Abstract
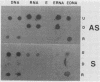
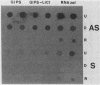
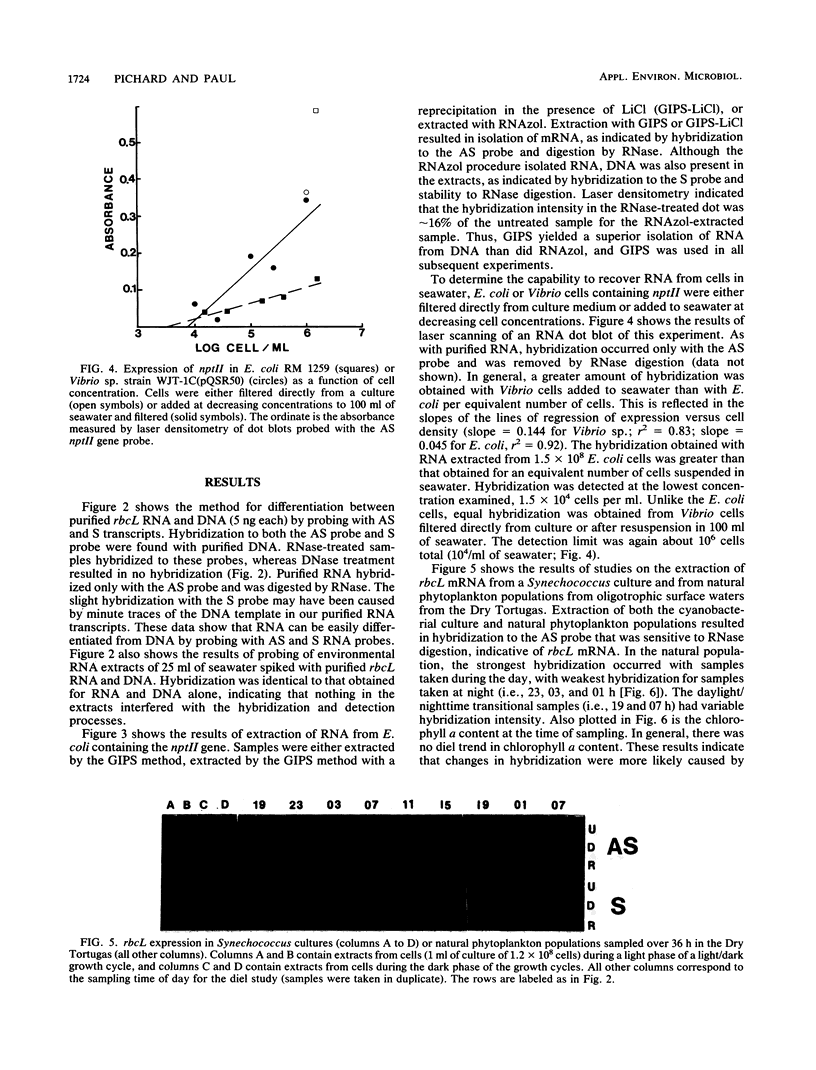
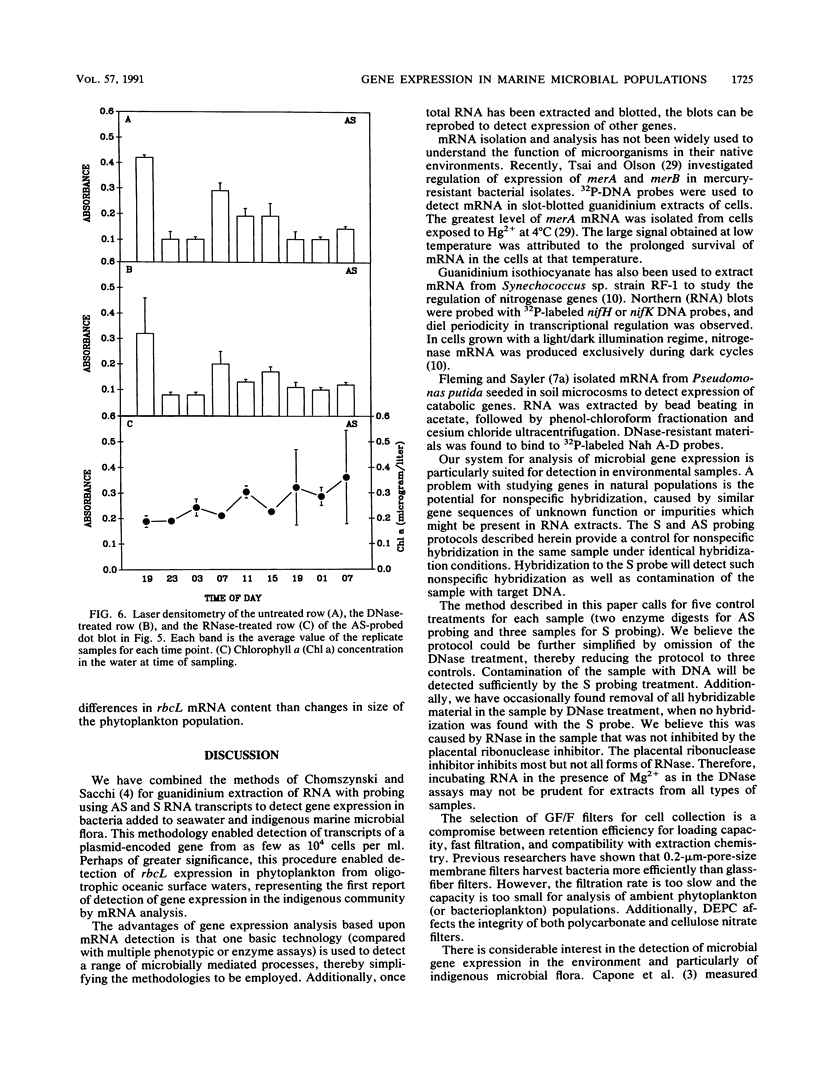
A simple method that combines guanidinium isothiocyanate RNA extraction and probing with antisense and sense RNA probes is described for analysis of microbial gene expression in planktonic populations. Probing of RNA sample extracts with sense-strand RNA probes was used as a control for nonspecific hybridization or contamination of mRNA with target DNA. This method enabled detection of expression of a plasmid-encoded neomycin phosphotransferase gene (nptII) in as few as 104Vibrio cells per ml in 100 ml of seawater. We have used this method to detect expression of the ribulose-1,5-bisphosphate carboxylase large-subunit gene (rbcL) in Synechococcus cultures and natural phytoplankton populations in the Dry Tortugas, Florida. During a 36-h diel study, rbcL expression of the indigenous phytoplankton was greatest in the day, least at night (1100, 0300, and 0100 h), and variable at dawn or dusk (0700 and 1900 h). These results are the first report of gene expression in natural populations by mRNA isolation and probing. This methodology should be useful for the study of gene expression in microorganisms released into the environment for agricultural or bioremediation purposes and indigenous populations containing highly conserved target gene sequences.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beck E., Ludwig G., Auerswald E. A., Reiss B., Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982 Oct;19(3):327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- Berry J. O., Nikolau B. J., Carr J. P., Klessig D. F. Transcriptional and post-transcriptional regulation of ribulose 1,5-bisphosphate carboxylase gene expression in light- and dark-grown amaranth cotyledons. Mol Cell Biol. 1985 Sep;5(9):2238–2246. doi: 10.1128/mcb.5.9.2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capone D. G., O'neil J. M., Zehr J., Carpenter E. J. Basis for Diel Variation in Nitrogenase Activity in the Marine Planktonic Cyanobacterium Trichodesmium thiebautii. Appl Environ Microbiol. 1990 Nov;56(11):3532–3536. doi: 10.1128/aem.56.11.3532-3536.