Abstract
The identification of the prokaryotic species which constitute marine bacterioplankton communities has been a long-standing problem in marine microbiology. To address this question, we used the polymerase chain reaction to construct and analyze a library of 51 small-subunit (16S) rRNA genes cloned from Sargasso Sea bacterioplankton genomic DNA. Oligonucleotides complementary to conserved regions in the 16S rDNAs of eubacteria were used to direct the synthesis of polymerase chain reaction products, which were then cloned by blunt-end ligation into the phagemid vector pBluescript. Restriction fragment length polymorphisms and hybridizations to oligonucleotide probes for the SAR11 and marine Synechococcus phylogenetic groups indicated the presence of at least seven classes of genes. The sequences of five unique rDNAs were determined completely. In addition to 16S rRNA genes from the marine Synechococcus cluster and the previously identified but uncultivated microbial group, the SAR11 cluster [S. J. Giovannoni, T. B. Britschgi, C. L. Moyer, and K. G. Field. Nature (London) 345:60-63], two new gene classes were observed. Phylogenetic comparisons indicated that these belonged to unknown species of alpha- and gamma-proteobacteria. The data confirm the earlier conclusion that a majority of planktonic bacteria are new species previously unrecognized by bacteriologists.
Full text
PDF

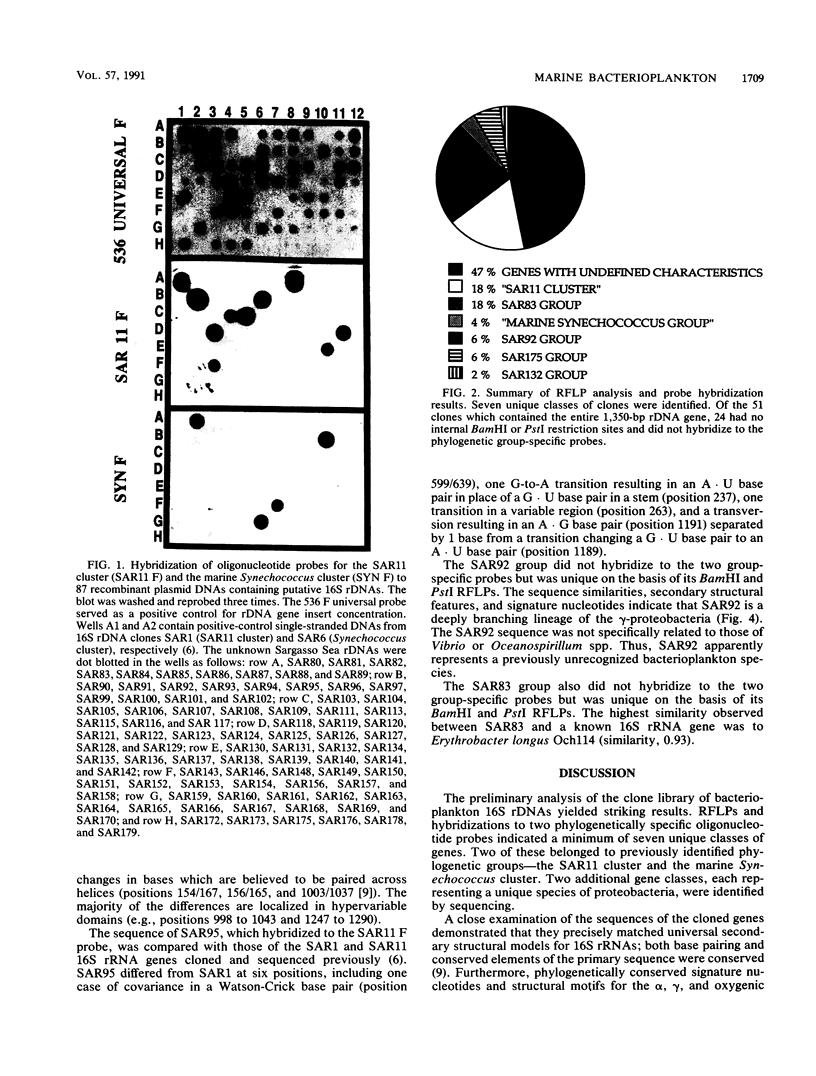
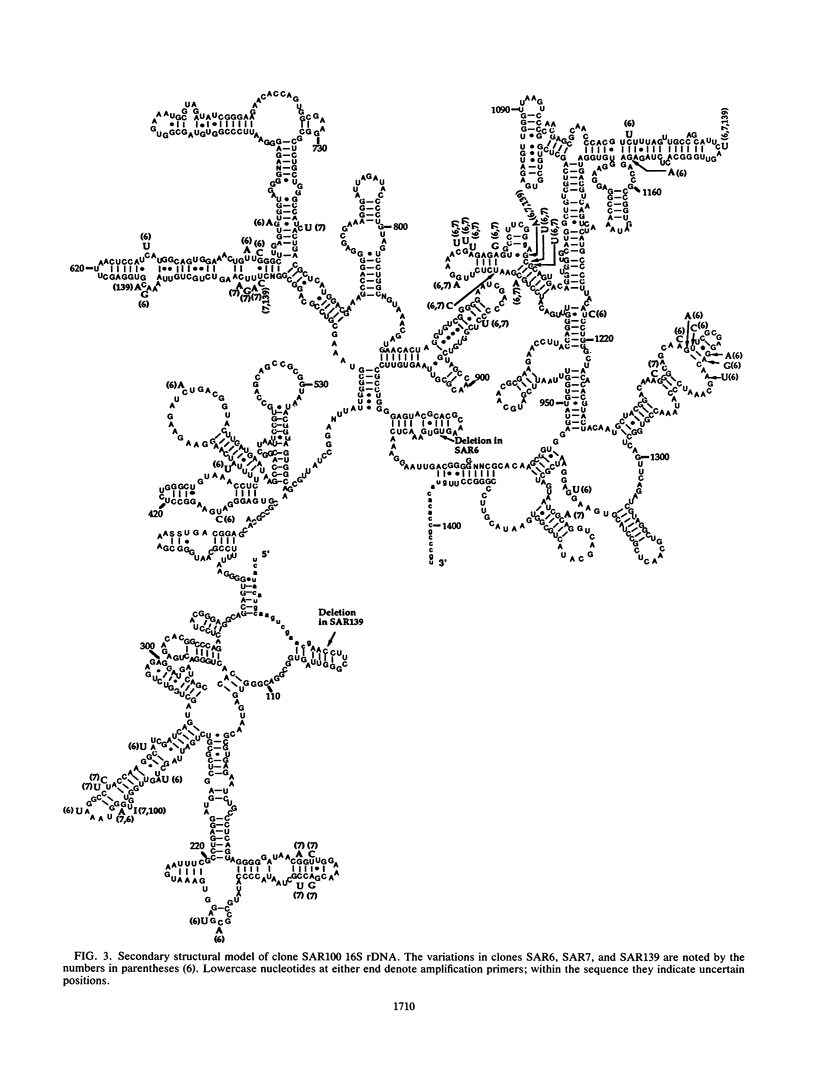
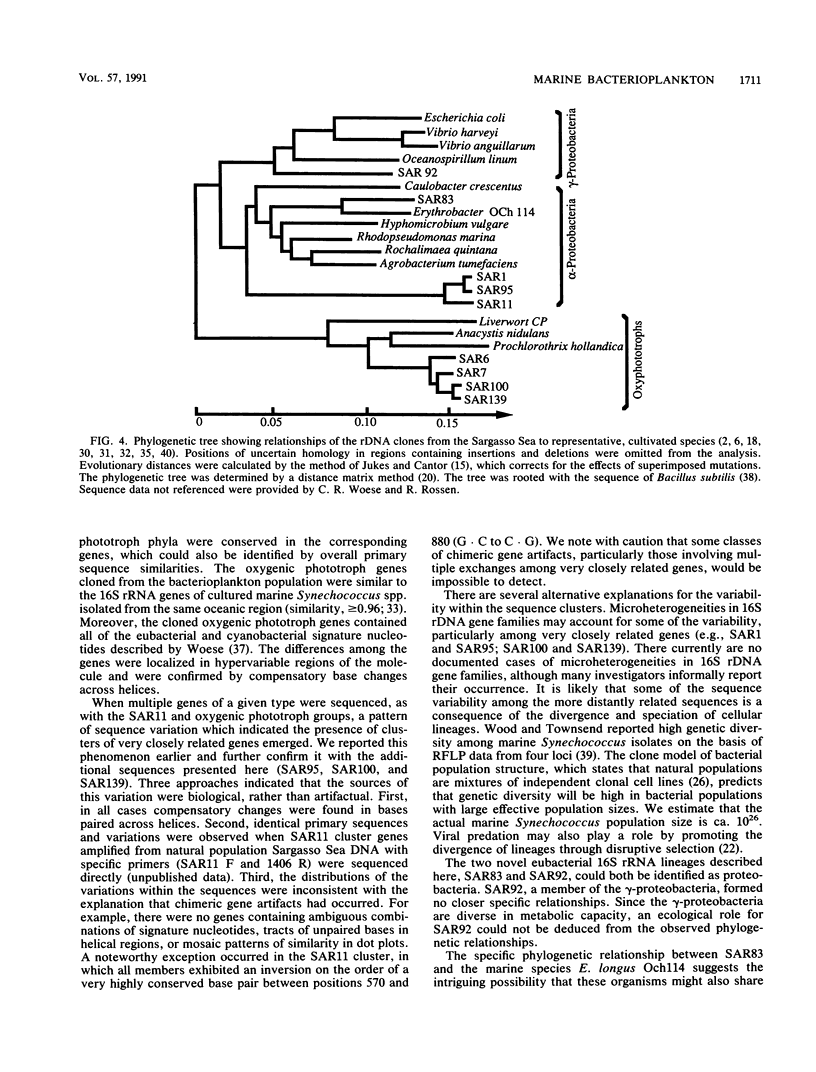


