands), tritiated thymidine incorporation into a cold trichloroacetic acid precipitate and bacterial cell production were measured simultaneously. Empirical conversion factors were determined by division of both parameters. They ranged from 0.25 × 1018 to 1.31 × 1018 cells mol of tritiated thymidine-1 (mean, 0.60 × 1018 cells mol of tritiated thymidine-1). In addition, DNA concentrations were measured by fluorometry with Hoechst 33258. The validity of this technique was confirmed. Down to a generation time of 0.67 day, bacterial DNA content showed little variation, with values of 3.8 to 4.9 fg of DNA cell-1. Theoretical conversion factors, which can be derived from DNA content under several assumptions, were between 0.26 × 1018 and 0.34 × 1018 cells mol of thymidine-1 (mean, 0.30 × 1018 cells mol of thymidine-1). Isotope dilution was considered the main factor in the observed discrepancy between the conversion factors. In all experiments, a tritiated thymidine concentration of 20 nM was used. Control experiments indicated maximum incorporation at this concentration. It was therefore concluded that the observed difference resulted from intracellular isotope dilution which cannot be detected by current techniques for isotope dilution analysis.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bloem J., Ellenbroek F. M., Bär-Gilissen M. J., Cappenberg T. E. Protozoan grazing and bacterial production in stratified lake vechten estimated with fluorescently labeled bacteria and by thymidine incorporation. Appl Environ Microbiol. 1989 Jul;55(7):1787–1795. doi: 10.1128/aem.55.7.1787-1795.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloem J., Starink M., Bär-Gilissen M. J., Cappenberg T. E. Protozoan grazing, bacterial activity, and mineralization in two-stage continuous cultures. Appl Environ Microbiol. 1988 Dec;54(12):3113–3121. doi: 10.1128/aem.54.12.3113-3121.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brittain A. M., Karl D. M. Catabolism of tritiated thymidine by aquatic microbial communities and incorporation of tritium into RNA and protein. Appl Environ Microbiol. 1990 May;56(5):1245–1254. doi: 10.1128/aem.56.5.1245-1254.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesarone C. F., Bolognesi C., Santi L. Improved microfluorometric DNA determination in biological material using 33258 Hoechst. Anal Biochem. 1979 Nov 15;100(1):188–197. doi: 10.1016/0003-2697(79)90131-3. [DOI] [PubMed] [Google Scholar]
- Davis C. L. Uptake and incorporation of thymidine by bacterial isolates from an upwelling environment. Appl Environ Microbiol. 1989 May;55(5):1267–1272. doi: 10.1128/aem.55.5.1267-1272.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R. M., Taggart R. T., Pettijohn D. E. Size and DNA content of purfied E. coli nucleoids observed by fluorencence microscopy. Nature. 1975 Jan 3;253(5486):60–62. doi: 10.1038/253060a0. [DOI] [PubMed] [Google Scholar]
- Jeffrey W. H., Paul J. H. Underestimation of DNA synthesis by [h]thymidine incorporation in marine bacteria. Appl Environ Microbiol. 1988 Dec;54(12):3165–3168. doi: 10.1128/aem.54.12.3165-3168.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman D., Ducklow H., Mitchell R. Estimates of bacterial growth from changes in uptake rates and biomass. Appl Environ Microbiol. 1982 Dec;44(6):1296–1307. doi: 10.1128/aem.44.6.1296-1307.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lark K. G. Initiation and control of DNA synthesis. Annu Rev Biochem. 1969;38:569–604. doi: 10.1146/annurev.bi.38.070169.003033. [DOI] [PubMed] [Google Scholar]
- McDonough R. J., Sanders R. W., Porter K. G., Kirchman D. L. Depth distribution of bacterial production in a stratified lake with an anoxic hypolimnion. Appl Environ Microbiol. 1986 Nov;52(5):992–1000. doi: 10.1128/aem.52.5.992-1000.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munro H. N. The determination of nucleic acids. Methods Biochem Anal. 1966;14:113–176. doi: 10.1002/9780470110324.ch5. [DOI] [PubMed] [Google Scholar]
- Paul J. H., Myers B. Fluorometric determination of DNA in aquatic microorganisms by use of hoechst 33258. Appl Environ Microbiol. 1982 Jun;43(6):1393–1399. doi: 10.1128/aem.43.6.1393-1399.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann B., Søndergaard M. Measurements of diel rates of bacterial secondary production in aquatic environments. Appl Environ Microbiol. 1984 Apr;47(4):632–638. doi: 10.1128/aem.47.4.632-638.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robarts R. D., Wicks R. J., Sephton L. M. Spatial and Temporal Variations in Bacterial Macromolecule Labeling with [methyl-H]Thymidine in a Hypertrophic Lake. Appl Environ Microbiol. 1986 Dec;52(6):1368–1373. doi: 10.1128/aem.52.6.1368-1373.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servais P., Martinez J., Billen G., Vives-Rego J. Determining [H]Thymidine Incorporation into Bacterioplankton DNA: Improvement of the Method by DNase Treatment. Appl Environ Microbiol. 1987 Aug;53(8):1977–1979. doi: 10.1128/aem.53.8.1977-1979.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits J. D., Riemann B. Calculation of cell production from [h]thymidine incorporation with freshwater bacteria. Appl Environ Microbiol. 1988 Sep;54(9):2213–2219. doi: 10.1128/aem.54.9.2213-2219.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C., Morowitz H. J. Genome size and evolution. Chromosoma. 1973;40(2):121–126. doi: 10.1007/BF00321457. [DOI] [PubMed] [Google Scholar]
- Weisblum B., Haenssler E. Fluorometric properties of the bibenzimidazole derivative Hoechst 33258, a fluorescent probe specific for AT concentration in chromosomal DNA. Chromosoma. 1974;46(3):255–260. doi: 10.1007/BF00284881. [DOI] [PubMed] [Google Scholar]