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Frischer M. E., Thurmond J. M., Paul J. H. Natural plasmid transformation in a high-frequency-of-transformation marine Vibrio strain. Appl Environ Microbiol. 1990 Nov;56(11):3439–3444. doi: 10.1128/aem.56.11.3439-3444.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S. R., Tabita F. R. Cloning and expression of the chloroplast-encoded rbcL and rbcS genes from the marine diatom Cylindrotheca sp. strain N1. Plant Mol Biol. 1989 Jul;13(1):69–79. doi: 10.1007/BF00027336. [DOI] [PubMed] [Google Scholar]
- Jorgensen R. A., Rothstein S. J., Reznikoff W. S. A restriction enzyme cleavage map of Tn5 and location of a region encoding neomycin resistance. Mol Gen Genet. 1979;177(1):65–72. doi: 10.1007/BF00267254. [DOI] [PubMed] [Google Scholar]
- Meyer R., Laux R., Boch G., Hinds M., Bayly R., Shapiro J. A. Broad-host-range IncP-4 plasmid R1162: effects of deletions and insertions on plasmid maintenance and host range. J Bacteriol. 1982 Oct;152(1):140–150. doi: 10.1128/jb.152.1.140-150.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul J. H. Use of hoechst dyes 33258 and 33342 for enumeration of attached and planktonic bacteria. Appl Environ Microbiol. 1982 Apr;43(4):939–944. doi: 10.1128/aem.43.4.939-944.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puissant C., Houdebine L. M. An improvement of the single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Biotechniques. 1990 Feb;8(2):148–149. [PubMed] [Google Scholar]
- Reith M., Cattolico R. A. Inverted repeat of Olisthodiscus luteus chloroplast DNA contains genes for both subunits of ribulose-1,5-bisphosphate carboxylase and the 32,000-dalton Q(B) protein: Phylogenetic implications. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8599–8603. doi: 10.1073/pnas.83.22.8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki Y., Sakihama T., Kamikubo T., Shinozaki K. Phytochrome-mediated regulation of two mRNAs, encoded by nuclei and chloroplasts of ribulose-1,5-bisphosphate carboxylase/oxygenase. Eur J Biochem. 1983 Jul 1;133(3):617–620. doi: 10.1111/j.1432-1033.1983.tb07507.x. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Sasaki Y., Sakihama T., Kamikubo T. Coordinate light-induction of two mRNAs, encoded in nuclei and chloroplasts, of ribulose 1,5-bisphosphate carboxylase/oxygenase. FEBS Lett. 1982 Jul 19;144(1):73–76. doi: 10.1016/0014-5793(82)80571-1. [DOI] [PubMed] [Google Scholar]