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baldari C., Cesareni G. Plasmids pEMBLY: new single-stranded shuttle vectors for the recovery and analysis of yeast DNA sequences. Gene. 1985;35(1-2):27–32. doi: 10.1016/0378-1119(85)90154-4. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Sleeter D. D., Noller H. F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981 May 15;148(2):107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- Ferguson R. L., Buckley E. N., Palumbo A. V. Response of marine bacterioplankton to differential filtration and confinement. Appl Environ Microbiol. 1984 Jan;47(1):49–55. doi: 10.1128/aem.47.1.49-55.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., Britschgi T. B., Moyer C. L., Field K. G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990 May 3;345(6270):60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Olsen G. J., Pace N. R. Phylogenetic group-specific oligodeoxynucleotide probes for identification of single microbial cells. J Bacteriol. 1988 Feb;170(2):720–726. doi: 10.1128/jb.170.2.720-726.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni S. J., DeLong E. F., Schmidt T. M., Pace N. R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990 Aug;56(8):2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Kogure K., Simidu U., Taga N. A tentative direct microscopic method for counting living marine bacteria. Can J Microbiol. 1979 Mar;25(3):415–420. doi: 10.1139/m79-063. [DOI] [PubMed] [Google Scholar]
- Lane D. J., Field K. G., Olsen G. J., Pace N. R. Reverse transcriptase sequencing of ribosomal RNA for phylogenetic analysis. Methods Enzymol. 1988;167:138–144. doi: 10.1016/0076-6879(88)67015-7. [DOI] [PubMed] [Google Scholar]
- Olsen G. J. Phylogenetic analysis using ribosomal RNA. Methods Enzymol. 1988;164:793–812. doi: 10.1016/s0076-6879(88)64084-5. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgaramella V., Khorana H. G. CXII. Total synthesis of the structural gene for an alanine transfer RNA from yeast. Enzymic joining of the chemically synthesized polydeoxynucleotides to form the DNA duplex representing nucleotide sequence 1 to 20. J Mol Biol. 1972 Dec 28;72(2):427–444. doi: 10.1016/0022-2836(72)90155-6. [DOI] [PubMed] [Google Scholar]