- Shinozaki K., Sugiura M. The gene for the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase is located close to the gene for the large subunit in the cyanobacterium Anacystis nidulans 6301. Nucleic Acids Res. 1983 Oct 25;11(20):6957–6964. doi: 10.1093/nar/11.20.6957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K., Yamada C., Takahata N., Sugiura M. Molecular cloning and sequence analysis of the cyanobacterial gene for the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase. Proc Natl Acad Sci U S A. 1983 Jul;80(13):4050–4054. doi: 10.1073/pnas.80.13.4050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita F. R. Molecular and cellular regulation of autotrophic carbon dioxide fixation in microorganisms. Microbiol Rev. 1988 Jun;52(2):155–189. doi: 10.1128/mr.52.2.155-189.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabita F. R., Small C. L. Expression and assembly of active cyanobacterial ribulose-1,5-bisphosphate carboxylase/oxygenase in Escherichia coli containing stoichiometric amounts of large and small subunits. Proc Natl Acad Sci U S A. 1985 Sep;82(18):6100–6103. doi: 10.1073/pnas.82.18.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y. L., Olson B. H. Effects of Hg, CH(3)-Hg, and Temperature on the Expression of Mercury Resistance Genes in Environmental Bacteria. Appl Environ Microbiol. 1990 Nov;56(11):3266–3272. doi: 10.1128/aem.56.11.3266-3272.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle E., Kobayashi H., Akazawa T. Transcriptional regulation of genes for plant-type ribulose-1,5-bisphosphate carboxylase/oxygenase in the photosynthetic bacterium, Chromatium vinosum. Eur J Biochem. 1988 May 2;173(3):483–489. doi: 10.1111/j.1432-1033.1988.tb14024.x. [DOI] [PubMed] [Google Scholar]
- Zehr J. P., Limberger R. J., Ohki K., Fujita Y. Antiserum to Nitrogenase Generated from an Amplified DNA Fragment from Natural Populations of Trichodesmium spp. Appl Environ Microbiol. 1990 Nov;56(11):3527–3531. doi: 10.1128/aem.56.11.3527-3531.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]