- Stahl D. A., Flesher B., Mansfield H. R., Montgomery L. Use of phylogenetically based hybridization probes for studies of ruminal microbial ecology. Appl Environ Microbiol. 1988 May;54(5):1079–1084. doi: 10.1128/aem.54.5.1079-1084.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomioka N., Sugiura M. The complete nucleotide sequence of a 16S ribosomal RNA gene from a blue-green alga, Anacystis nidulans. Mol Gen Genet. 1983;191(1):46–50. doi: 10.1007/BF00330888. [DOI] [PubMed] [Google Scholar]
- Turner S., Burger-Wiersma T., Giovannoni S. J., Mur L. R., Pace N. R. The relationship of a prochlorophyte Prochlorothrix hollandica to green chloroplasts. Nature. 1989 Jan 26;337(6205):380–382. doi: 10.1038/337380a0. [DOI] [PubMed] [Google Scholar]
- Ward D. M., Weller R., Bateson M. M. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990 May 3;345(6270):63–65. doi: 10.1038/345063a0. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Woese C. R., Dobson M. E., Weiss E. A common origin of rickettsiae and certain plant pathogens. Science. 1985 Nov 1;230(4725):556–558. doi: 10.1126/science.3931222. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Debrunner-Vossbrinck B. A., Oyaizu H., Stackebrandt E., Ludwig W. Gram-positive bacteria: possible photosynthetic ancestry. Science. 1985;229:762–765. doi: 10.1126/science.11539659. [DOI] [PubMed] [Google Scholar]
- Yang D., Oyaizu Y., Oyaizu H., Olsen G. J., Woese C. R. Mitochondrial origins. Proc Natl Acad Sci U S A. 1985 Jul;82(13):4443–4447. doi: 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]